Abstract
The appearance of increasing drug resistance in apicomplexan intracellular Plasmodium falciparum presents a significant challenge. P. falciparum infection results in cerebral malaria (CM), causing irreversible damage to the brain leading to high mortality cases. To enhance the clinical outcome of the disease, further research is required to identify new molecular targets involved in disease manifestations. Presently, the role of non-coding microRNAs (miRNAs) derived from different cells implicated in CM pathogenesis is still barely understood. Despite the absence of miRNA machinery in Plasmodium, host-parasite interactions can lead to disease severity or impart resistance to malaria. Cytoadherence and sequestration of parasitized RBCs dysregulate the miRNA profile of brain endothelial cells, leukocytes, monocytes, and platelets, disrupting blood–brain barrier integrity and activating inflammatory signaling pathways. The abundance of miRNA in blood plasma samples of CM patients directly correlates to cerebral symptoms compared to non-CM patients and healthy individuals. Moreover, the differential host-miRNA signatures distinguish P. falciparum from P. vivax infection. Here, we review the diverse functions of host-miRNA, either protective, pathogenic, or a combination of the two, which may act as prognostic markers and novel antimalarial drug targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malaria is a known inflammatory parasitic disease, with cerebral malaria (CM) being one of its most severe forms responsible for neuro-inflammation and disruption of the blood–brain barrier caused by Plasmodium falciparum. CM is an essential concern in the modern world, accounting for significant mortality rates, especially in children (Brewster et al. 1990; John et al. 2008; WHO 2020). In compliance with the world malaria report 2020, there were an estimated 229 million malaria cases globally, with about 94% of cases reported in the WHO African Region (WHO 2020). The high burden of malaria and death still prevails among pregnant women and children (Makenga et al. 2020; SIMON-OKE et al. 2019). However, a more pressing situation would be accelerating current drug inefficacy, leading to clinical resistance and asymptomatic peripheral parasitemia in highly endemic regions (Laishram et al. 2012; Raman et al. 2020). Concordantly, the prevalence of malaria coinfection with other vector-borne parasites, HIV, and opportunistic pathogens curbs the drug efficacy (Deen 2021; Salam et al. 2018). Current biological threats like parasite deletions of pfhrp2/3 genes make RDTs (rapid diagnostic tests) based on HRP2 ineffectual (Thomson et al. 2019; WHO 2017). Mutations in PfKelch13 creating partial resistance to artemisinin (ART) and ART partner drugs (Birnbaum et al. 2020; Das et al. 2019; Nsanzabana 2019). Emerging vector resistance to insecticides has been a growing concern regarding malaria transmission (Gnanguenon et al. 2015; Ingham et al. 2017; Yewhalaw et al. 2011). The dream for a malaria vaccine is no longer unattainable. Among many proposed vaccines for malaria, including pre-erythrocytic vaccines, blood-stage vaccines, and transmission-blocking vaccines, RTS,S vaccine has been the front runner candidate that began its pilot implementation project in three African countries in 2018 (Coelho et al. 2017). However, the latest studies reveal that the efficacy of the RTS,S vaccine is partial and depends on host immunologic and genetic factors (Dobano et al. 2019; Khan et al. 2020; Nielsen et al. 2018). This situation calls for novel ways to diagnose and treat the infection, and miRNAs might provide us with a promising standpoint for tackling this current predicament.
MicroRNAs (miRNAs) are small non-coding RNA molecules with no biological function, approximately 22 nucleotides in length. They regulate mRNA expression via a process known as translational repression by binding to their putative mRNA target through complementary base pairing, as demonstrated in Fig. 1. Thus, leading to cleavage of mRNA or decreases translational efficiency by causing improper ribosomal loading followed by mRNA destabilization. (Bartel 2009; Bartel 2018; Bartel 2004; Fabian et al. 2010). Additionally, miRNAs in high concentrations intervene at a transcriptional level by hypermethylation of genes encoding target RNAs resulting in reduced transcription (Khraiwesh et al. 2010). Metazoan microRNAs maintain a characteristic profile under normal physiological conditions where they regulate tissue-specific functions in which they are uniquely expressed (Liang et al. 2007; Ludwig et al. 2016; van de Bunt et al. 2013). Dysregulation of miRNA profile is observed in response to disrupting normal metabolic processes due to external stimuli like infiltration of a pathogen into a host cell (Ruiz-Tagle et al. 2020; Zhou et al. 2018). Since many pathogens prefer a specific tissue to infest, some miRNAs uniquely expressed in those tissues characteristically dysregulate in response, thereby altering the expression levels of their target genes. Such incidences reinforce miRNAs clinical potential as next-generation medicine and diagnostic tool (Chakraborty et al. 2017).
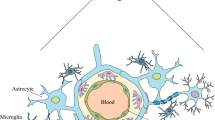
Interestingly, these alterations might cause a protective effect or aid in disease progression. P. falciparum completes its asexual life-cycle in human red blood cells (RBCs) as a host. The RBCs harbor few miRNAs and modulate the infectivity process by serving as clinical biomarkers of specific disease conditions (Sun et al. 2018; Sun et al. 2020). Considerable work has shown that intracellular malarial parasites lack miRNA biogenesis, and changes in host-miRNAs control parasite development, as shown in Fig. 2 (Rathjen et al. 2006; Xue et al. 2008). However, minimal information on miRNA in malaria pathogeny has been established and remains elusive to date. Our review’s main aim is to understand better how this dysregulation of specific miRNAs during plasmodial infection affects host-parasite interactions and provide insight on ways to exploit them for clinical benefit. A complete list of the functions and pathways targeted by each miRNA during Plasmodium infection is represented in Table 1.
Role of miR-451 in Plasmodium infection
The miR-451 family originates from a bicistronic gene cluster—premiR-144/451 that encodes for miR-144 and miR-451, which are highly conserved genes. miR-451 are profoundly located inside RBCs and are primarily involved in normal human erythropoiesis (Masaki et al. 2007; Wang et al. 2019) and tumor progression (Bai and Wu 2019). An in vitro analysis has shown that miR-451a is found in abundance in the cell conditioning media of human brain endothelial (HBE) cells infected with cytoadhering strain of P. falciparum FCR3 serving as a model for cerebral malaria (Gupta et al. 2021). A study outlined the examination of extracellular vesicles (EVs) released by parasitized-RBCs (pRBCs) during a blood-stage P. falciparum infection that revealed miR-451a to be highly concentrated inside those EVs (Babatunde et al. 2018). Moreover, these EVs containing miR-451a RISC complex with Ago2 integrated with endothelial cells and downregulated CAV-1 and ATF-2, causing endothelial barrier dysfunction and apoptosis (Mantel et al. 2016).
Similarly, another study demonstrated a high accumulation of hAgo2-miRNA complex inside EVs released from pRBCs, which were transferred into the parasites where miR-451 downregulated the var gene of parasite virulence factor PfEMP1, essentially providing resistance (Wang et al. 2017). This particular miRNA is found inside sickle cell erythrocytes and variants, blocking Plasmodium mRNA translation and conferred malaria resistance (Feliciano 2012; LaMonte et al. 2012). Furthermore, a notable dichotomy was observed in the dysregulation of miR-451 when comparing the plasma miRNA profiles of patients suffering from P. vivax and P. falciparum. Significant downregulation of miR-451 was noted during a P. vivax infection which negatively correlated to parasitemia. On the other hand, miR-451 showed marked upregulation in a P. falciparum infection (Chamnanchanunt et al. 2015). The collected evidence shows the potential of this miRNA family as an ideal biomarker in distinguishing different forms of malarial disease and a possible candidature (miR-451) in the treatment of P. falciparum malaria. An in vivo study suggested that miR-451 negatively regulated parasitemia by limiting CD4+ proliferation by directly targeting the Myc gene during infection (Chapman et al. 2017). Myc gene is a known regulator of cell cycle progression and tissue proliferation (Henriksson and Luscher 1996), and CD4+ T-cells have been demonstrated to be critical in malaria clearance in humans (Pombo et al. 2002). It has been evidenced by inoculating P. yoelii XNL into WT and miR 451−/− mice. The miR 451−/− mice showed faster parasite clearance in blood and an increased leukocyte response, particularly CD4+ T-cells. The reason explained was partly due to the suppression of Myc-regulated genes that affected T-cell proliferation. Based on the findings, we reasoned that nonoverlapping evidence of miR-451 between in vivo and in vitro systems attributes to the differences in metabolic regulations and biological pathways.
Role of miR-155 in Plasmodium infection
Certain miRNAs do not show significant dysregulation during a specific infection but could position themselves as a critical player in therapeutics, such as the case of miR-155 in the course of cerebral malaria infection. miR-155 has widely been accepted as an oncomiR significantly upregulated and modulates tumor progression in breast cancer (Iorio et al. 2005; Jiang et al. 2010; Neilsen et al. 2013). Besides its oncogenic properties, miR-155 is also involved in regulating inflammation (Hu et al. 2014; Tili et al. 2011), with SHIP1 and SOCS1 being its primary targets (O'Connell et al. 2009; Qayum et al. 2016). Bioinformatic analysis exhibited miR-155 to moderate CD36, TLR4, IFN-γ, and PRR15 responsible for the progression of cerebral malaria (Rangel 2017). Furthermore, a study revealed that inhibiting miR-155 leads to increased survivability due to reduced vascular leakage and conservation of the “blood–brain barrier” despite causing a higher inflammatory response. It was experimentally shown by infecting miR-155−/− and WT mice with P. berghei ANKA (Barker et al. 2017). The study also substantiated that miR-155 inhibition with Antagomir155 reduced vascular leakage induced by sera samples collected from Ugandan children with CM in an ex vivo endothelial microvessel model (Barker et al. 2017). Another study displayed a significant increase of miR-155 in Kupffer cells of mice infected with injections of genetically attenuated parasites (GAP) of P. berghei uis3(-) to induce immunity, and ectopic administration of miR-155 using adeno-associated virus 8 (AAV8) vectors reduced the number of injections required to induce sterile immunity in the liver from 3 to 1(Hentzschel et al. 2014).
In other apicomplexan parasites, a study reported that intravenously injected Leishmania donovani (LV82) amastigotes in miR-155 knockout (KO) mice showed an inadequate immune response a higher parasite count in the spleen and liver. The recovery of miR-155 KO mice from parasitemia is twice as slow as its WT counterpart. This study indicates miR-155 as an enhancer of immunity by modulating IFN-γ, IL-4, and CCR2 but is inessential in resolving/curing infection since both mutant and WT mice eventually cleared the parasites (Varikuti et al. 2019). Another investigation showed that T. gondii (RH) infected dendritic cells (DC2) released exosomes with high miR-155-5p levels. These exosomes were taken up by macrophage cells (RAW264.7) where miR-155-5p targeted SOCS1 to activate the NF-κB pathway, triggering pro-inflammatory cytokines (IL-6, TNFα, iNOS). This condition resulted in M1 polarization of the macrophages leading to inhibition of T. gondii proliferation. Transfection of miR-155-5p mimic into RAW264.7 cells under in vitro conditions displayed a similar effect (Jiang et al. 2021). The availability of such compelling evidence indicates miR-155 as a possible target for treating cerebral malaria as an adjunct therapy.
Role of miR-27a in Plasmodium infection
miR-27a is a crucial component of the host-miRNA profile of many apicomplexan parasitic infections. This family of miRNA is highly expressed in endothelial cells, involved in angiogenesis, and the central nervous system controlling apoptosis (Urbich et al. 2012) and neuroinflammation during oxidative stress in the brain (Narasimhan et al. 2012). Increased expression of miR-27a was observed in the brain tissue of mice infected with P. berghei ANKA linked to increased TNF expression (El-Assaad et al. 2011). Additionally, miR-27a-5p had elevated levels in the brain tissue of CM-infected mice (Martin-Alonso et al. 2018). Interestingly, a study on Thai patients with P. falciparum infection showed that miRSNPs in miR-27a and miR-146a did not alter CM pathogenesis (Wah et al. 2019). It indicates that miR-27a retains its function and target specificity even upon mutation marking this small non-coding RNA to have clinical significance in malarial pathogenesis. These findings solidify the prospect of miR-27a as a CM fingerprint. A study elucidated the upregulation of miR-27a/b blocked ABCA1 gene expression (Zhang et al. 2014). Inhibition of ABCA1 conferred protection from CM as ABCA1 KO mice reported complete resistance to P. berghei ANKA infection (Combes et al. 2005). Notably, it was observed that Tanshinone IIA (Tan) inhibited LPS induced inflammatory damage to human bronchial epithelial cells (BEAS-2B) by repressing miR-27a, causing inactivation of PI3K/AKT and JNK pathways (Liu and Meng 2018). It is noteworthy to look at the role of miRNAs during inflammation in severe P. falciparum infection. The therapeutic capability of miR-27a has been explored in other parasitic diseases. A significantly elevated level of miR-27a was observed in the intestinal tissue of Kazakh sheep resistant to Echinococcus granulosus infection, suggesting the involvement of miR-27a mediated resistance (Jiang et al. 2016). In Leishmania major infected macrophages, miR-27a mimic acted synergistically with miR-340, resulting in reduced macrophage infectivity by downregulation of IL-10 and TGF-β1 expression (Hamidi et al. 2021). These inflammatory pathways are also involved in the progression of cerebral malaria, indicating that further studies warrant exploration of this crucial lead.
Role of miR-150 in Plasmodium infection
miR-150 is abundantly found in the brain tissue of mice induced with CM infection (Cohen et al. 2018; El-Assaad et al. 2011; Martin-Alonso et al. 2018). This particular miRNA is highly abundant in monocytes, and its increased levels in brain tissue are due to the sequestration of monocytes during disease progression (El-Assaad et al. 2011). miR-150 is expressed majorly in mature lymphocytes and is known to negatively regulate transcription factor c-Myb controlling different stages of lymphocyte development, specifically B cell differentiation (Xiao et al. 2007). A study revealed that mutant mice lacking miR-150 had decreased mature NK cells (Bezman et al. 2011), affecting parasite clearance. It was further evidenced where human NK cells were directly stimulated by Leishmania promastigotes or their lipophosphoglycan (LPG) to produce IFN-γ, which further activated macrophages and curtailed the progression of early-stage infection in culture (Bogdan 2012). Conferring with prior reports, miR-150 aggregation in the brain tissue during CM possibly hampers NK cell development and aid in the pathogenesis of cerebral malaria. Interestingly, erythrocytes loaded with chemically synthesized miR-150-3p and miR-197-5p hindered parasite invasion and growth. Moreover, the miRNA-loaded pRBCs downregulated the expression of Plasmodium apicortin resulting in reduced secretion of apical membrane antigen1 (AMA1) (Chakrabarti et al. 2020). Injecting miR-150 alone or combining antimalarial agents in malaria patients could effectively reduce the parasitic burden as a new therapeutic intervention. Conversely, a study linked miR-150-5p as a marker to non-thrombocytopenic P. vivax infections by conducting bioinformatic analyses using patient samples (Santos et al. 2020). The revealed data holds as miR-150 is a known modulator of platelet biogenesis and activity (Gatsiou et al. 2012; Pordzik et al. 2018). Further investigations should open up diagnostic prospects of miR-150 in the case of P. vivax malaria concerning severe thrombocytopenia.
Role of miR-146 in Plasmodium infection
The miR-146 family has shown great potential in moderating cerebral malaria biogenesis. This family has been involved in innate immunity via regulation of TLR signaling resulting in cytokine response (Sonkoly et al. 2008). miR-146 and miR-155 induce pro-inflammatory stimuli like IL-1, TNFα, and TLRs (Sheedy and O'Neill 2008) and are widely studied as oncogenic modulators (Testa et al. 2017). They also have been verified to modulate genes (CD36, TLR4, IFN-γ, and PRR15) responsible for cerebral malaria (Rangel 2017). Furthermore, increased levels of miR-146a in plasma microvesicle of CBA mice infected with P. berghei ANKA were confirmed. The increase was attributed to the triggering of factors such as IL-1 and TNF, causing suppression of inflammation-inducing genes and those involved in toll-like receptor pathways like TLR2 and TLR4 (Cohen et al. 2018). We previously discussed miRSNPs do not alter miRNA function and target specificity in CM (Wah et al. 2019). The data is consistent regarding another community study that elucidated miR-146a polymorphism was not associated with P. falciparum, P. vivax, or mixed infection in southern India (van Loon et al. 2020). However, another study from the same group demonstrated that miR-146a SNP (rs2910164) promoted the manifestation of P. falciparum malaria, especially in Ghanaian women suffering from pregnancy-associated malaria (PAM), by causing failure of TLR activity leading to altered expression in IRK-1 and TRAF6 levels (van Loon et al. 2019).
Additionally, miR-146a has shown a manipulative role in other parasitic infections. Elevated expression of miR-146a during Leishmania major infection in mice blocks TGF-β signaling, causing the diminution of the parasite inside the macrophages (Nimsarkar et al. 2020). However, in a Toxoplasma infection mice model, increasing miR-146a and miR-155 correlated with increased infection. Ablation of miR-146a expression in infected mice showed decreased IFN-γ levels and played a protective role during early infection of Toxoplasma in the gut (Cannella et al. 2014). These findings point to the fact that each parasite-host interaction responds differently in the presence or absence of the same microRNA. This implies that the role of a specific microRNA is functionally different in every apicomplexan infection, even if it shows similar dysregulation among all of them.
Role of let-7 in Plasmodium infection
The let-7 family of miRNA is important to assess when discussing CM. This family of miRNA regulates all the three genes of the RAS domain in humans (Johnson et al. 2005) and is embroiled in negatively regulating TLR4 expression, the major immune receptor of microbial lipopolysaccharide during protozoan infection (Androulidaki et al. 2009; Hu et al. 2009). Significantly elevated levels of Let-7i were noted in the brain tissue of CBA mice suffering from CM. The role of TLRs during P. falciparum infection is argued, but increased levels of this miRNA in CM infection leading to TLR4 activation might be due to the difference in host genetic factors (El-Assaad et al. 2011). On the other hand, let-7i contributes to Cholangiocyte immune responses against Cryptosporidium parvum infection by regulating TLR4 (Chen et al. 2007). A general trend of decrease of let-7i upon microbial infection is evidenced by an experiment that showed NFκB p50 and C/EBPβ mediating let-7i silencing following C. parvum infection or LPS treatment (Chen et al. 2007). It is hypothesized that infection leads to the formation of a repressor (NFκB p50-C/EBPβ silencer complex) that binds to the let-7i promoter region and promotes histone H3 deacetylation (Chen et al. 2007; O'Hara et al. 2010).
The protective role of the let-7 family is further strengthened from research which demonstrated accelerated liver generation due to upregulation of a multitude of members of the family (let-7a-5p, let-7b-5p, let-7c-5p, let-7d-5p, let-7f-5p, let-7 g-5p, let-7i-5p) among others (miR-27a included) in Balb/c mice protected by vaccination during a crisis of Plasmodium chabaudi blood-stage malaria (Dkhil et al. 2016). Surprisingly, a member of the let-7 family is involved in regulating gene expression in P. falciparum. Let-7a, accompanied by two more host RBC miRNAs (miR-15a and miR-144), are imported into the parasite along with components of miRISC (hAgo2) that target Plasmodium mRNAs encoding putative Rad54, L/S symporter genes, and Mal8p1.29, which provide additional stability and regulatory network to parasite mRNAs (Dandewad et al. 2019). Furthermore, it is observed that hAgo2 is imported in each of the intraerythrocytic stages of P. falciparum and its levels increase as the parasite progresses from ring to schizont stage (Dandewad et al. 2019), thus serving as a possible indicator in disease progression.
Role of miRNA signature (cohort) in Plasmodium infection
It is also imperative to look at sets of miRNAs as individual markers and define microRNAs that work synergistically to provide a more detailed account of disease progression and a prognostic marker or a medium for treatment. A study reported dichotomous miR expression in whole blood miRNA profile of CHMI volunteers with falciparum malaria on three occasions, i.e., prior to Day 4 and Day 7. A 3-miR signature consisting of miR 15a-3p, miR 30c-5p, and miR30e-5p is differentially expressed between high miR responders versus low miR responders. The signature negatively correlated with parasite burden and is considered a potential peripheral blood biomarker controlling blood-stage infection (Burel et al. 2017). Upon acute P. berghei ANKA infection, differentiation of monocyte-derived dendritic cells resulted in dysregulation of miR-16-5p and miR-491-5p as a signature that potentially targets neuroinflammation and dendritic cell maturation (Assis et al. 2020). Consistent with the above findings, the expression profile of peripheral whole blood miRNA during the blood stage of adult imported falciparum malaria (AIFM) in patients showed marked upregulation of five miRNAs (miR-6780b-5p, miR-3135b, miR-1246, miR-6126, and miR-3613-5p) (Li et al. 2018).
Further in silico analysis established the diverse roles of these miRNAs in immune response and as a biomarker in the early detection of P. falciparum malaria. Subsequently, in whole blood samples of complicated P. vivax malaria, five miRNAs (hsa-miR-7977, hsa-miR-28-3p, hsa-miR-378-5p, hsa-miR-194-5p, hsa-miR-3667-5p) were significantly upregulated. In silico analysis exemplified that high levels of miR-7977 may play a putative role in complicated P. vivax through UBA52 or TGF-beta signaling pathway. This makes miR-7977 a robust potential candidate as a biomarker for differentiating complicated P. vivax malaria from uncomplicated type for effective prognosis and treatment (Kaur et al. 2018). Another study conducted using a longitudinal pediatric cohort in Burkina Faso demonstrated that diminution of immune cells during P. falciparum infection was attributed to the upregulation of miR-15a-5p, miR-16-5p and miR-181c-5p by targeting anti-apoptotic gene BCL2 and induce apoptosis (Dieng et al. 2020). Lymphocyte depletion was hypothesized to be caused due to internalization of pRBCs enriched with the upregulated miR signature.
Additionally, there is mounting evidence to show that P. chabaudi infection in female C57BL/6 mice demonstrated sustained expression of hepatic miRNAs signature after repeated infection imparted protection by regulating the epigenetic modifications in genes. The reprogramming of the distinct liver-miRNA species in immune mice resulted in the self-healing of P. chabaudi infection (Delic et al. 2011). Same investigators further reported the effect of protective vaccinations on differential miRNA expression in the liver of Balb/c mice against P. chabaudi challenge infections. They observed that vaccinations induced liver-miRNA signatures were consistent and able to self-heal and survive mice to lethal infections (Dkhil et al. 2016). Moreover, another study revealed a similar effect on liver-miRNA signature upon P. chabaudi infection in response to testosterone (Al-Quraishy et al. 2012). Presently, limited studies fail to provide a detailed mechanistic action on the self-healing potential of dysregulated liver-miRNA species during reinfection, and further studies are required to understand their protective role.
Conclusion and future perspectives
miRNAs remain an attractive novel target in alleviating the disease condition, especially when the Plasmodium spp. have developed clinical resistance to almost all front-line antimalarial drugs. This review comprehensively covered various facets involving miRNAs interactions with host-signaling pathways during Plasmodium infections. miRNA therapy is recommended as a standalone or adjunctive therapy for numerous diseases in various clinical trials. Therefore, suppressing detrimental miRNAs and restoring suppressed miRNAs could be a feasible approach in arresting the growth of Plasmodium and other inflammatory pathways. Also, this review stresses that a thorough understanding is required of the paradoxical functions of miRNA, their targetome interactions and signaling pathways. Due to the absence of miRNA biogenesis in Plasmodium, controlling the host cell-induced miRNAs regulatory pathway provides an alternative treatment strategy in malaria infections. In support of this notion, coadministration of chemically modified antisense oligonucleotides in a stable nanocarrier system along with antimalarial agents may significantly reduce cerebral-malarial symptoms. Usage of miRNA sponges will help decipher the loss- or gain-of-function of listed miRNAs involved in pathogenesis as an alternative tool. Aberrant expression of malaria-specific miRNAs in plasma could be a possible indicator in diagnostics as a non-invasive, early detection marker. There is limited information on miRNA-regulatory pathways in malaria which warrants further studies in clinical use. Altogether, it seems reasonable to suggest that miRNAs in Plasmodium infection can be considered a potential druggable target and biomarker tool.
References
Al-Quraishy S, Dkhil MA, Delic D, Abdel-Baki AA, Wunderlich F (2012) Organ-specific testosterone-insensitive response of miRNA expression of C57BL/6 mice to Plasmodium chabaudi malaria. Parasitol Res 111(3):1093–1101. https://doi.org/10.1007/s00436-012-2937-3
Androulidaki A et al (2009) The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31(2):220–231. https://doi.org/10.1016/j.immuni.2009.06.024
Assis PA et al (2020) Integrative analysis of microRNA and mRNA expression profiles of monocyte-derived dendritic cells differentiation during experimental cerebral malaria. J Leukoc Biol 108(4):1183–1197. https://doi.org/10.1002/JLB.1MA0320-731R
Babatunde KA et al (2018) Malaria infected red blood cells release small regulatory RNAs through extracellular vesicles. Sci Rep 8(1):884. https://doi.org/10.1038/s41598-018-19149-9
Bai H, Wu S (2019) miR-451: a novel biomarker and potential therapeutic target for cancer. Onco Targets Ther 12:11069–11082. https://doi.org/10.2147/OTT.S230963
Barker KR et al (2017) miR-155 modifies inflammation, endothelial activation and blood-brain barrier dysfunction in cerebral malaria. Mol Med 23:24–33. https://doi.org/10.2119/molmed.2016.00139
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Bartel DP (2018) Metazoan microRNAs. Cell 173(1):20–51. https://doi.org/10.1016/j.cell.2018.03.006
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. https://doi.org/10.1016/s0092-8674(04)00045-5
Bezman NA, Chakraborty T, Bender T, Lanier LL (2011) miR-150 regulates the development of NK and iNKT cells. J Exp Med 208(13):2717–2731. https://doi.org/10.1084/jem.20111386
Birnbaum J et al (2020) A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 367(6473):51–59. https://doi.org/10.1126/science.aax4735
Bogdan C (2012) Natural killer cells in experimental and human leishmaniasis. Front Cell Infect Microbiol 2:69. https://doi.org/10.3389/fcimb.2012.00069
Brewster DR, Kwiatkowski D, White NJ (1990) Neurological sequelae of cerebral malaria in children. Lancet 336(8722):1039–1043. https://doi.org/10.1016/0140-6736(90)92498-7
Burel JG et al. (2017) Dichotomous miR expression and immune responses following primary blood-stage malaria. JCI Insight 2(15). https://doi.org/10.1172/jci.insight.93434
Cannella D et al (2014) miR-146a and miR-155 delineate a microRNA fingerprint associated with Toxoplasma persistence in the host brain. Cell Rep 6(5):928–937. https://doi.org/10.1016/j.celrep.2014.02.002
Chakrabarti M, Garg S, Rajagopal A, Pati S, Singh S (2020) Targeted repression of Plasmodium apicortin by host microRNA impairs malaria parasite growth and invasion. Dis Model Mech 13(6). https://doi.org/10.1242/dmm.042820
Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS (2017) Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids 8:132–143. https://doi.org/10.1016/j.omtn.2017.06.005
Chamnanchanunt S et al (2015) Downregulation of plasma miR-451 and miR-16 in Plasmodium vivax infection. Exp Parasitol 155:19–25. https://doi.org/10.1016/j.exppara.2015.04.013
Chapman LM, Ture SK, Field DJ, Morrell CN (2017) miR-451 limits CD4(+) T cell proliferative responses to infection in mice. Immunol Res 65(4):828–840. https://doi.org/10.1007/s12026-017-8919-x
Chen XM, Splinter PL, O’Hara SP, LaRusso NF (2007) A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem 282(39):28929–28938. https://doi.org/10.1074/jbc.M702633200
Coelho CH, Doritchamou JYA, Zaidi I, Duffy PE (2017) Advances in malaria vaccine development: report from the 2017 malaria vaccine symposium. NPJ Vaccines 2:34. https://doi.org/10.1038/s41541-017-0035-3
Cohen A, Zinger A, Tiberti N, Grau GER, Combes V (2018) Differential plasma microvesicle and brain profiles of microRNA in experimental cerebral malaria. Malar J 17(1):192. https://doi.org/10.1186/s12936-018-2330-5
Combes V et al (2005) ABCA1 gene deletion protects against cerebral malaria: potential pathogenic role of microparticles in neuropathology. Am J Pathol 166(1):295–302. https://doi.org/10.1016/S0002-9440(10)62253-5
Dandewad V, Vindu A, Joseph J, Seshadri V (2019) Import of human miRNA-RISC complex into Plasmodium falciparum and regulation of the parasite gene expression. J Biosci 44(2). https://doi.org/10.1007/s12038-019-9870-x
Das S, Manna S, Saha B, Hati AK, Roy S (2019) Novel pfkelch13 gene polymorphism associates with artemisinin resistance in Eastern India. Clin Infect Dis 69(7):1144–1152. https://doi.org/10.1093/cid/ciy1038
Deen J (2021) Coinfections and Malaria. In: Hommel M, Kremsner PG (eds) Encyclopedia of malaria. Springer, New York, pp 1–10
Delic D, Dkhil M, Al-Quraishy S, Wunderlich F (2011) Hepatic miRNA expression reprogrammed by Plasmodium chabaudi malaria. Parasitol Res 108(5):1111–1121. https://doi.org/10.1007/s00436-010-2152-z
Dieng MM et al (2020) Integrative genomic analysis reveals mechanisms of immune evasion in P. falciparum malaria. Nat Commun 11(1):5093
Dkhil MA, Al-Quraishy SA, Abdel-Baki AS, Delic D, Wunderlich F (2016) Differential miRNA expression in the liver of Balb/c mice protected by vaccination during crisis of Plasmodium chabaudi blood-stage malaria. Front Microbiol 7:2155. https://doi.org/10.3389/fmicb.2016.02155
Dobano C et al (2019) Concentration and avidity of antibodies to different circumsporozoite epitopes correlate with RTS, S/AS01E malaria vaccine efficacy. Nat Commun 10(1):2174. https://doi.org/10.1038/s41467-019-10195-z
El-Assaad F et al (2011) Differential microRNA expression in experimental cerebral and noncerebral malaria. Infect Immun 79(6):2379–2384. https://doi.org/10.1128/IAI.01136-10
Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79:351–379. https://doi.org/10.1146/annurev-biochem-060308-103103
Feliciano P (2012) miRNAs and malaria resistance. Nat Genet 44(10):1079–1079. https://doi.org/10.1038/ng.2428
Gatsiou A, Boeckel JN, Randriamboavonjy V, Stellos K (2012) MicroRNAs in platelet biogenesis and function: implications in vascular homeostasis and inflammation. Curr Vasc Pharmacol 10(5):524–531. https://doi.org/10.2174/157016112801784611
Gnanguenon V et al (2015) Malaria vectors resistance to insecticides in Benin: current trends and mechanisms involved. Parasit Vectors 8:223. https://doi.org/10.1186/s13071-015-0833-2
Gupta H et al (2021) Plasma microRNA profiling of Plasmodium falciparum biomass and association with severity of malaria disease. Emerg Infect Dis 27(2):430–442. https://doi.org/10.3201/eid2702.191795
Hamidi F et al (2021) Inhibition of anti-inflammatory cytokines, IL-10 and TGF-beta, in Leishmania major infected macrophage by miRNAs: a new therapeutic modality against leishmaniasis. Microb Pathog 153:104777. https://doi.org/10.1016/j.micpath.2021.104777
Henriksson M, Luscher B (1996) Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res 68:109–182. https://doi.org/10.1016/s0065-230x(08)60353-x
Hentzschel F et al (2014) AAV8-mediated in vivo overexpression of miR-155 enhances the protective capacity of genetically attenuated malarial parasites. Mol Ther 22(12):2130–2141. https://doi.org/10.1038/mt.2014.172
Hu G et al (2009) MicroRNA-98 and let-7 confer cholangiocyte expression of cytokine-inducible Src homology 2-containing protein in response to microbial challenge. J Immunol 183(3):1617–1624. https://doi.org/10.4049/jimmunol.0804362
Hu R et al (2014) miR-155 promotes T follicular helper cell accumulation during chronic, low-grade inflammation. Immunity 41(4):605–619. https://doi.org/10.1016/j.immuni.2014.09.015
Ingham VA, Pignatelli P, Moore JD, Wagstaff S, Ranson H (2017) The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae. BMC Genomics 18(1):669. https://doi.org/10.1186/s12864-017-4086-7
Iorio MV et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16):7065–7070. https://doi.org/10.1158/0008-5472.CAN-05-1783
Jiang D, Wu S, Xu L, Xie G, Peng H-J (2021) Anti-Infection Roles of mir155-5p Packaged in Exosomes Secreted by Dendritic Cells Infected with Toxoplasma Gondii. https://doi.org/10.21203/rs.3.rs-287510/v1
Jiang S, Li X, Wang X, Ban Q, Hui W, Jia B (2016) MicroRNA profiling of the intestinal tissue of Kazakh sheep after experimental Echinococcus granulosus infection, using a high-throughput approach. Parasite 23:23. https://doi.org/10.1051/parasite/2016023
Jiang S et al (2010) MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res 70(8):3119–3127. https://doi.org/10.1158/0008-5472.CAN-09-4250
John CC et al (2008) Cerebral malaria in children is associated with long-term cognitive impairment. Pediatrics 122(1):e92–e99. https://doi.org/10.1542/peds.2007-3709
Johnson SM et al (2005) RAS is regulated by the let-7 microRNA family. Cell 120(5):635–647. https://doi.org/10.1016/j.cell.2005.01.014
Kaur H, Sehgal R, Kumar A, Sehgal A, Bansal D, Sultan AA (2018) Screening and identification of potential novel biomarker for diagnosis of complicated Plasmodium vivax malaria. J Transl Med 16(1):272. https://doi.org/10.1186/s12967-018-1646-9
Khan S et al (2020) Immune escape and immune camouflage may reduce the efficacy of RTS, S vaccine in Malawi. Hum Vaccin Immunother 16(2):214–227. https://doi.org/10.1080/21645515.2018.1560772
Khraiwesh B et al (2010) Transcriptional control of gene expression by microRNAs. Cell 140(1):111–122. https://doi.org/10.1016/j.cell.2009.12.023
Laishram DD et al (2012) The complexities of malaria disease manifestations with a focus on asymptomatic malaria. Malar J 11:29. https://doi.org/10.1186/1475-2875-11-29
LaMonte G et al (2012) Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe 12(2):187–199. https://doi.org/10.1016/j.chom.2012.06.007
Li JJ et al (2018) Identification of potential whole blood MicroRNA biomarkers for the blood stage of adult imported falciparum malaria through integrated mRNA and miRNA expression profiling. Biochem Biophys Res Commun 506(3):471–477. https://doi.org/10.1016/j.bbrc.2018.10.072
Liang Y, Ridzon D, Wong L, Chen C (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8:166. https://doi.org/10.1186/1471-2164-8-166
Liu X, Meng J (2018) Tanshinone IIA ameliorates lipopolysaccharide-induced inflammatory response in bronchial epithelium cell line BEAS-2B by down-regulating miR-27a. Biomed Pharmacother 104:158–164. https://doi.org/10.1016/j.biopha.2018.05.021
Ludwig N et al (2016) Distribution of miRNA expression across human tissues. Nucleic Acids Res 44(8):3865–3877. https://doi.org/10.1093/nar/gkw116
Makenga G et al (2020) Prevalence of malaria parasitaemia in school-aged children and pregnant women in endemic settings of sub-Saharan Africa: a systematic review and meta-analysis. Parasite Epidemiol Control 11:e00188. https://doi.org/10.1016/j.parepi.2020.e00188
Mantel PY et al (2016) Infected erythrocyte-derived extracellular vesicles alter vascular function via regulatory Ago2-miRNA complexes in malaria. Nat Commun 7:12727. https://doi.org/10.1038/ncomms12727
Martin-Alonso A et al (2018) Differentially expressed microRNAs in experimental cerebral malaria and their involvement in endocytosis, adherens junctions, FoxO and TGF-beta signalling pathways. Sci Rep 8(1):11277. https://doi.org/10.1038/s41598-018-29721-y
Masaki S, Ohtsuka R, Abe Y, Muta K, Umemura T (2007) Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis. Biochem Biophys Res Commun 364(3):509–514. https://doi.org/10.1016/j.bbrc.2007.10.077
Narasimhan M, Patel D, Vedpathak D, Rathinam M, Henderson G, Mahimainathan L (2012) Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PLoS ONE 7(12):e51111. https://doi.org/10.1371/journal.pone.0051111
Neilsen PM et al (2013) Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 32(24):2992–3000. https://doi.org/10.1038/onc.2012.305
Nielsen CM, Vekemans J, Lievens M, Kester KE, Regules JA, Ockenhouse CF (2018) RTS, S malaria vaccine efficacy and immunogenicity during Plasmodium falciparum challenge is associated with HLA genotype. Vaccine 36(12):1637–1642. https://doi.org/10.1016/j.vaccine.2018.01.069
Nimsarkar P, Ingale P, Singh S (2020) Systems studies uncover miR-146a as a target in Leishmania major infection model. ACS Omega 5(21):12516–12526. https://doi.org/10.1021/acsomega.0c01502
Nsanzabana C (2019) Resistance to artemisinin combination therapies (ACTs): do not forget the partner drug! Trop Med Infect Dis 4(1). https://doi.org/10.3390/tropicalmed4010026
O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D (2009) Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci U S A 106(17):7113–7118. https://doi.org/10.1073/pnas.0902636106
O’Hara SP et al (2010) NFkappaB p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. J Biol Chem 285(1):216–225. https://doi.org/10.1074/jbc.M109.041640
Pombo DJ et al (2002) Immunity to malaria after administration of ultra-low doses of red cells infected with Plasmodium falciparum. Lancet 360(9333):610–617. https://doi.org/10.1016/S0140-6736(02)09784-2
Pordzik J et al (2018) The potential role of platelet-related microRNAs in the development of cardiovascular events in high-risk populations, including diabetic patients: a review. Front Endocrinol (lausanne) 9:74. https://doi.org/10.3389/fendo.2018.00074
Qayum AA et al (2016) IL-10-induced miR-155 targets SOCS1 to enhance IgE-mediated mast cell function. J Immunol 196(11):4457–4467. https://doi.org/10.4049/jimmunol.1502240
Raman J et al (2020) High levels of imported asymptomatic malaria but limited local transmission in KwaZulu-Natal, a South African malaria-endemic province nearing malaria elimination. Malar J 19(1):152. https://doi.org/10.1186/s12936-020-03227-3
Rangel G et al (2017) Prediction of miRNA that modulate significant host response genes as potential biomarkers in cerebral malaria infection
Rathjen T, Nicol C, McConkey G, Dalmay T (2006) Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett 580(22):5185–5188. https://doi.org/10.1016/j.febslet.2006.08.063
Ruiz-Tagle C, Naves R, Balcells ME (2020) Unraveling the role of microRNAs in Mycobacterium tuberculosis infection and disease: advances and pitfalls. Infect Immun 88(3). https://doi.org/10.1128/IAI.00649-19
Salam N, Mustafa S, Hafiz A, Chaudhary AA, Deeba F, Parveen S (2018) Global prevalence and distribution of coinfection of malaria, dengue and chikungunya: a systematic review. BMC Public Health 18(1):710. https://doi.org/10.1186/s12889-018-5626-z
Santos ML et al (2020) A distinct fingerprint of inflammatory mediators and miRNAs in Plasmodium vivax severe thrombocytopenia. bioRxiv. https://doi.org/10.1101/2020.08.20.260463
Sheedy FJ, O’Neill LA (2008) Adding fuel to fire: microRNAs as a new class of mediators of inflammation. Ann Rheum Dis 67(Suppl 3):iii50-5. https://doi.org/10.1136/ard.2008.100289
SIMON-OKE IA, Ogunseem MF, Afolabi OJ, Awosolu O (2019) 2019:6 %J Journal of Biomedicine and Translational Research. https://doi.org/10.14710/jbtr.v5i1.3711
Sonkoly E, Stahle M, Pivarcsi A (2008) MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol 18(2):131–140. https://doi.org/10.1016/j.semcancer.2008.01.005
Sun L et al (2018) Different erythrocyte microRNA Profiles in low- and high-altitude individuals. Front Physiol 9:1099. https://doi.org/10.3389/fphys.2018.01099
Sun L, Yu Y, Niu B, Wang D (2020) Red blood cells as potential repositories of microRNAs in the circulatory system. Front Genet 11:442. https://doi.org/10.3389/fgene.2020.00442
Testa U, Pelosi E, Castelli G, Labbaye C (2017) miR-146 and miR-155: two key modulators of immune response and tumor development. Noncoding RNA 3(3). https://doi.org/10.3390/ncrna3030022
Thomson R et al (2019) pfhrp2 and pfhrp3 gene deletions that affect malaria rapid diagnostic tests for Plasmodium falciparum: analysis of archived blood samples from 3 African countries. J Infect Dis 220(9):1444–1452. https://doi.org/10.1093/infdis/jiz335
Tili E et al (2011) Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proc Natl Acad Sci U S A 108(12):4908–4913. https://doi.org/10.1073/pnas.1101795108
Urbich C et al (2012) MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 119(6):1607–1616. https://doi.org/10.1182/blood-2011-08-373886
van de Bunt M et al (2013) The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS ONE 8(1):e55272. https://doi.org/10.1371/journal.pone.0055272
van Loon W, Gai PP, Hamann L, Bedu-Addo G, Mockenhaupt FP (2019) MiRNA-146a polymorphism increases the odds of malaria in pregnancy. Malar J 18(1):7. https://doi.org/10.1186/s12936-019-2643-z
van Loon W et al (2020) MiRNA-146a polymorphism was not associated with malaria in Southern India. Am J Trop Med Hyg 102(5):1072–1074. https://doi.org/10.4269/ajtmh.19-0845
Varikuti S et al. (2019) MicroRNA 155 contributes to host immunity against Leishmania donovani but is not essential for resolution of infection. Infect Immun 87(8). https://doi.org/10.1128/IAI.00307-19
Wah ST, Hananantachai H, Patarapotikul J, Ohashi J, Naka I, Nuchnoi P (2019) MicroRNA-27a and microRNA-146a SNP in cerebral malaria. Mol Genet Genomic Med 7(2):e00529. https://doi.org/10.1002/mgg3.529
Wang T, Wu F, Yu D (2019) miR-144/451 in hematopoiesis and beyond. ExRNA 1(1):16. https://doi.org/10.1186/s41544-019-0035-8
Wang Z et al (2017) Red blood cells release microparticles containing human argonaute 2 and miRNAs to target genes of Plasmodium falciparum. Emerg Microbes Infect 6(8):e75. https://doi.org/10.1038/emi.2017.63
World Health Organization (2017) False-negative RDT results and implications of new reports of P. falciparum histidine-rich protein 2/3 gene deletions. https://apps.who.int/iris/bitstream/10665/258972/1/WHO-HTM-GMP-2017.18-eng.pdf
World Health Organization (2020) World malaria report 2020: 20 years of global progress and challenges. In World malaria report 2020: 20 years of global progress and challenges. https://apps.who.int/iris/rest/bitstreams/1321872/retrieve
Xiao C et al (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131(1):146–159. https://doi.org/10.1016/j.cell.2007.07.021
Xue X, Zhang Q, Huang Y, Feng L, Pan W (2008) No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J 7:47. https://doi.org/10.1186/1475-2875-7-47
Yewhalaw D et al (2011) Multiple insecticide resistance: an impediment to insecticide-based malaria vector control program. PLoS ONE 6(1):e16066. https://doi.org/10.1371/journal.pone.0016066
Zhang M et al (2014) MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis 234(1):54–64. https://doi.org/10.1016/j.atherosclerosis.2014.02.008
Zhou X, Li X, Wu M (2018) miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct Target Ther 3:14. https://doi.org/10.1038/s41392-018-0006-9
Funding
The V.R. lab is supported by the research grant from Department of Science & Technology (DST), INSPIRE-Faculty Project (DST/INSPIRE/04/2018/003541), Ministry of Science and Technology, Government of India.
Author information
Authors and Affiliations
Contributions
A.M. and V.R. contributed equally towards literature survey, data interpretation, and preparation of the figures. V.R. contributed to the overall supervision, design of the review, critical analysis of the intellectual content, revision of the manuscript, and funding acquisition. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Kevin S.W. Tan
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abhinab Mohanty and Vinoth Rajendran are joint first author - contributed equally to this paper.
Rights and permissions
About this article
Cite this article
Mohanty, A., Rajendran, V. Mammalian host microRNA response to plasmodial infection: role as therapeutic target and potential biomarker. Parasitol Res 120, 3341–3353 (2021). https://doi.org/10.1007/s00436-021-07293-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-021-07293-7