Abstract
Sphaeromyxa azevedoi n. sp. is described from the gall bladder of the goby Gobioides grahamae (Gobiidae) captured on the Paracauari River in Salvaterra, on Marajó Island, northern Brazil. A total of 50 G. grahamae specimens were analysed, and 15 (30%) were parasitised by the plasmodia and myxospore of Sphaeromyxa azevedoi n. sp. Large plasmodia were observed floating in the bile. These plasmodia were flat, rounded, oval or elongated, and of varying sizes. The mature myxospores, found singly or in pairs, were 27.1 ± 2.7 (20.5–30.1) μm Length and 3.8 ± 0.2 (3.5–4.4) μm Width in the valvular view. The myxospore has two polar capsules of equal size, 8.1 ± 0.6 (7.4–9.4) μm in length and 2.9 ± 0.2 (2.3–3.3) μm in width. A polar tubule was observed in each capsule, arranged perpendicularly to the principal axis, with three or four coils. The histological analysis showed that the plasmodia and myxospore are located in the lumen of the gall bladder, arranged in pairs, and the epithelium of the gall bladder presented multifocal necrosis. The SSU rDNA of Sphaeromyxa azevedoi n. sp. clusters in the ‘balbianii’ group of the Sphaeromyxa clade. The morphological characteristics and molecular phylogeny of Sphaeromyxa azevedoi n. sp. support its classification as a new species of the genus Sphaeromyxa, which represents an important advancement in the understanding of the diversity of the myxozoan parasite fauna of Brazilian fishes, especially considering that the new species may be detrimental to the host, a commercially important Brazilian fish species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The goby Gobioides grahamae, Palmer and Wheeler, 1955 (Perciformes) is a member of the family Gobiidae, and is found in the western Atlantic Ocean, in coastal areas and estuaries ranging from the Guianas to northern Brazil (Murdy 1998). In Brazilian Amazonia, G. grahamae is typical of estuarine environments and, while it is not used as a human food, it is an important prey of a number of estuarine fish species, including Arius parkeri and Arius couma (Ariidae), as well as the freshwater species Brachyplatystoma vaillantii and Brachyplatystoma flavicans. Given this, G. grahamae is considered to be excellent live bait for large game fish, and artisanal fishers capture these fish and transport them alive to use as bait or to sell to the crews of larger vessels (Barthem 1990; Mendes and Barthem 2010). Parasites may have negative impacts on the growth and behaviour of the host, reduce its resistance to stress factors and increase its susceptibility to bacterial infection and its vulnerability to predators, which may ultimately result in mortality (Brassard et al. 1982; Lom and Dyková 1992; Violante-González et al. 2009; Miller et al. 2014; Nekouei et al. 2018). Hepatic steatosis has been found in association with Microsporidium sp. infections (Videira et al. 2014) and Kudoa sp. provoked the degeneration of the epithelium of the palate and multifocal areas of necrosis (Videira et al. 2020) in specimens of G. grahamae collected in the same region as the present study.
In Amazonia, a considerable diversity of microparasites has been found in a range of fish hosts (Mathews et al. 2015; Azevedo et al. 2016; Zatti et al. 2017; Matos et al. 2018) including gobiids, such as Gobioides broussonnetti (Velasco et al. 2012; Azevedo et al. 2013). In the present study, microparasites were detected in the gall bladders of G. grahamae specimens collected in estuarine waters of the Amazon region, specifically the Paracauari River, on Marajó Island. These parasites presented characteristics typical of the myxosporean genus Sphaeromyxa Thélohan, 1892, which is currently known to have approximately 50 species that parasitise the coelozoic cavity of the gall bladder and bile ducts of fish from both marine and estuarine environments (Karlsbakk et al. 2013; Whipps and Zhao 2015; Chen et al. 2020). The results of the morphological and phylogenetic analyses of the parasites indicated the existence of a new Sphaeromyxa species in G. grahamae, the first known recorded occurrence of this genus in a fish host from the Amazon region.
Materials and methods
Fifty specimens of G. grahamae were captured between August 2017 and February 2018 from the Paracauari River (00°45ʹ S, 48°30ʹ W) in the municipality of Salvaterra, on Marajó Island, in Pará state, northern Brazil. The fish were caught using standard fishing equipment, and they were transported alive, in aerated plastic bags containing river water to the Carlos Azevedo Research Laboratory at the Federal Rural University of Amazonia (UFRA) in Belém, where they were maintained in aerated aquaria containing river water (up to 2 days). The collection of the specimens was authorised by the UFRA Ethics Committee for the use of Animals in Research (CEUA: 013/2014) and the Brazilian Institute for the Environment and Renewable Natural Resources (IBAMA) through SISBIO/ICMBio license 271,191.
The fish specimens were anaesthetized and then euthanized with tricaine methanesulfonate (MS–222 SIGMA) at a concentration of 50 mg/L, and then necropsied. The external surface and the internal organs of the fish specimens were examined for the presence of parasites and tegumentary lesions. Myxosporean plasmodia and myxospore were found in the gall bladder and documented using a Zeiss Primo Star light microscope attached to a Zeiss AxioCamERc 5 s camera, with AxioVision 5.1 software, with images being obtained using a Zeiss Axioscope A1 differential interference contrast (DIC) microscope. The morphometry of the fresh myxospores (n = 20) was determined using the approach of Lom and Arthur (1989). All measurements are given in micrometres (μm), and they were obtained from the first infected host. The gallbladder with the highest parasite load was selected for the quantitative analysis of the fresh myxospores, as well as their histology, and molecular biology. All the images were also obtained from this gallbladder.
Histology
For the histological analysis, the infected gallbladder was fixed in Davidson solution (95% alcohol, formaldehyde and acetic acid, distilled water) for 24 h, dehydrated in an increasing series of ethanol and embedded in paraffin for the extraction of histological Sects. 5 μm thick. These sections were processed and stained with Ziehl–Neelsen (Luna 1968). The stained slides were photographed under the Zeiss Primo Star light microscope attached to a Zeiss AxioCamERc 5 s camera, with AxioVision 5.1 software.
Molecular biology
For the molecular analysis, plasmodia and myxospores floating in the bile were collected from the gallbladder of the G. grahamae specimen. The plasmodia and myxospores were fixed in 80% ethanol. The DNA was extracted using the PureLink® Genomic DNA mini kit (Invitrogen, USA), following the protocol for the extraction of ‘Mammalian Tissue and Mouse/Rat Tail Lysate’ provided by the manufacturer. The DNA sample was quantified in a Nanodrop 1000 spectrophotometer (Thermo Scientific), and PCRs were run to obtain the partial sequence of small subunit ribosomal DNA (SSU rDNA) of the myxospores. This sequence was amplified using the standard primers for the molecular characterisation of the Myxozoa. The initial amplification used eukaryotic universal forward primer 18e (Hillis and Dixon 1991) and reverse primer 18R (Whipps et al. 2003), which was followed by a nested PCR with the MC5/MC3 primers (Molnár et al. 2002).The PCRs were run in a final volume of 25 μL, containing 1 × ReddyMix PCR Master Mix (Thermo Scientific, USA), 75 mM Tris–HCl (pH 8.8), 20 mM of KCl, 0.1 (v/v) of Nonidet P-40, 1.5 mM of MgCl2, 0.2 mM of each nucleotide triphosphate (Thermo Scientific, USA), 10 pmol of each primer, 1.25 U of Taq DNA polymerase (Thermo Scientific, USA) and the DNA template (10–50 ng/μL). The reaction protocol for the primers 18e and 18R consisted of 95 °C for 5 min, followed by 35 cycles of 95 °C for 60 s, 50 °C (annealing) for 90 s and 72 °C for 120 s, with a final extension step of 10 min at 72 °C. For the nested PCR, the reaction protocol was 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 56 °C (annealing) for 30 s and 72 °C for 60 s, with a final extension step of 72 °C for 10 min. Subsequently, 3 µL of the PCR mix was electrophoresed in 1% agarose gel with 1X Tris–Borate-EDTA (TBE), stained with SYBR® Safe (Invitrogen, USA) and visualised under blue light. The PCR products were purified with GFX™ PCR DNA and a Gel Band Purification kit (GE Healthcare, UK), according to the manufacturer’s instructions. The PCR products were sequenced separately for primers 18e, 18R and MC5. The sequencing reactions were conducted with the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, USA), following the manufacturer’s instructions, in an ABI 3100 Genetic Analyser (Applied Biosystems, USA).
The PCR products were sequenced in an ABI 3100 Genetic Analyser, using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, USA), following the manufacturer’s instructions. The nucleotide sequences were assembled in the BioEdit software (Hall 1999) and the ambiguous bases were clarified using the respective chromatograms. The GenBank sequences of the SSU rDNA of the related species obtained through the BLAST search were aligned in Clustal X 1.8 (Thompson et al. 1997), at the default setting, to determine their phylogenetic relationships with the new species. The SSU rDNA sequences of the Sphaeromyxa species deposited in GenBank were used for the phylogenetic analysis, together with sequences of species related to the genus Sphaeromyxa (Bartošová-Sojková et al. 2015; Karlsbakk et al. 2013). The sequences of the SSU rDNA of Kudoa amamiensis Hervio et al. (1997) and Kudoa alliaria Whipps and Diggles (2006) were included as the outgroup. Bayesian Inference was implemented in MrBayes, version 3.2.6 (Ronquist et al. 2012), with GTR + I + G evolutionary model selected by jModelTest 2.1.10 (Darriba et al. 2012) under Bayesian Information Criterion (BIC). Markov Chain Monte Carlo (MCMC) searches of two simultaneous runs of four chains of 5,000,000 generations, with every 500th tree being sampled (Ronquist and Huelsenbeck 2003). The burn-in length (500,000 generations) was determined in Tracer v1.6 (Rambaut and Drummond 2007). The phylogenetic tree was edited in FigTree v.1.4.0 (Rambaut 2012). Genetic distances were computed in PAUP* 4.0b1 (Swofford 2003) using the default p parameter for the SSU rDNA.
Results
Prevalence
Fifty specimens of Gobioides grahamae were examined, of which 15 (30%) were infected by plasmodia and myxospore with morphological characteristics typical of the genus Sphaeromyxa.
Plasmodia
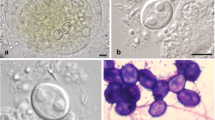
Large plasmodia were observed macroscopically through the wall of the gall bladder, floating in the bile. These plasmodia varied in shape (flat, rounded, oval or elongated) and size (Fig. 1a, b). A large, rounded plasmodium, measuring 134.0 × 145.6 μm, was observed, in which the ectoplasm was clearly visible and transparent (Fig. 1b), while the endoplasm was characterised by a network of vacuoles of varying sizes (Fig. 1c).
Plasmodia of Sphaeromyxa azevedoi n. sp. in the gall bladder of G. grahamae. A Plasmodia (arrows) of varying sizes, flat-shaped and rounded. Scale bar = 100 μm. B Large, rounded plasmodium (*), showing the clearly demarcated, transparent ectoplasm (arrows). Scale bar: 20 μm. C Plasmodium, showing the ectoplasm and the endoplasm (*) with of a vacuoles of varying sizes. Scale bar: 20 μm
Myxospores
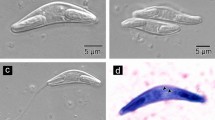
Mature myxospores were found floating in the bile (Fig. 2a, b). Some myxospores were found floating in pairs in the bile (Fig. 2b). The myxospores had superficial striations and were elongate in the frontal view, fusiform, straight to convex (Fig. 2c), with rounded extremities. In the valvular view, the myxospore was 27.1 ± 2.7 (20.5–30.1) μm length and 3.8 ± 0.2 (3.5–4.4) μm width (Table 1). The myxospore has two polar capsules of equal size 8.1 ± 0.6 (7.4–9.4) μm in length and 2.9 ± 0.2 (2.3–3.3) μm in width (Table 1). Both polar capsules have an elongated oval shape and are located in the opposite extremities of the spore, separated by the binucleated sporoplasm (Fig. 2c). Each polar capsule contains a polar tubule, which is arranged perpendicularly to its principal axis, with three or four coils (Fig. 2c). The histological analysis confirmed that the myxospore is present in the lumen of the gall bladder (Fig. 2d). The new species was drawn schematically (Fig. 3) to provide an overview of its morphological observations.
Myxospore of Sphaeromyxa azevedoi n. sp. in the gall bladder of G. grahamae. A Mature myxospore (arrows). Scale bar: 10 μm. B Mature myxospore (arrows) arranged in pairs, showing the polar capsules arranged at the extremities of the spore, and polar capsules of the degraded myxospore (arrowheads). Scale bar = 10 μm. C Myxospore of Sphaeromyxa azevedoi n. sp., in which the polar tubule (PF) can be observed within the polar capsule (PC), perpendicular to its principal axis. Scale bar: 5 μm. D Gall bladder stained with Ziehl–Neelsen, showing myxospore (arrows) of Sphaeromyxa azevedoi n. sp. in the lumen (arrow). Scale bar: 20 μm
Description of the taxon
Phylum Cnidaria Hatschek, 1888
Class Myxosporea Bütschli, 1881
Order Bivalvulida Shulman, 1959
Family Sphaeromyxidae Lom and Noble, 1984
Genus Sphaeromyxa Thélohan, 1892
Species Sphaeromyxa azevedoi n. sp.
Taxonomic summary
Type host: Gobioides grahamae (Palmer and Wheeler, 1955).
Site of infection: gall bladder. No inflammatory infiltration was found in association with the parasitic infection.
Type locality: Paracauari River (00°45ʹ S, 48°30ʹ W), municipality of Salvaterra, Marajó Island, Pará, Brazil.
Type specimens: A glass slide with a stained, 5-µm histological section containing the syntype of the new myxosporan species was deposited at the Zoology Museum of the National Institute of Amazonian Research (INPA) in Manaus, Amazonas, Brazil, under catalogue number CNIDARIA—INPA 039.
GenBank accession number: MK573247.
Etymology: The species Sphaeromyxa azevedoi sp. n. is named in honour of Prof. Dr. Carlos Azevedo, an eminent researcher in the field of Protozoology at the Department of Cellular Biology of the Abel Salazar Institute of Biomedical Sciences at the University of Porto, Portugal.
Phylogenetic analysis
The sequencing of the myxospore of Sphaeromyxa azevedoi sp. n. resulted in a partial sequence of 1919 bps of the SSU rDNA (For multi-species alignment were used a total of 1569 bp in the final dataset). One clade, which obtained high nodal support, was formed by all the Sphaeromyxa species that have DNA sequences deposited in GenBank. The molecular phylogeny indicates that the genus Sphaeromyxa is monophyletic, forming clusters arranged according to the morphological characteristics of the myxospores. There is one major clade, with high nodal (posterior probability) support, which groups all the Sphaeromyxa species for which molecular data are available. This clade is divided into three subclades, denominated the ‘balbianii’, ‘incurvata’ and ‘limocapitis’ groups. Sphaeromyxa azevedoi n. sp. is part of the ‘balbianii’ group, a subclade formed by Sphaeromyxa artedielli (Karlsbakk et al. 2013), Sphaeromyxa longa (Bartošová-Sojková et al. 2015), Sphaeromyxa zaharoni (Diamant et al. 2004), Sphaeromyxa horrida (Miller et al. 2018) and Sphaeromyxa balbianii (Karlsbakk et al. 2013). Sphaeromyxa azevedoi n. sp. is the basal species of S. artedielli and S. longa, which parasitise the marine fish Artediellus atlanticus Jordan and Evermann, 1989 and Trisopterus minutus Linnaeus, 1758, respectively.
When only the SSU rDNA sequences of Sphaeromyxa species were considered in the analysis, the smallest p distance was 5.5%, between Sphaeromyxa azevedoi n. sp. and S. artedielli (KF 135220), while the greatest p distance was 10.7%, between Sphaeromyxa azevedoi n. sp. and S. clini (KM201336).
Discussion
The species of the genus Sphaeromyxa can be separated into three groups based on the morphology of the myxospores, an arrangement supported by phylogenetic studies (Whipps and Font 2013; Kristmundsson and Freeman 2013; Bartošová-Sojková et al. 2015). The first two groups (‘incurvata’ and ‘balbianii’) were proposed by Laird (1953). The morphology of the ‘incurvata’ group is characterised by arched myxospore with pyriform polar capsules, while the ‘balbianii’ group has straight, slightly curved and fusiform or ovoid myxospores, with ovoid polar capsules. The third group (‘limocapitis’) was proposed by Bartošová-Sojková et al. (2015), based on phylogenetic analyses, which indicate that Sphaeromyxa limocapitis has fusiform myxospore with pointed extremities, which represents a distinct lineage within the genus Sphaeromyxa. This is one of the few myxozoan genera in which the morphological classification is consistent with the estimated phylogeny, although only a few species have been analysed phylogenetically (Bartošová-Sojková et al. 2015).
The species of the genus Sphaeromyxa are typical of both marine and estuarine environments (Karlsbakk et al. 2013). Sphaeromyxa azevedoi n. sp. was found in the region of the Amazon estuary, where its host was G. grahamae (Gobiidae). Three other Sphaeromyxa species have been described from gobiid hosts; Sphaeromyxa pultai (Tripathi, 1953) which was found in Odontamblyopus rubicundus, in India, and Sphaeromyxa sevastopoli (Naidenova, 1970), which was found in Neogobius fluviatilis, in the Azov Sea, in Russia, are morphologically consistent with the ‘balbianii’ group, although both species differ from Sphaeromyxa azevedoi n. sp. due to its smaller and more truncated myxospore (Table 1). The other species is S. kenti (Whipps and Font, 2013), which was found in Gobiosoma bosc (Gobiidae) from the estuarine waters of Lake Pontchartrain in Louisiana, in the USA, and belongs to the ‘incurvata’ group, which includes species with arched myxospores, which are distinct from Sphaeromyxa azevedoi n. sp.
The straight or slightly curved myxospores of Sphaeromyxa azevedoi n. sp. are also consistent with the characteristics of the ‘balbianii’ group (Laird, 1953). Among the myxospores of Sphaeromyxa spp., the morphometry of Sphaeromyxa azevedoi n. sp. is similar to those of S. lomi (Moser and Noble, 1977) and S. pultai (Tripathi, 1953) (Table 1). The extremities of the myxospore of most of the species of the ‘balbianii’ group are truncated, as observed in the valvular and sutural views, as in S. artedielli, S. longa, S. zaharoni, S. horrida and S. balbianii, even though this characteristic was not observed in the mature myxospore of Sphaeromyxa azevedoi n. sp., which have rounded extremities (Fig. 2b, c).
The plasmodia of Sphaeromyxa azevedoi n. sp. are flat in shape, rounded, oval or elongated, with different sizes, well-defined ectoplasm and endoplasm containing a network of vacuoles of varying sizes (Fig. 1a, b). The plasmodia of the new species are typical of the genus Sphaeromyxa, and have characteristics that are consistent with those described in other Sphaeromyxa species, including pansporoplasts surrounding the vacuoles of the maturing myxospore (Lom 1969, 2004; Lom and Dyková 1992; Diamant et al. 2004; Karlsbakk et al. 2013).
In most cases, infections by Sphaeromyxa species do not appear to cause abnormalities of the tissue. However, infection by Sphaeromyxa is known to cause damage to the gall bladder and bile ducts in some cases, such as Kalavati and Vaidehi (1991), who described damage to the gall bladder associated with infection by S. ganapatii; Sears et al. (2011), who reported hepatic pathologies and damage to the bile ducts associated with infection by S. cannolii; and Bartošová-Sojková et al. (2015), who verified, in histological sections, complete or partial occlusion of the bile ducts caused by the dilatation of S. clini plasmodia.
Molecular phylogenetic analyses confirm that Sphaeromyxidae form a monophyletic group, where clusters are formed based mainly on the morphological characteristics of myxospore (Kristmundsson and Freeman, 2013; Chen et al., 2020). Sphaeromyxa azevedoi n. sp. was assigned to the ‘balbianii’ group, with five other species. Sphaeromyxa artedielli and S. longa were assigned to the same subclade as Sphaeromyxa azevedoi n. sp. in the phylogenetic analysis (Fig. 4). However, the two species are much less similar, morphologically (Table 1). The morphological characteristics of the myxozoans are important for the identification of new species, although the complementary analysis of molecular traits provides a much more reliable diagnosis of a new species (Fiala et al., 2015). Sphaeromyxa azevedoi n. sp. is phylogenetically closest to the marine species S. artedielli and S. longa, and less closely related to S. kenti, a gobiid parasite of an estuarine fish host. While no Sphaeromyxa species has been described based only on molecular data in Brazilian host, the molecular phylogeny presented here indicated that neither the habitat nor the genus of the host contributes to the arrangement of the Sphaeromyxa clades.
Cladogram of the partial SSU rRNA gene sequences of Sphaeromyxa azevedoi n. sp. and closely related myxosporeans, generated by Bayesian Inference (BI). The GenBank accession numbers are shown next to the species names. The numbers at the nodes are the posterior probabilities calculated by the BI. The new species is highlighted in bold type
In fact, Sphaeromyxa azevedoi n. sp. is the first member of the ‘balbianii’ group that includes the molecular analysis, known to have myxospore with rounded extremities. It is important to note, however, that few genetic data are available for the species of the genus Sphaeromyxa, and SSU rDNA sequences are available for only 14 taxa (Fig. 4). As most of the descriptions of Sphaeromyxa species are based on morphological characteristics, it is important to confirm the existence of new species based on molecular analyses.
Sphaeromyxa azevedoi n. sp. represents the first description of a new species of this genus from Brazil, based on molecular data. Pinto (1928) recorded the occurrence of S. balbianii in Brazil, from the gall bladder of the shovelhead shark Sphyrna tiburo (Linnaeus, 1758), although it has smaller myxospore than Sphaeromyxa azevedoi n. sp., with truncated extremities (Table 1). The combined morphological and molecular (SSU rDNA sequences) analyses indicate that Sphaeromyxa azevedoi n. sp. is a member of the ‘balbianii’ group, and is the first Sphaeromyxa species described in G. grahamae, a gobiid host from Brazilian Amazonia.
References
Azevedo C, Rocha S, Matos E, Oliveira E, Matos P, Al-Quraishy S, Casal G (2016) Ultrastructural and phylogenetic description of Kudoa orbicularis n. sp. (Myxosporea: Multivalvulida): a parasite infecting the muscle of the fish Chaetobranchopsis orbicularis (Teleostei: Cichlidae) in the Amazon region. J Eukaryot. Microbiol. 63(1), 27–36. https://doi.org/10.1111/jeu.12244
Azevedo C, Videira M, Casal G, Matos P, Oliveira E, Al-quraishy S, Matos E (2013) Fine structure of the plasmodia and myxospore of Ellipsomyxa gobioides n. sp. (Myxozoa) found in the gallbladder of Gobioides broussonnetii (Teleostei: Gobiidae) from the lower Amazon River. J Eukaryot Microbiol 60(5): 490–496. https://doi.org/10.1111/jeu.12056
Barthem RB (1990) Descrição da pesca da piramutaba (Brachyplatystoma vaillantii. Pimelodidae) no estuário e na calha do rio Amazonas. Bol Mus Para Emlio Goeldi sér.Antropol 6(1):117–13
Bartošová-Sojková P, Kodádková A, Pecková H, Kuchta R, Reed CC (2015) Morphology and phylogeny of two new species of Sphaeromyxa Thélohan, 1892 (Cnidaria: Myxozoa) from marine fish (Clinidae and Trachichthyidae). Parasitology 142(5):660–674. https://doi.org/10.1017/S0031182014001723
Brassard P, Rau ME, Curtis MA (1982) Parasite-induced susceptibility to predation in diplostomiasis. Parasitology 85:495–501. https://doi.org/10.1017/S0031182000056274
Chen W, Yang C, Whipps CM, Peng Z, Zhao Y (2020) Taxonomy on three novel species of Sphaeromyxa Thélohan 1892 (Myxozoa, Bivalvulida, Sphaeromyxidae) with insight into the evolution of the genus. Parasitol Res 119(5):1493–1503. https://doi.org/10.1007/s00436-020-06656-w
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772. https://doi.org/10.1038/nmeth.2109
Diamant A, Whipps CM, Kent ML (2004) A new species of Sphaeromyxa (Myxosporea: Sphaeromyxina: Sphaeromyxidae) in devil firefish, Pterois miles (Scorpaenidae), from the northern red sea: morphology, ultrastructure, and phylogeny. J Parasitol 90(6):1434–1442. https://doi.org/10.1645/GE-336R
Hall TA (1999) BioEdit: a user–friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hillis DM, Dixon MT (1991) Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 66(4):411–453
Kalavati C, Vaidehi J (1991) Two new species of myxosporidians from fishes of Chilika Lake: genus Sphaeromyxa Thelohan and genus Zschokkella Auerbach. Uttar Pradesh Journal of Zoology 11(2):146–150
Karlsbakk E, Einen AC, Bartosová P (2013) Sphaeromyxa artedielli sp. n. (Myxozoa: Sphaeromyxidae) a parasite of sculpins (Cottidae) in northern Norway. Folia Parasitol 60(5):425–432. https://doi.org/10.14411/fp.2013.045
Kristmundsson A, Freeman MA (2013) Sphaeromyxids form part of a diverse group of myxosporeans infecting the hepatic biliary systems of a wide range of host organisms. Parasit Vectors 6:51. https://doi.org/10.1186/1756-3305-6-51
Laird M (1953) The protozoa of New Zealand intertidal zone fishes. Transactions of the Royal Society of New Zealand 81:79–143
Lom J, Arthur JR (1989) A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 12(2):151–156. https://doi.org/10.1111/j.1365-2761.1989.tb00287.x
Lom J (1969) Notes on the ultrastructure and sporoblast development in fish parasitizing myxosporidian of the genus Sphaeromyxa. Z Zellforsch 97(3):416–437. https://doi.org/10.1007/BF00968848
Lom J (2004) Morphology and ultrastructure of Sphaeromyxa noblei sp. n. (Myxozoa), parasite of Heteroclinus whiteleggii (Pisces) from Australian New South Wales coast. Folia Parasitologica 51: 19–26. https://doi.org/10.14411/fp.2004.003
Lom J, Dyková I (1992) Protozoan parasites of fishes. Elsevier, Amsterdam, p 315
Luna LG (1968) Manual of histologic staining methods of the armed forces institute of pathology. McGraw-Hill, New York
Mathews PD, Silva MR, Maia AA, Adriano EA (2015) Ultrastructure and ssrRNA sequencing of Myxidium amazonense n. sp. a myxosporean parasite of Corydoras melini from the Rio Negro river, Amazonas state, Brazil. Parasitol Res 114 (12): 4675–4683. https://doi.org/10.1007/s00436-015-4715-5
Matos PS, Silva DT, Hamoy I, Matos E (2018) Morphological features and molecular phylogeny of Hoferellus azevedoi n. sp. (Myxozoa: Myxobilatidae) found in Chaetobranchus flavescens Heckel, 1840 (Teleostei: Cichlidae) from Marajó Island, northern Brazil. Parasitol Res 117(4): 1087-1093. https://doi.org/10.1007/s00436-018-5785-y
Mendes FLS, Barthem RB (2010) Hábitos alimentares de bagres marinhos (Siluriformes: Ariidae) do estuário amazônico. Amazônia: Ci & Desenv 5 (10).
Miller TL, Barnett SK, Seymour JE, Jenkins TP, Mcnamara M, Adlard RD (2018) Biliary tract-infecting myxosporeans from estuarine and reef stonefish (Scorpaeniformes: Synanceiidae) off eastern Australia, with descriptions of Sphaeromyxa horrida n. sp. and Myxidium lapipiscis n. sp. (Myxosporea: Bivalvulida). J Parasitol 104(3): 254–261. https://doi.org/10.1645/17-79
Miller KM, Teffer A, Tucker S, Li S, Schulze AD, Trudel M, Juanes F, Tabata A, Kaukinen KH, Ginther NG, Ming TJ, Cooke SJ, Hipfner JM, Patterson DA, Hinch SG (2014) Infectious disease, shifting climates, and opportunistic predators: cumulative factors potentially impacting wild salmon declines. Evol Appl 7:812–855. https://doi.org/10.1111/eva.12164
Molnár K, Eszterbauer E, Szckely C, Dán A, Harrach B (2002) Morphological and molecular biological studies on intramuscular Myxobolus spp. of cyprinid fish. J Fish Dis 25:643–652. https://doi.org/10.1046/j.1365-2761.2002.00409.x
Moser M, Noble ER (1977) Three genera of myxosporida (Protozoa) in macrourid fishes. Int J Parasitol 7(2):93–96. https://doi.org/10.1016/0020-7519(77)90072-8
Murdy EO (1998) A review of the genus Gobioides. Ichthyol Res 45(2):121–133. https://doi.org/10.1007/BF02678554
Naidenova NN (1970) Parasitofauna of fishes from family Gobiidae from the Azov Sea. Biologiya Morya 20:84–113 ((In Russian))
Nekouei O, Vanderstichel R, Ming T, Kaukinen KH, Thakur K, Tabata A, Laurin E, Tucker S, Beacham TD, Miller KM (2018) Detection and assessment of the distribution of infectious agents in juvenile Fraser River Sockeye Salmon, Canada, in 2012 and 2013. Frontiers in Microbiology 9:3221. https://doi.org/10.3389/fmicb.2018.03221
Pinto C (1928) Myxosporideos e outros protozoários intestinais de peixes observados na América do Sul. Arch Inst Biol 1:102–136
Rambaut A (2012) FigTree v1.4.0. University of Oxford, Oxford, UK. http://tree.bio.ed.ac.uk/software/figtree.
Rambaut A, Drummond AJ (2007) Trace v1.4, available from http://beast.bio.ed.ac.uk/Tracer
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 63(3): 539–42. https://doi.org/10.1093/sysbio/sys029
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Sears BF, Anderson P, Greiner EC (2011) A new species of myxosporean (Sphaeromyxidae), a parasite of lined seahorses, Hippocampus erectus, from the Gulf of Mexico. J Parasitol 97(4):713–716. https://doi.org/10.1645/GE-2565.1
Tripathi YR (1953) Studies on parasites of Indian fishes. I. Protozoa Myxosporidia together with a check list of parasitic protozoa described from Indian fishes. Records of the Indian Museum 50:63–88
Swofford DL PAUP* (2003) Phylogenetic analysis using parsimony (*and other methods). v. 4.0 beta 10. Sinauer associates, Sunderland.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL–X window interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882. https://doi.org/10.1093/nar/25.24.4876
Velasco M, Matos P, Sanches O, São Clemente SC, Videira M, Santos P, Matos E (2012) Necrotizing myositis associated with parasitism by Myxobolus sp. (Myxozoa) in the palate of the violet goby, Gobioides broussonnetii (Gobiidae), from Marajó Island. Brazil Aquaculture 358–359:129–131. https://doi.org/10.1016/j.aquaculture.2012.06.033
Videira M, Velasco M, Matos P, São Clemente SC, Sanches O, Matos E (2014) Hepatic steatosis associated with microsporidiosis in teleost fishes from Marajó Island. Brazil an Acad Bras Ciênc 86(3):1347–1350. https://doi.org/10.1590/0001-3765201420130147
Videira M, Velasco M, Sanches O, Matos P , Santos PS, Matos E (2020) First report of Kudoa sp. in the palate and pharyngeal musculature of Gobioides grahamae Palmer and Wheeler, 1955 (Perciformes, Gobiidae) from Marajó Island, Brazil. Arq. Bras Med Vet. Zootec 72(2):517–522. https://doi.org/10.1590/1678-4162-11081
Violante-González J, García-Varela M, Rojas-Herrera A, Guerrero SG (2009) Diplostomiasis in cultured and wild tilapia Oreochromis niloticus in Guerrero State, Mexico. Parasitol Res 105:803–807. https://doi.org/10.1007/s00436-009-1458-1
Zatti SA, Atkinson SD, Bartholomew JL, Maia AAM, Adriano EA (2017) Amazonian waters harbour an ancient freshwater Ceratomyxa lineage (Cnidaria: Myxosporea). Acta Trop 169:100–106. https://doi.org/10.1016/j.actatropica.2017.02.006
Whipps CM, Adlard RD, Bryant MS, Lester RJG, Findlay V, Kent ML (2003) First report of three Kudoa species from eastern Australia: Kudoa thyrsites from mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from sweeper (Pempheris ypsilychnus). J Eukaryot Microbiol 50(3): 215–219. https://doi.org/10.1111/j.1550-7408.2003.tb00120.x
Whipps CM, Font WF (2013) Interaction of two myxozoan parasites from naked goby Gobiosoma bosc, in Lake Pontchartrain, Louisiana. J Parasitol 99(3): 441–447.https://doi.org/10.1645/12-49.1
Whipps CM (2015 Oct) Zhao Y (2015) Synopsis of the species of the genus Sphaeromyxa Thélohan, 1892 (Myxosporea: Bivalvulida: Variisporina: Sphaeromyxidae). Syst Parasitol 92(2):81–99. https://doi.org/10.1007/s11230-015-9591-y
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Sascha L Hallett
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Carreira, K.C.V., da Silva, D.T., de Carvalho Sanches, O. et al. Sphaeromyxa azevedoi n. sp. (Myxozoa: Sphaeromyxidae) infecting the gall bladder of Gobioides grahamae (Perciformes: Gobiidae) in the Amazon region. Parasitol Res 121, 867–875 (2022). https://doi.org/10.1007/s00436-022-07443-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07443-5