Abstract
There is always controversy regarding identification of different species in the genus Eurytrema. Identification has been based mainly on morphology, which can be misleading and subject to differing interpretation among the scientists. Therefore, the aim of this study was to identify Eurytrema flukes both by morphology and molecular properties on the basis of 18-subunit ribosomal RNA (18S rRNA) gene as well as internal transcribed spacer 2 (ITS2) to clarify their phylogenetic status. Among six different agroecological areas of Bangladesh, 22 Eurytrema flukes were recovered from the bile ducts of 22 cattle in Bandarban, a hill district. The flukes were identified as Eurytrema cladorchis through morphometric and morphological studies. Phylogenetic analyses were conducted by neighbor-joining phylogram inferred from both 18S rRNA (1784 bp) gene and ITS2 (229 bp) sequences. A monophyletic clade was constructed by the E. cladorchis from Bangladesh; however, the clade was distinct from those formed by Eurytrema pancreaticum and Eurytrema coelomaticum. This study first described the existence of E. cladorchis from Bangladesh and may provide useful information for both morphological and molecular properties that may further help to clarify phylogenetic relationships within the genus Eurytrema and also for other digeneans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the genus Eurytrema usually parasitize the pancreatic ducts and bile ducts of domestic and wild ruminants and humans. Eurytrema flukes are often associated with the epithelial hyperplasia and hypertrophy of pancreatic duct and bile duct, and periductal fibrosis leading to death of the animal in heavy infestations (Tang 1950). So far, 12 species of Eurytrema from ruminants have been reported to be distributed globally. These are Eurytrema pancreaticum Janson, 1889; Eurytrema coelomaticum Giard & Billet, 1892; Eurytrema dajii Bhalerao, 1924; Eurytrema media Chertkova, 1957; Eurytrema ovis Tubangui, 1925; Eurytrema parvum Senoo, 1907; Eurytrema tonkinense Gillard & Ngu, 1941; Eurytrema escuderoi Eduardo, Manuel & Tongson, 1976; Eurytrema cladorchis Chin, Li and Wei, 1965; Eurytrema hydropotes Tang & Tang, 1975; Eurytrema fukienensis Tang & Tang, 1978; and Eurytrema sphaeriorchis Tang, Lin & Lin, 1978. But, there is a controversy regarding the validity for most of the type species (Jones 1985). Eurytrema pancreaticum, E. coelomaticum and E. cladorchis are the three valid species (Jones 1985; Zheng et al. 2006) and considered important for the domestic ruminants. E. cladorchis was first described from pancreatic duct of wild deer in the mountain area of Guinzhou Province in China (Jin et al. 1965), and thereafter, epidemiology, biology, and life history of this species have been studied intensively in China (Tang and Tang 1977; Tang et al. 1978; Tang and Lin 1980). The species was also reported to infect the duodenum of ox, abomasum of cow, and peritoneal fluid and veins surrounding the stomach of goat (Jones 1985). Morphological identification of E. cladorchis from Nepal was conducted by Jones (1985), which is the first report of the fluke outside China. In this study, the morphometric and morphological properties of Eurytrema flukes from Bangladesh were compared with the findings of E. cladorchis (Jones 1985; Tang and Tang 1978) as well as E. pancreaticum (Wiroreno et al. 1987) and E. coelomaticum (Kumar 1998).
The sequences of 18S rRNA gene were found to be an effective marker for inferring phylogenetic relationships among the families of digeneans (Blair and Barker 1993). In addition, internal transcribed spacer (ITS) regions of the ribosomal DNA may provide useful information for elucidating interspecies and intraspecies relationships among closely related genera in many eukaryotes (Orosova et al. 2010; Yamada et al. 2012). So far, there is no report on precise characterization of E. cladorchis both by morphology and molecular properties. Therefore, in this study, the 18S rRNA genes and internal transcribed spacer 2 (ITS2) regions of Eurytrema flukes from Bangladesh were sequenced along with morphometric and morphological studies for precise characterization of the flukes. Phylogenetic relationships among the different species of the genus Eurytrema were also inferred from the sequences of 18S rRNA and ITS2 to elucidate relationships among the species in the genus.
Materials and methods
Sample collection
Eurytrema flukes were recovered from the bile duct of cattle at a slaughterhouse in Bandarban district, a mountainous area located in the Southeast part of Bangladesh bordering Myanmar in the East. A total of 22 flukes were collected from 22 cattle (1 fluke from each host) in September, 2012. The flukes were washed with physiological saline and preserved in 70 % ethanol and were transferred to the laboratory for morphological and molecular studies.
Morphological studies
The fixed flukes were subjected to morphological studies. First, flukes’ length, maximum body width, distance from anterior extremity (AE) to maximum body width, distance from AE to upper end of testes, internal and external diameter of oral sucker were measured under an optical microscope, and then, tissues from the anterior part (from both the shoulders) of the flukes were removed for DNA extraction. The remaining parts of the flukes were stained with hematoxylin–carmine solution, and detailed morphological and morphometric analyses were conducted. Internal and external diameter of the ventral sucker, length and width of the cirrus sac, length and width of both testes, length and width of the ovary, length of the vitelline glands, and long and short diameter of mature eggs were measured on each stained specimen. The ratio of body length to body width, oral sucker to ventral sucker, oral sucker to body width, ventral sucker to body width, and oral sucker to ventral sucker were also calculated. These specimens were deposited in the National Science Museum, Tokyo, Japan (NSMT–PI 6142 ~ 6163).
DNA analyses
Total DNA was extracted from the anterior part (from both the shoulders) of 22 flukes to avoid reproductive organs with an aim to exclude genetic materials from other flukes by using High Pure PCR template preparation kit (Roche, Mannheim, Germany) following manufacturer instructions, and was stored at −20 °C for future use. We used tissues from fresh flukes (preserved in 70 % ethanol, not stained and mounted) for DNA extraction with an objective to obtain long unbroken chain of DNA. DNA fragments of 18S rRNA and ITS2 regions were amplified by polymerase chain reaction (PCR) in a total volume of 25 μl containing 1.25 U Taq polymerase (Promega, Madison, USA), 2 μl template DNA, 0.2 mM each dNTP, 0.1 μM each primer set (50-pmol/μl reaction mixture), and PCR buffer. 18S rRNA was divided into three parts by three primer sets, Primer A (Cai et al. 2012), 1R (5′-ACACCCGTTGAAAGGCATCGAGA-3′), 2 F (5′-TCGTGGTTGTGCAGCCTTTTTGC-3′), 2R (5̕′-AGAAGTAAACACCTGCGCAGCAC-3′), 3 F (5̕′-GTGAAATTCTTGGATCGCCGCCA-3′), and primer D (Cai et al. 2012). For 18S rRNA, the reaction cycle consisted of an initial denaturation at 94 °C for 3 min, followed by 35 cycles at 94 °C for 45 s, 55 °C (first part: primer A and 1R) for 1 min, 72 °C for 1 min, with a final extension at 72 °C for 1 min. The annealing temperature for the second part (2 F and 2R) and third part (3 F and primer D) was 60 and 62 °C, respectively, whereas for amplification of ITS2 regions, the primer sets used were ITS2-F and ITS2-R (Itagaki and Tsutsumi 1998). In the case of ITS2, a reaction cycle consisting of an initial denaturation at 94 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min, with a final extension at 72 °C for 10 min. The PCR reaction was performed using GenAmp PCR system 2700 (Applied Biosystems, Tokyo, Japan). PCR products were separated by 1.2 % agarose gel electrophoresis, stained by ethidium bromide, and subsequently visualized by using an ultraviolet transilluminator.
PCR amplicons for both 18S rRNA and ITS2 were sequenced directly from both directions by using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, USA). The thermal program was 25 cycles at 96 °C for 10 s, 55 °C for 5 s, and 60 °C for 4 min for ITS2. For 18S rRNA, the cycle conditions were the same except for annealing temperature that was 55 °C for the first part (primers A and 1R), 60 °C for the second part (2 F and 2R) while 62 °C for the third part (3 F and primer D). Amplicons were precipitated with ethylenediaminetetra acetic acid (EDTA)/ethanol for purification. The resultant sequencing ladders were determined using a 3500-Genetic Analyzer (Applied Biosystems, Foster City, USA). Data obtained from each sequence was analyzed using GENETYX version 10.0.2 software (Fluxus Technology Ltd., Suffolk, England).
Phylogenetic analyses
Both the sequences of 18S rRNA (1784 bp) and ITS2 (229 bp) were aligned by Clustal X version 2.0 software, and the haplotypes were distinguished. Neighbor-joining (Nj) phylogram inferred from both the sequences of 18S rRNA and ITS2 were constructed by PAUP 4.0b10 (Swofford 2001). 18S rRNA sequence of Fasciola gigantica (accession no. AJ011942) was used as an outgroup for constructing a tree inferred from 18S rRNA sequences. Additionally, 18S rRNA sequences of E. pancreaticum (DQ401034), E. coelomaticum (DQ401035), Fj–Ca–E. cladorchis (Cai et al. 2012), Fj–Ca–Eurytrema fukienensis (Cai et al. 2012) and Dicrocoelium dendriticum (Y11236) were used as references. ITS2 sequence of Fasciola gigantica (AJ557569) was used as an outgroup for constructing a tree inferred from ITS2 sequences. ITS2 sequences of E. pancreaticum (LC012790), D. dendriticum (KC774515), D. chinensis (AB367790) and D. hospes (EF102026) were used as references. Bootstrap analyses were conducted using 1000 replicates for the trees. Estimates of evolutionary divergence between the 18S rRNA sequences were calculated by MEGA version 6.06 (Tamura et al. 2013). Analyses were conducted using the maximum composite likelihood model. The analysis involved nucleotide sequences of D. dendriticum, E. pancreaticum, E coelomaticum, Fj–Ca–E. cladorchis, Fj–Ca–E. fukienensis, and 11 genotypes of Eurytrema flukes from Bangladesh in the case of 18S rRNA, while 4 genotypes of Eurytrema flukes from Bangladesh, E. pancreaticum, D. dendriticum, D. chinensis and D. hospes were subjected for ITS2. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated.
Results
Morphological and morphometric description
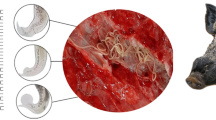
The representative fluke is shown in Fig. 1. The fluke body is flattened, elongated to oval with smooth margins (not ruffled) being tapered at both the extremities and lacking caudal appendages. Maximum width of the fluke is in front of the middle half of the body (1.9–4.2 mm from the anterior extremity). Oral sucker is subterminal and smaller than the ventral sucker, which is located at the middle to anterior one third of the body. Pharynx is small, esophagus is short, and the caeca narrow and bifurcated at the level of genital pore reaching posteriorly along the body margin beyond the posterior limit of the vitelline glands but remaining short from the posterior extremity of the body. Testes are large, 0.81–1.79 mm × 0.65–1.96 mm, heavily branched, symmetrical (average distance from the anterior extremity to the upper end of testis is 1.66 and 1.65 mm for left and right testis, respectively), posterolateral to ventral sucker. Cirrus sac is elongated (0.34–1.0 mm × 0.18–0.42 mm) containing the seminal vesicle inside, and situated between the caecal bifurcation and acetabulum. Genital pore is median. Ovary is lobulated (4–10 lobes), post testicular, and submedian to left or right from the midline. Vitelline glands are long, arranged in groups, extracaecal, and extending posteriorly from the caudal margin of the testes, keeping short from the end point of the caeca. Most of the hind body is occupied by the uterus, being mostly intercaecal; the ascending limb extends anteriorly between the testes, passing over the ventral sucker/acetabulum and then forming a coil reaching in front of the acetabulum prior to opening into the genital pore. Uterus contains numerous dark brown, operculated eggs. Overall dimension of mature eggs is 0.041–0.052 mm × 0.030–0.033 mm (Table 1). In this study, along with other species of the genus Eurytrema, the Eurytrema flukes from Bangladesh were also morphologically compared with E. cladorchis reported previously (Tang and Tang 1978; Jones 1985). The most notable morphological feature of the flukes is big testes, which are located symmetrically posterolateral to the acetabulum and are heavily branched. The ventral sucker (acetabulum) is distinctively larger (average 0.70 mm) than the oral sucker (average 0.53 mm). From these morphological and morphometric properties (Table 1), 22 Eurytrema flukes from Bangladesh were identified as E. cladorchis.
Representative entire fluke (ventral view) of E. cladorchis from Bangladesh. OS oral sucker, P pharynx, C caecum, CS cirrus sac, VS ventral sucker, OV ovary, MGl Mehlis’ gland, T testis, V vitelline follicles, EB excretory bladder, EP excretory pore. Actually, this fluke was not included in this study but displayed identical morphology to the flukes studied. The broken lines indicate that the portions of the flukes were used for DNA extraction
18S rRNA and ITS2 sequence analyses
The nucleotide sequences (1784 bp) of 18S rRNA from the 22 flukes demonstrated 8 variable sites yielding 11 genotypes represented by Ec–A to Ec–K (accession nos. LC005981–LC005991), whereas the nucleotide sequences (229 bp) of ITS2 regions from the Eurytrema flukes displayed 4 substitution sites yielding 4 genotypes represented by Ec–1 to Ec–4 (accession nos. LC006029–LC006032). Estimation of the evolutionary divergence between the genotypes of E. cladorchis from Bangladesh suggests a low discrimination; the values range from 0.000 (identical) to 0.002 (99.8 % similar) for 18S rRNA while the value was 0.000 for ITS2 genotypes. In case of 18S rRNA, estimate of evolutionary divergence between the E. cladorchis from Bangladesh and E. pancreaticum (DQ401034) showed 99.4–99.6 % similarity while 98.9–99 % similarity was observed between the E. cladorchis and E. coelomaticum (DQ401035). The genetic identity of E. cladorchis from Bangladesh with Fj-Ca–E. cladorchis and Fj–Ca–E. fukienensis (Cai et al. 2012) was 98.1–98.3 and 98.4–98.6 %, respectively. But, the genetic identity of Fj–Ca–E. cladorchis (Cai et al. 2012) was 98.1–98.3 and 98.4–98.6 % with E pancreaticum and E. coelomaticum, respectively, whereas in ITS2, 96.2 % similarity was observed between E. cladorchis from Bangladesh and E. pancreaticum (LC012790).
Phylogenetic analyses
Both 18S and ITS2 genotypes of E. cladorchis from Bangladesh formed a monophyletic clade in the phylograms (Figs. 2 and 3). In the phylogram constructed from 18S rRNA, E. coelomaticum, and Fj–ca–E. cladorchis and Fj–Mm–E. fukienensis from China (Cai et al. 2012) formed one cluster which is different from the cluster formed by the E. cladorchis from Bangladesh while D. dendriticum was distinct from the genus Eurytrema (Fig. 2). Similarly, in the phylogram inferred from ITS2 sequences, E. pancreaticum was sister to the E. cladorchis clade while the members of the genus Dicrocoelium formed a distinct clade (Fig. 3).
Discussion
E. cladorchis Chin, Li and Wei, 1965 is one of the valid species in the genus Euryrema (Jones 1985), and the detailed life cycle of this species has been studied in China (Tang and Tang 1977; Tang et al. 1978; Tang and Lin 1980). Existence of the flukes was first reported from Nepal outside China where E. cladorchis was first identified, and detailed morphology of the fluke was studied (Jones 1985). E. cladorchis inhabits the pancreatic duct of domestic ruminants and wild deer (Muntiacus muntjak and Hydropotes inermis) (Cai et al. 2012) while the flukes were also reported as aberrant (duodenum of ox, abomasum of cow, and veins around stomach and peritoneal fluids of goat) (Jones 1985). In the present study, the flukes were recovered from the bile duct of cattle. These widespread locations of the flukes in the final host indicate that they have no predilection sites favoring them with superior survivability and adaptability. The flukes were first reported from the wild musk deer (Moschus chinensis) in the mountain areas of Guizhou Province, China (Jin et al. 1965). Afterward, the flukes were reported from both wild deer (Muntiacus muntjak, Hydropotes inermis) and domestic ruminants in the mountainous area of Fujian, Zhejiang, Jiangxi and Anhui Provinces, China (Cai et al. 2012) and also from domestic ruminants in Nepal (Jones 1985). In this study, the flukes were recovered from domestic cattle in the mountainous area of Bangladesh bordering Myanmar, suggesting that the flukes might be basically parasites of wild ruminants, gradually adapted to domestic ruminants in the forest and mountain area. The distribution of the flukes might occur from China to Nepal and Bangladesh through infected barking deer (Muntiacus muntjak) which is popularly called Maya deer in Bangladesh.
Neighbor-joining phylogram inferred from 18S rRNA (1784 bp) displayed that all haplotypes of E. cladorchis from Bangladesh formed a monophyletic clade where other members of the genus Eurytrema are sister to E. cladorchis (Fig. 2). On the other hand, the cluster formed by E. cladorchis from Bangladesh is quite distinct from that formed by Fj–Ca–E. cladorchis and Fj–Mm–E. fukienensis reported from China (Cai et al. 2012). The estimates of evolutionary divergence value suggest that the genetic distance between E. cladorchis from Bangladesh and Fj–Ca–E. cladorchis reported from China is 1.7–1.9 %. These results indicate that E. cladorchis in these two countries are genetically quite distinct. The genetically distinct E. cladorchis from Bangladesh and China with identical morphology suggest a cryptic species complex. Again, Fj–Ca–E. cladorchis and Fj–Mm–E. fukienensis (Cai et al. 2012) are included into the monophyletic clade in the tree constructed from the 18S rRNA gene sequences. These results prompt us to interpret that there might be misapprehension regarding the identification of Fj–Ca–E. cladorchis. Althouh Cai et al. (2012) mentioned that they identified E. cladorchis by morphology based on the description of Tang & Tang (1978), they did not include the morphological observation in the report. Therefore, we could not compare the morphology of Fj–Ca–E. cladorchis with E. cladorchis from Bangladesh in this study. The evolutionary divergence data showed that the intraspecific variation among the E. cladorchis from Bangladesh was up to 0.2 % only. The genetic distance (intraspecific variation) among E. cladorchis flukes is reported in this article for the first time, whereas the interspecific variation between 18S rRNA sequence of E. cladorchis and E. pancreaticum was 0.4–0.6 %. On the other hand, 1.0–1.1 % variation was observed between 18S rRNA sequences of E. cladorchis and E. coelomaticum. These interspecific distances within different genera in the family Dicrocoeliidae are usual (Zheng et al. 2007). In the neighbor-joining (Nj) phylogram constructed from ITS2 sequences (229 bp), E. cladorchis from Bangladesh and E. pancreaticum formed a monophyletic clade from a common ancestor with the high bootstrap value (100.0) (Fig. 3). Members of the genus Dicrocoelium are distinct from the genus Eurytrema in the tree. We could not compare the ITS2 sequences with other members of the genus Eurytrema due to unavailability of data in GenBank or elsewhere.
References
Blair D, Barker SC (1993) Affinities of the Gyliauchenidae: utility of the 18S rRNA genes for the phylogenetic inference in the Digenea (Platyhelminthes). Int J Parasitol 23:527–532
Cai Z, Zhang Y, Ye X (2012) Phylogenetic relationships of the genus Eurytrema from domestic and wild animal based on 18S rRNA sequences. Parasitol Res 111:1637–1644
Itagaki T, Tsutsumi K (1998) Triploid form of Fasciola in Japan: genetic relationships between Fasciola hepatica and Fasciola gigantica determined by ITS–2 sequence of nuclear rDNA. Int J Parasitol 28:777–781
Jin DX, Li GZ, Wei CF (1965) A new species of Eurytrema in animal in Guizhou Province. Acta Parasitol Sinica 2:28–35
Jones A (1985) Eurytrema cladorchis Chin, Li and Wei, 1965 (Trematoda: Dicrocoelidae), a little known species from China and Nepal. Syst Parasitol 7:43–45
Kumar V (1998) Trematode infections and diseases of man and animals. Eurytremiasis. Kluwer Academic Publisher, Dordrecht, pp 230–233
Orosova M, Ivica KH, Eva B, Marta S (2010) Chromosomal characteristics of multiple rDNA clusters and intragenomic variability of ribosomal ITS2 in Caryophyllaeides fennica (Cestoda). Parasitol Int 59:351–357
Swofford DL (2001) PAUP: Phylogenetic analysis using parsimony and other methods ver. 4.0beta. Sanderland, Massachussetts: Sinauer Associates
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolution genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tang CC (1950) Studies on the life history of Eurytrema pancreaticum Janson, 1889. J Parasitol 36:559–574
Tang CT, Lin TM (1980) Investigations of Eurytrematosis of cattle and goats in mountainous regions of North Fu–jian. Acta Zool Sin 26:42–51 (In Chinese )
Tang CC, Tang CT (1977) The biology and epidemiology of Eurytrema coelomaticum (Giard and Billet, 1892) and Eurytrema pancreaticum (Jansen, 1889) in cattle and sheep in China. Acta Zool Sin 23:267–282 (In Chinese )
Tang CC, Tang CT (1978) Investigation on trematodes belonging to the family Dicrocoelidae Odhner, 1911. J Xiamen Univ 4:64–80
Tang CT, Lin TM, Lin HM (1978) On the life cycle of Eurytrema cladorchis of cattle and goat in Pucheng, North Fujian. Xiamen Daxue Xuebao 14:104–117 (In Chinese)
Wiroreno W, Carney WP, Ansori M (1987) Description and growth pattern of Eurytrema pancreaticum from Bos indicus from East Java. Proc Helminthol Soc Wash 54:73–77
Yamada S, Yoshida A, Yoshida K, Kuraishi T, Hattori S, Kai C, Nagai Y, Sakoda T, Tatara M, Abe S, Fukumoto SI (2012) Phylogenetic relationships of three species within the family Heligmonellidae (Nematoda; Heligmosomoidea) from Japanese rodents and a lagomorph based on the sequences of ribosomal DNA internal transcribed spacers, ITS–1 and ITS–2. Jpn J Vet Res 60:15–21
Zheng YD, Luo XN, Shi CH, Zong RQ, Jing ZZ, Cai XP (2006) Molecular relationship of Eurytrema coelomaticum inferred from 18S rRNA sequence. Chin J Parasitol Parasit Dis 24:345–348 (in Chinese)
Zheng Y, Luo X, Jing Z, Hu Z, Cai X (2007) Comparison of 18S ribosomal gene sequences of Eurytrema coelomaticum and Eurytrema pancreaticum. Parasitol Res 100:645–646
Acknowledgments
This study was supported in part by a Grant-in-Aid for Science Research (B) and (C) (nos. 23405044, 24580420, 22405037) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments comply with the current laws of the country.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mohanta, U.K., Ichikawa-Seki, M., Hayashi, K. et al. Morphological and molecular characterization of Eurytrema cladorchis parasitizing cattle (Bos indicus) in Bangladesh. Parasitol Res 114, 2099–2105 (2015). https://doi.org/10.1007/s00436-015-4398-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-015-4398-y