Abstract
The roe deer (Capreolus capreolus) has been identified as an intermediate host for six known Sarcocystis species, S. capreolicanis, S. entzerothi, S. gracilis, S. linearis, S. oviformis, and S. silva. In this study, we identified Sarcocystis species in the diaphragm and tongue muscles from the Lithuanian and Spanish roe deer, respectively, on the basis of a microscopic examination and DNA analysis. A total of 43 and 27 sarcocysts were isolated and characterized from the Lithuanian and Spanish roe deer, respectively. Overall six Sarcocystis species were identified in roe deer from Lithuania, and only three of them, S. gracilis, S. linearis, and S. silva were found to have infecting animals from Spain. The current paper represents first molecular results of Sarcocystis species in the Spanish roe deer. Furthermore, transmission electron microscopy examination revealed specific wall structure of sarcocysts studied, S. linearis was characterized by ribbon-like villar protrusions (vp) (type 8a), and S. oviformis was distinguished by elongated vp resembling spades or mushroom-like structures (type 39). Based on 18S rDNA and cox1 sequences, Sarcocystis species from the roe deer showed considerable intraspecific genetic variability. However, similar values of intraspecific genetic variation were estimated at both genes analysed. The highest variability was observed for S. capreolicanis and S. linearis in both genes and for S. silva at cox1. Consequently, the level of genetic variability of Sarcocystis from the roe deer varied depending on species rather than on gene analysed or geographical area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parasites of the genus Sarcocystis are characterized by an obligatory two-host life cycle (Dubey et al. 2016). Members of the family Cervidae may act as intermediate hosts for numerous Sarcocystis species. Overall, six Sarcocystis species infecting the European roe deer (Capreolus capreolus), Sarcocystis capreolicanis, Sarcocystis entzerothi, Sarcocystis gracilis, Sarcocystis linearis, Sarcocystis oviformis, and Sarcocystis silva, were adequately described by means of microscopical and molecular methods (Dahlgren and Gjerde 2009; Gjerde 2012; Gjerde et al. 2017; Prakas et al. 2017).
Pioneer European Sarcocystis infection surveys were carried out based on morphological methods (Erber et al. 1978; Schramlová and Blazek 1978; Entzeroth 1982; Santini et al. 1997; Kutkienė 2001; Spickschen and Pohlmeyer 2002; López et al. 2003); both scanning and transmission electron microscopy (SEM/TEM) analyses were conducted for sarcocysts of S. capreolicanis, S. gracilis and S. silva; however, only SEM data are available for S. linearis and S. oviformis and TEM for S. entzerothi (Dahlgren and Gjerde 2009; Gjerde et al. 2017; Prakas et al. 2017). Hence, further studies using electron microscopy would contribute to a better characterization of Sarcocystis species in the roe deer. Over the past decade, the necessity to use DNA markers to discriminate Sarcocystis parasites infecting different cervids, including the roe deer, arose (Dahlgren and Gjerde 2009; Calero-Bernal et al. 2015; Prakas et al. 2016). The 18S rDNA and cox1 markers have been shown to be suitable for differentiation between closely related Sarcocystis species using ruminants as intermediate hosts (Gjerde 2013, 2016). Based on these markers, the composition for presence of Sarcocystis species in roe deer has been revealed in several European countries, such as Italy, Lithuania, Poland, and Norway (Kolenda et al. 2014; Dubey et al. 2016; Prakas et al. 2017; Gjerde et al. 2017). Thus, more detailed researches are needed to elucidate species richness and genetic variability in distant areas and distinct ecosystems.
Previous studies on parasites of roe deer in Spain revealed a very high prevalence (99%) of Sarcocystis spp. infection (Pérez-Creo et al. 2013); however, no molecular analyses were performed. In the present study, we identified the Sarcocystis species in roe deer samples from Lithuania and Spain, ultrastructurally characterized sarcocysts of S. linearis and S. oviformis, and revealed differences in intraspecific genetic variability of Sarcocystis species found in distant countries.
Material and methods
Samples and light microscope (LM) examination
Diaphragm muscles of 40 roe deer from Lithuania and 41 tongues of roe deer from 10 provinces in Spain were collected between 2015 and 2018. About 50–100 g of diaphragms and 100–150 g of tongues were collected from animals examined. All studied animals had been legally hunted for trophy and meat consumption purposes, and samples, harvested the same day of hunt, were kept frozen until shipment to the corresponding laboratories. The morphological analysis of sarcocysts was performed in thawed-squashed preparations. Under LM, sarcocysts were isolated from muscle fibres and screened according to the size and shape of sarcocysts and the structure of the cyst wall (Kirillova et al. 2018). Isolated sarcocysts were separately preserved in ethanol for further DNA extraction and molecular analysis. Sarcocystis infection intensity from Lithuanian animals was evaluated in stained muscle samples by counting sarcocysts per gram of tissue (Prakas et al. 2019).
Histological examination
Histological examination was performed with Spanish specimens only. Muscle samples were cut into 1 cm3 pieces, fixed in 10% buffered formalin, embedded in paraffin, and sectioned 5 μm thick; sections were stained with haematoxylin and eosin (H-E) and observed under the microscope. Intensity of infection was estimated as the number of tissue cysts per section (2.5 cm2).
Transmission electron microscopy (TEM) analysis
Two excised sarcocysts, which by means of LM were similar to S. linearis and S. oviformis, were fixed in 2.5% glutaraldehyde and post-fixed in 1% buffered osmium tetroxide; thereafter sections were cut on Leica UC6 ultramicrotome and contrasted with 4% uranyl acetate and 3% lead citrate, as suggested by Trupkiewicz et al. (2016). Sections were stained with toluidine blue and examined under LM for the presence of cysts. Finally, ultra-thin sections were examined at the Spanish National Centre for Electron Microscopy (Madrid, Spain) using the JEOL JEM 1400 Plus device at 80 kW.
Molecular analysis
Genomic DNA was extracted from 70 individual sarcocysts (43 sarcocysts from 28 individual Lithuanian roe deer and 27 sarcocysts from 19 Spanish roe deer) using QIAamp® DNA Micro Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Partial 18S rDNA sequences were amplified with SarAF/SarBR and SarCF/SarDR primer pairs (Kutkienė et al. 2010), while mitochondrial cytochrome c oxidase subunit I gene (cox1) sequences were amplified with SF1 forward primer in combination with one of the following reverse primers, SR8D/SR9/SR5/SR12H depending on Sarcocystis species (Gjerde 2013; Gjerde 2014; Prakas et al. 2017). PCR reactions were performed using DreamTaq PCR Master Mix (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). The cycling conditions started for 5 min at 95 °C, followed by 35 cycles of 45 s at 94 °C, 60 s at 54–60 °C depending on the primer pair, and 80 s at 72 °C and ended with 10 min at 72 °C. PCR products were evaluated using 1.5% agarose gel electrophoresis and purified with exonuclease ExoI and alkaline phosphatase FastAP (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). Most of the samples (n = 170) were sequenced directly with the 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using the same forward and reverse primers as for PCR, except for 18S rDNA fragments of S. capreolicanis and S. linearis that resulted in double peaks and poly signals within 18S rDNA. Thus, to obtain unambiguous sequences, the amplicons of S. capreolicanis and S. linearis (n = 36) were cloned into plasmids with the help of CloneJET PCR Cloning Kit (Thermo Fisher Scientific Baltics, Vilnius, Lithuania). Cloning reactions were chilled on ice for 1 min and transformed into 100 μl of chemo-competent E. coli DH10B cells. Clones were selected on LB agar plates supplemented with 50 μg/ml ampicillin. The recombinant clones were screened to obtain the insert of a desired size verified by colony PCR amplification. Individual colony was suspended in liquid LB media and used for colony PCR. Each PCR was performed in a final 50 μl volume consisting of 0.5 μM of vector primers pJET1.2 forward and reverse, dNTP mix (0.2 mM of each), 10 × DreamTaq buffer with 20 mM MgCl2, 1.25 U DreamTaq polymerase (Thermo Fisher Scientific Baltics, Vilnius, Lithuania), and 1 μl of the chosen clone colony suspended in liquid media and nuclease-free water. PCR fragments were analysed by agarose gel electrophoresis, and the remaining portion of the positive colony was used to inoculate liquid LB media with ampicillin for plasmid miniprep extraction. DNA plasmids were extracted from E. coli DH10B cells using GeneJet Plasmid Miniprep Kit (Thermo Fisher Scientific Baltics, Vilnius, Lithuania) according to the manufacturer’s instructions and afterwards sequenced using pJET1.2 forward and reverse vector primers. The number of segregating sites (S), the number of haplotypes (h) and haplotype diversity (Hd), and the average number of nucleotide differences (K) and nucleotide diversity (π) were calculated for the whole dataset and separate populations using DnaSP v6 (Rozas et al. 2017). The sequences used for the analysis of intraspecific genetic variation are given in Table S1 of the Supplementary material.
Results
Sarcocysts were detected in 95% (38/40) of diaphragm muscles and 100% (41/41) of tongues from Lithuanian and Spanish roe deer, respectively. Three types of microscopic sarcocysts having hair-like, finger-like, or no visible villar protrusions (vp) and one type of macroscopic sarcocyst were detected (Table 1). Based on molecular results, sarcocysts with no visible vp under LM belonged to S. gracilis or S. linearis. The sarcocysts of these species slightly differed in size as cysts of S. linearis were smaller to those of S. gracilis; however, the dimensions overlapped (Table 1). Molecular results confirmed that sarcocysts having hair-like vp belonged to S. capreolicanis, while sarcocysts with finger-like vp corresponded to S. entzerothi or S. silva, whose morphological differentiations were described in more detail by Prakas et al. (2017). Macroscopic, oval-shaped sarcocysts were identified as S. oviformis.
In Spanish animals, 100% of H-E-stained tissue sections were positive; there was an absence of myositis foci, and the average parasite load was estimated in 42.7 cysts/section (17.1 cysts/cm2); parasite load records were established in four categories: 1–10 cysts (19.5% of animals), 11–50 cysts (48.8%), 51–100 cysts (22.0%), and more than 100 cysts (9.7%). The maximum load (194 cysts/section) was observed in the roe deer Cc25 collected in Burgos province. No secondary cyst wall was discovered in 1751 cysts examined, and two morphological types were observed: type-A, thin-walled (< 1 μm in thickness) with vp apparently absent, and type-B, thick-walled (> 5 μm) and finger-like 6–8 μm long vp; distribution of observed morphological types was 65.9% of type A and 2.4% of type B and 31.7% of mixed infections (Fig. 1). Mature bradyzoites but no metrocytes were observed in septated sarcocysts. In the infected Lithuanian animals, the average parasite load of 197.0 cysts/g (median 86.5 cysts/g) of muscle was observed in methylene blue-dyed samples. Parasite load records were divided into the same above-described categories to evaluate the intensity of infection: 1–10 cysts (7.9%), 11–50 cysts (26.3%), 51–100 cysts (21.1%), and more than 100 cysts (44.7%).
Micrographs of Sarcocystis sarcocysts from the roe deer. (a-d) H-E-stained sections. Arrowheads point to cyst wall. Note, bradyzoites (br). (e, f) Toluidine blue-stained sections. Arrowheads point to cyst wall. Note, br, host cells (hc), and septae (se). (g, h) TEM micrographs. Note, br, hc, ground substance (gs), peduncle (pe), and villar protrusions (vp). Bars: a-c = 25 μm; d = 10 μm; e, f = 25 μm; g, h = 0.5 μm. (a) Thin-walled sarcocyst type A resembling S. linearis or S. gracilis. (b-d) Thick-walled sarcocysts, with finger-like vp resembling S. silva. (e) Evident thin-walled sarcocyst, identified later as S. linearis. (f) Evident thick-walled sarcocyst, identified later as S. oviformis. (g) Detailed view of the thin-walled S. linearis. Note, blebs (arrowheads) between vp and tapered ends of vp (arrows). (h) Details of the vp of S. oviformis. Note, pe and electrondense granules (arrow)
Two sarcocysts were processed for TEM analysis (Table 2); a thin-walled (0.5 μm wide) cyst was characterized by sectioned ribbon-like vp with tapered ends resembling tau crosses and corresponded to S. linearis (type 8a by Dubey et al. (2016)). Thick-walled (6.5 μm wide) cyst had elongated vp, which mostly appeared sectioned with condensation of electrondense material at the base and 1.2 μm long peduncle, resembling spades or mushroom-like structures and corresponded to S. oviformis (type 39 by Dubey et al. (2016)).
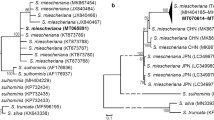
In the present study, the obtained sequences of Sarcocystis species from the roe deer are deposited in GenBank with accession numbers MN334245–MN334330 and MN339281–MN339350. Molecular analysis led to the identification of six Sarcocystis species in Lithuanian roe deer, S. capreolicanis (n = 8), S. entzerothi (n = 6), S. gracilis (n = 9), S. linearis (n = 5), S. oviformis (n = 5), and S. silva (n = 10) (Table 3). Three Sarcocystis species, S. gracilis (n = 18), S. linearis (n = 7), and S. silva (n = 2) were confirmed in roe deer from Spain. All isolates of the same Sarcocystis species were obtained from different animals. Intra-isolate sequence differences at 18S rDNA region were analysed for S. capreolicanis and S. linearis from Lithuania and for S. linearis from Spain. A total of 12 clones from three isolates of S. capreolicanis and 24 clones from six isolates of S. linearis were processed and sequenced. Sequence differences between the clones from the same isolate of S. capreolicanis and S. linearis from Lithuania and S. linearis from Spain were 0.43–1.45%, 0.49–1.52%, and 0.44–1.68%, respectively. Similarly, the range of genetic variation between clones from different isolates of the analysed species was broader, but with the overlapping values. Sequence differences of S. capreolicanis and S. linearis from Lithuania and Spain accounted for 0.22–2.20%, 0–1.63%, and 0.27–1.74%, respectively. In general, having evaluated intraspecific genetic variability in Lithuania and Spain, the highest DNA variability for S. capreolicanis was estimated at 18S rDNA, for S. silva at cox1 and for S. linearis at both genes analysed (Table 4). A comparison of intraspecific genetic variations amongst S. gracilis and S. linearis species from both countries demonstrated a slightly higher variability in Lithuania.
Overall comparisons of intraspecific genetic variability at studied loci of Sarcocystis species from roe deer sampled in Lithuania and Spain, as well as Italy, Norway, and Poland, were made. The highest intraspecific genetic variability was detected in S. capreolicanis and S. linearis in both genes and in S. silva at cox1, while some less variability was revealed in S. gracilis at cox1 and S. silva at 18S rDNA, whereas very low or none genetic variability was detected for S. entzerothi and S. oviformis at both loci examined and for S. gracilis at 18S rDNA. In general, similar genetic variability was observed within same Sarcocystis species from different countries. No relation between the level of intraspecific genetic variability and the sampling size was noticed.
Discussion
In the present study, very high infection prevalence was established in roe deer in Lithuania (95%) and Spain (100%). On the basis of morphological and molecular results, six Sarcocystis species, S. capreolicanis, S. entzerothi, S. gracilis, S. linearis, S. oviformis, and S. silva, were identified in Lithuanian roe deer, while only three species (S. gracilis, S. linearis, and S. silva) were found in the animals from Spain (Tables 1 and 3). Sarcocystis gracilis and S. silva have been molecularly identified in the roe deer from Italy (n = 4 animals), Poland (n = 4), Norway (n = 9), Spain (n = 41), and Lithuania (n = 46) (Dahlgren and Gjerde 2009; Gjerde 2012; Kolenda et al. 2014; Gjerde et al. 2017; Prakas et al. 2017; this study). These two are probably the most common Sarcocystis species employing the roe deer as an intermediate host. By means of molecular methods, sarcocysts of S. capreolicanis were confirmed in Italy, Lithuania, and Norway; however, they were not detected in Poland and Spain. Most likely this species was not detected in Poland due to a small sample size. Pérez-Creo et al. (2013) examined oesophagus, diaphragm, and heart of 101 roe deer hunted in north-western Spain and detected sarcocysts of S. capreolicanis, S. gracilis, S. silva, and Sarcocystis sp. by means of LM and/or TEM. During the present study, S. capreolicanis was not found in Spain, probably due to different muscles examined or because such species is rare in the investigated provinces. There is no data indicating that S. linearis is present in Norway or Poland, but since the species was only recently discovered (Gjerde et al. 2017), future researches are desirable, whereas S. entzerothi was confirmed only in Lithuanian roe deer, sika deer and fallow deer (Prakas et al. 2017; Rudaitytė-Lukošienė et al. 2018, 2020). The sika deer is native to East Asia and was introduced into different countries including Lithuania. It is possible that the roe deer is not a natural intermediate host of S. entzerothi as it was suggested by Gjerde et al. (2017), and the species could have spread in the area through the introduced sika deer. Macrocysts of S. oviformis were not found in Italy or Spain. We suppose that either S. oviformis was overlooked in Spanish roe deer and not detected in Italy due to a small number of animals examined or definitive hosts of S. oviformis are not prevalent in Southern Europe. Definitive hosts of S. oviformis are not revealed; however, based on phylogenetic placement of S. oviformis, corvid birds are assumed to be their final hosts (Gjerde et al. 2017).
In this study, LM morphological analysis was insufficient to discriminate sarcocysts of S. gracilis from those of S. linearis. The size ranges of these two Sarcocystis species corresponded to those reported in the previous study (Gjerde et al. 2017). Sarcocysts of S. linearis and S. oviformis were morphologically analysed using TEM. The ultrastructural description of sarcocysts provides valuable information when correlates with molecular data. In the present article, S. linearis and S. oviformis sarcocysts are described by TEM for the first time and supplement the previously published descriptions of these species based on SEM (Dahlgren and Gjerde 2009; Gjerde et al. 2017). The current S. linearis TEM pics represent ribbon-like vp with the tapered ends, and these vp are separated by up to 1.3 μm wide segments of well-defined blebs; this was clearly visible in Fig. 4C provided by Gjerde et al. (2017). Several Sarcocystis sp. of similar ultrastructure have been reported in roe deer from Germany, Italy, and Spain, and sarcocysts of this unnamed species are likely to have belonged to S. linearis (Entzeroth 1982; Santini et al. 1997; López et al. 2003; Pérez-Creo et al. 2013). On the other hand, sarcocyst of S. oviformis displayed 6.5–10 μm long vp having the appearance of a sea anemone; this was initially shown by Dahlgren and Gjerde (2009) and Gjerde (2012) in Norwegian roe deer. TEM pics showed sectioned vp with thin peduncle and condensate material at the base of vp. In some micrographs, un-sectioned vp morphology resembled those tongue-shaped vp of S. ovalis from the moose (Alces alces). Bradyzoites of both species were unsuitable for description due to preservation of cysts.
In this study, Sarcocystis species examined revealed significant differences in intraspecific genetic variability. When compared with the molecular data of Sarcocystis species from the roe deer currently available from five European countries, the level of interspecific genetic variability depended more on the species rather than on locus or the geographical area examined. Our results are consistent with the findings of Kolenda et al. (2014) who observed high similarity in the level of genetic variability of a population of the same Sarcocystis species from roe deer living in different geographic regions.
The present survey provided new data on the complex epidemiology of Sarcocystis species infecting wild ungulates, especially on S. oviformis and S. linearis; in addition, the necessity to carry out further ultrastructural-molecular-combined investigations is emphasized in order to link previous research and current knowledge by adequately describing parasite species as an aid to unravel the role of possible definitive hosts.
References
Calero-Bernal R, Verma SK, Cerqueira-Cézar CK, Schafer LM, Van Wilpe E, Dubey JP (2015) Sarcocystis mehlhorni, n. sp. (Apicomplexa: Sarcocystidae) from the black-tailed deer (Odocoileus hemionus columbianus). Parasitol Res 114:4397–4403. https://doi.org/10.1007/s00436-015-4679-5
Dahlgren SS, Gjerde B (2009) Sarcocystis in Norwegian roe deer (Capreolus capreolus): molecular and morphological identification of Sarcocystis oviformis n. sp. and Sarcocystis gracilis and their phylogenetic relationship with other Sarcocystis species. Parasitol Res 104:993–1003. https://doi.org/10.1007/s00436-008-1281-0
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2016) Sarcocystosis of animals and humans, 2nd edn. CRC Press, Boca Raton
Entzeroth R (1982) A comparative light and electron microscope study of the cysts of Sarcocystis species of roe deer (Capreolus capreolus). Z Parasitenkd 66:281–292. https://doi.org/10.1007/bf00925345
Erber M, Boch J, Barth D (1978) Drei Sarkosporidienarten des Rehwildes. Berl Münch Tierärztl Wochenschr 91:482–486.
Gjerde B (2012) Morphological and molecular characterization and phylogenetic placement of Sarcocystis capreolicanis and Sarcocystis silva n. sp. from roe deer (Capreolus capreolus) in Norway. Parasitol Res 110:1225–1237. https://doi.org/10.1007/s00436-011-2619-6
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579–591. https://doi.org/10.1016/j.ijpara.2013.02.004
Gjerde B (2014) Sarcocystis species in red deer revisited: with a redescription of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. https://doi.org/10.1017/S0031182013001819
Gjerde B (2016) Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis). Parasitol Res 115:1473–1492. https://doi.org/10.1007/s00436-015-4881-5
Gjerde B, Giacomelli S, Bianchi A, Bertoletti I, Mondani H, Gibelli LR (2017) Morphological and molecular characterization of four Sarcocystis spp., including Sarcocystis linearis n. sp., from roe deer (Capreolus capreolus) in Italy. Parasitol Res 116:1317–1338. https://doi.org/10.1007/s00436-017-5410-5
Kirillova V, Prakas P, Calero-Bernal R, Gavarāne I, Fernández-García JL, Martínez-González M, Rudaitytė-Lukošienė E, Martínez-Estéllez MÁH, Butkauskas D, Kirjušina M (2018) Identification and genetic characterization of Sarcocystis arctica and Sarcocystis lutrae in red foxes (Vulpes vulpes) from Baltic States and Spain. Parasit Vectors 11:173. https://doi.org/10.1186/s13071-018-2694-y
Kolenda R, Ugorski M, Bednarski M (2014) Molecular characterization of Sarcocystis species from Polish roe deer based on ssu rRNA and cox1 sequence analysis. Parasitol Res 113:3029–3039. https://doi.org/10.1007/s00436-014-3966-x
Kutkienė L (2001) The species composition of European roe deer (Capreolus capreolus) Sarcocystis in Lithuania. Acta Zool Lituanica 11:97–101. https://doi.org/10.1080/13921657.2001.10512363
Kutkienė L, Prakas P, Sruoga A, Butkauskas D (2010) The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis). Parasitol Res 107:879–888. https://doi.org/10.1007/s00436-010-1945-4
López C, Panadero R, Bravo A, Paz A, Sánchez-Andrade R, Díez-Baños P, Morrondo P (2003) Sarcocystis spp. infection in roe deer (Capreolus capreolus) from the north-west of Spain. Z Jagdwiss 49:211–218. https://doi.org/10.1007/bf02189739
Pérez-Creo A, Panadero R, López C, Díaz P, Vázquez L, Díez-Baños P, Morrondo P (2013) Prevalence and identity of Sarcocystis spp. in roe deer (Capreolus capreolus) in Spain: a morphological study. Res Vet Sci 95:1036–1040. https://doi.org/10.1016/j.rvsc.2013.08.003
Prakas P, Butkauskas D, Rudaitytė E, Kutkienė L, Sruoga A, Pūraitė I (2016) Morphological and molecular characterization of Sarcocystis taeniata and Sarcocystis pilosa n. sp from the sika deer (Cervus nippon) in Lithuania. Parasitol Res 115:3021–3032. https://doi.org/10.1007/s00436-016-5057-7
Prakas P, Rudaitytė E, Butkauskas D, Kutkienė L (2017) Sarcocystis entzerothi n. sp. from the European roe deer (Capreolus capreolus). Parasitol Res 116:271–279. https://doi.org/10.1007/s00436-016-5288-7
Prakas P, Kirillova V, Calero-Bernal R, Kirjušina M, Rudaitytė-Lukošienė E, Habela MÁ, Gavarāne I, Butkauskas D (2019) Sarcocystis species identification in the moose (Alces alces) from the Baltic States. Parasitol Res 118:1601–1608. https://doi.org/10.1007/s00436-019-06291-0
Rozas J, Ferrer-Mata A, Sánchez-Del Barrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Rudaitytė-Lukošienė E, Prakas P, Butkauskas D, Kutkienė L, Vepštaitė-Monstavičė I, Servienė E (2018) Morphological and molecular identification of Sarcocystis spp. from the sika deer (Cervus nippon), including two new species Sarcocystis frondea and Sarcocystis nipponi. Parasitol Res 117:1305–1315. https://doi.org/10.1007/s00436-018-5816-8
Rudaitytė-Lukošienė E, Prakas P, Strazdaitė-Žielienė Ž, Servienė E, Januškevičius V, Butkauskas D (2020) Molecular identification of two Sarcocystis species in fallow deer (Dama dama) from Lithuania. Parasitol Int 75:102044. https://doi.org/10.1016/j.parint.2019.102044
Santini S, Mancianti F, Nigro M, Poli A (1997) Ultrastructure of the cyst wall of Sarcocystis sp. in roe deer. J Wildl Dis 33:853–859. https://doi.org/10.7589/0090-3558-33.4.853
Schramlová J, Blazek K (1978) Ultrastruktur der Cystenwand der Sarkosporidien des Rehes (Capreolus capreolus L.). Z Parasitenkd 55:43–48.
Spickschen C, Pohlmeyer K (2002) Untersuchung zum Vorkommen von Sarkosporidien bei Reh-, Rot- und Muffelwild in zwei unterschiedlichen Naturräumen des Bundeslandes Niedersachsen. Z Jagdwiss 48:35–48. https://doi.org/10.1007/bf02285355
Trupkiewicz JG, Calero-Bernal R, Verma SK, Mowery J, Davison S, Habecker P, Georoff TA, Ialeggio DM, Dubey JP (2016) Acute, fatal Sarcocystis calchasi-associated hepatitis in roller pigeons (Columba livia f. dom.) at Philadelphia zoo. Vet Parasitol 216:52–58. https://doi.org/10.1016/j.vetpar.2015.11.008
Funding
This work was supported by the Research Council of Lithuania (grant number S-MIP-17-45) and Open Access to research infrastructure of the Nature Research Centre under Lithuanian open access network initiative.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Daniel K. Howe
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Rudaitytė-Lukošienė, E., Delgado de las Cuevas, G.E., Prakas, P. et al. Sarcocystis spp. diversity in the roe deer (Capreolus capreolus) from Lithuania and Spain. Parasitol Res 119, 1363–1370 (2020). https://doi.org/10.1007/s00436-020-06603-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-020-06603-9