Abstract
Based on morphology and morphometry of gametocytes in blood and molecular phylogenetic analysis, we described a new species of hemoparasite from the genus Haemogregarina isolated from Lepidosiren paradoxa in the eastern Amazon region. Haemogregarina daviesensis sp. nov. is characterized by monomorphic gametocytes of varying maturity stage and their dimensions were 16 ± 0.12 μm (range 13–18) in length and 6 ± 0.97 μm (range 5–8) in width. The morphological and morphometric data were not identical with other haemogregarine species from fish. All specimens of L. paradoxa analyzed were infected by H. daviesensis sp. nov. and the parasitemia level was moderate (1–28/2000 blood erythrocytes). Two sequences were obtained from L. paradoxa, and these constituted a monophyletic sister clade to the Haemogregarina species. In addition, H. daviesensis sp. nov. detected here grouped with Haemogregarina sp. sequences isolated from chelonian Macrochelys temminckii, with 99% bootstrap support. This study provides the first data on the molecular phylogeny of an intraerythrocytic haemogregarine of freshwater fish and highlights the importance of obtaining additional information on aspects of the general biology of these hemoparasites in fish populations, in order to achieve correct taxonomic classification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lepidosiren paradoxa Fitzinger, 1837 (lungfish) is a monotype of Lepidosirenidae, which is a family of Dipnoi fish with distribution restricted to South America. In Brazil, its distribution is only in the Amazon and Paraná River systems (Machado et al. 2010; Nelson et al. 2016). This species is primarily a carnivorous fish, feeding on arthropods and snails, and it inhabits hypoxic shallow swamps and lakes. Lungfish are obligate air-breathers, but during the dry season, they are able to burrow into the mud, undergo physiological changes, and decrease their metabolic rates, thereby inducing aestivation (Almeida-Val et al. 2010).
Haemogregarina lepidosirenis Jepps, 1927 was described by Jepps (1927) in L. paradoxa in the Gran Chaco, in Paraguay. Haemogregarinidae Léger, 1911 is a family of protozoa comprising three genera: Haemogregarina Danilewsky, 1885; Cyrillia Lainson, 1981; and Desseria Siddall, 1995. The species in these genera are intraerythrocytic and have heteroxenous life cycles (Siddall 1995). Currently, these genera are regarded as differing in oocyst formation and sporozoite numbers in invertebrate hosts (vectors) and vertebrate hosts (fish and chelonians). The vectors for haemogregarines of fish are usually isopods of the family Gnathiidae (Davies and Smit 2001) and leeches of the families Glossiphoniidae and Piscicolidae (Lainson 1981; Siddall and Desser 1993). Transmission of these intracellular parasites occurs through sporozoite inoculation during vector hematophagy (Davies 1995; Siddall 1995; Davies and Johnston 2000).
In Brazil, the only existing records of haemogregarines in fish are Desseria mugili (Carini, 1932) Siddall, 1995, in Mugil liza Cuvier and Valenciennes, 1836; Haemogregarina platessae Lebailly, 1905, in Paralichthys orbignyanus Valenciennes, 1839; and Cyrilia lignieresi Lainson, 1981, in Synbranchus marmoratus Bloch, 1795 (Siddall 1995; Davies et al. 2008; Eiras et al. 2012); and Cyrilia sp. in cururu stingray (Potamotrygon cf. histrix) and Potamotrygon wallacei, from Mariuá Archipelago, Negro River, in the Brazilian Amazon Basin (Magro et al. 2016; Oliveira et al. 2017).
These descriptions of intracellular parasites were made using only the morphological aspects of the hemoparasites; however, the morphological descriptions are improper and uncertain because it is difficult to differentiate (Desser 1993). This is a potentially important gap in the knowledge of these parasites, because there is reason to suspect that haemogregarines could be highly diversified, especially Haemogregarina spp. (Úngari et al. 2018). Although a recently study provided the first molecular data of Haemogregarina bigemina from marine fish (Hayes and Smit 2019), no studies using molecular tools to determine the species of Haemogregarina in freshwater fish have been conducted and reported. There is a need to use molecular markers to delimit the species of haemogregarines and thus distinguish the taxa and clarify the biodiversity of this group, so as to set future taxonomic work on a firm footing.
Hence, the aim of the present study was to describe a new species of Haemogregarina in L. paradoxa, from the eastern Amazon region of Brazil, using the features of its morphology and molecular biology.
Materials and methods
Authorization
This study was developed according to the principles adopted by the Brazilian College of Animal Experimentation (COBEA) and with the participation of the Embrapa Amapá Animal Use Committee (#004-CEUA/CPAFAP) and ICMBio (Chico Mendes Institute of Biodiversity; SISBIO number: 37846-1).
Site of collection, fish capture, and haemogregarine survey procedures
In June and July 2016 and February 2017, four specimens of L. paradoxa (three males and one female) were caught using hooks and dip nets in the Igarapé Fortaleza basin, in the municipality of Santana, state of Amapá, in the eastern Amazon region of Brazil (0° 02′ 10.91″ S; 51° 09′ 39.42″ W). Blood samples of euthanized specimens by means of medullary sectioning were taken by puncturing the ventral pulmonary artery using insulin syringes containing 10% EDTA. These samples were used to prepare blood smear slides, which were air-dried, fixed using absolute methanol for 3 min, and stained with 10% Giemsa for 30 min. The blood smear slides were examined under optical microscopy using a calibrated ocular micrometer at × 1000 magnification under oil immersion for morphological and morphometric characterization of the haemogregarines. The following measurements were analyzed: (1) length and width of parasites; (2) dimensions of infected and non-infected erythrocytes; and (3) dimensions of the nuclei of infected and non-infected erythrocytes. The parasitemia level was estimated based on examination of 2000 erythrocytes, in 20 replicates of 100 erythrocytes per field examined, on one slide from each fish from which blood samples were obtained (Godfrey Jr et al. 1987, 1990).
Molecular and phylogenetic analysis
Genomic DNA from two samples that had been found to be positive for Haemogregarina sp. through blood smears was isolated from 200 μL of blood, using the illustra blood genomicPrep Mini Spin Kit (GE Healthcare), following the manufacturer’s instructions.
DNA samples were screened for the presence of Haemogregarina spp., using the primers HepF300 and Hep900, which amplify 600 bp of the 18S rRNA gene (Ujvari et al. 2004). The PCR conditions were 94 °C for 3 min, 35 cycles at 94 °C for 45 s, 56 °C for 1 min, 72 °C for 1 min, and a final extension step at 72 °C for 7 min. For additional sequencing and phylogenetic analysis, the two positive samples in the first PCR were also amplified using the 4558/2733 primer pair (Mathew et al. 2000), which target a larger fragment (1120 bp) of the 18S rRNA gene. The PCR conditions of the secondary PCR consisted of a pre-PCR step at 94 °C for 3 min, followed by 40 cycles of 94 °C for 1 min, 55 °C for 2 min, an extension at 72 °C for 2 min, and a final extension at 72 °C for 10 min. A negative control (distilled water) and a positive control (Hepatozoon canis DNA isolated from naturally infected dogs) were used in each reaction. The PCR products were purified and sequenced using the BigDye™ Terminator v.3.1 cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA, USA) and the ABI 3500 genetic analyzer (Applied Biosystems, Foster City, CA, USA).
The DNA sequences obtained in this study were edited using the BioEdit software v7.2.5 (Hall 1999) and were compared for similarity to the sequences available in GenBank® using BLAST. Multiple alignment, for the two sequences obtained here and 25 sequences retrieved from GenBank®, was performed using the MUSCLE algorithm and the GENEIOUS v.7.1.3 software. Only the sequences with at least 1000 bp were used for the multiple alignment. The best evolutionary model for maximum likelihood analysis was identified using the jModelTest v.2.1.10 software (Darriba et al. 2012). The best-fitting model based on the Akaike information criterion (AIC) was GTR + I + G. The maximum likelihood tree was implemented in PhyML v.3.0 (Guindon et al. 2010). A bootstrap with 1000 replicates was used to estimate the internal nodes of the tree. An alignment with 1045 bp from Haemogregarina daviesensis sp. nov. sequences obtained in this study and seven Haemogregarina spp. sequences available on GenBank® were used to estimate the uncorrected pairwise distance (p distance). This analysis was performed on the MEGA6 software (Tamura et al. 2013).
Results
Prevalence and parasitemia level
Intraerythrocytic gametocytes of Haemogregarina sp. were found in 100% of the L. paradoxa specimens examined. The mean parasitemia level was 6 parasites/2000 blood erythrocytes (range 1–28).
Morphometric data of parasites
The sizes of the gametocytes of Haemogregarina sp. were 16 ± 0.12 μm (range 13–18) in length and 6 ± 0.97 μm (range 5–8) in width. The sizes of the infected erythrocytes and non-infected erythrocytes were 48 ± 0.54 μm (range 35–57) in length and 36 ± 0.73 μm (range 23–47) in width; and 47 ± 0.35 μm (range 35–49) in length and 42 ± 0.37 μm (range 34–49) in width, respectively. The sizes of the non-infected erythrocyte nucleus and infected erythrocyte nucleus were 2.4 ± 0.24 μm (range 2–2.8) in length and 1.7 ± 0.4 μm (range 1.5–2.1) in width and 2.5 ± 0.41 μm (range 2–3.7) in length and 1.4 ± 0.28 μm (range 1–2.1) in width, respectively.
Molecular and phylogenetic analysis
Two specimens of L. paradoxa that were positive for Haemogregarina sp. in thin blood smears were also positive according to PCR. The sequences isolated in this study were deposited in GenBank® (MH503891 and MH503892). The nucleotide divergence (p distance) among Haemogregarina sp. detected here and Haemogregarina spp. around the world ranged from 1.9 to 5.1% (Table 1). Based on the morphological features and genetic divergence among Haemogregarina sp. isolated in this study and other Haemogregarina species, we considered Haemogregarina sp. to be a new species, named Haemogregarina daviesensis sp. nov.
A phylogenetic tree (Fig. 2), inferred from a 1020 bp fragment of the 18S rRNA gene from 27 sequences, showed that H. daviesensis sp. nov. obtained in our study grouped with Haemogregarina sp. sequences isolated from a turtle and formed a unique clade that was strongly supported with 99% bootstrapping. In addition, the H. daviesensis sp. nov. sequences obtained from L. paradoxa formed a sister clade with other Haemogregarina species and were positioned in distinct clades from Hemolivia spp. and Hepatozoon spp.
Taxonomic summary
Phylum Apicomplexa Levine, 1970
Subphylum Conoidasida Levine, 1988
Class Coccidia Leuckart, 1879
Order Eucoccidiorida Léger and Duboscq, 1910
Suborder Adeleorina Léger, 1911
Family Haemogregarinidae Léger, 1911
Genus Haemogregarina Danilewsky, 1885
Haemogregarina daviesensis sp. nov
Type hostLepidosiren paradoxa Fitzinger, 1837 (Lepidosirenidae), piramboia, South American lungfish.
Type locality Free living environment: Igarapé Fortaleza basin (0° 02′ 10.91″ S; 51° 09′ 39.42″ W), municipality of Santana, Amapá, Brazil.
Type material Hapantotype, four blood smears from Lepidosiren paradoxa at the Institute for Scientific and Technological Research of the State of Amapá (IEPA), Amapá, AP, Brazil (n° 155, 156, 157, 158).
Site of infection Blood erythrocytes.
Prevalence All 4 L. paradoxa (100%) analyzed were positive for haemogregarines.
Parasitemia The mean parasitemia level was 6 parasites/2000 blood erythrocytes (range 1–28).
Etymology The species is named after Angela Josephine Davies (1947–2013), to honor her contribution to the knowledge of parasitic protozoa.
Vector Unknown.
Gene sequence The 18S ribosomal gene sequences were deposited in GenBank under accession numbers MH503891 and MH503892.
Description
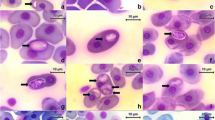
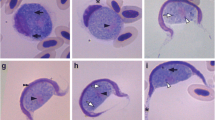
Gametocytes: All the stages observed were gametocytes, and these were generally monomorphic. However, they differed in width and length, thus indicating the presence of several stages of maturity. Gametocytes stained with Giemsa were found only in erythrocytes within a vacuole in cytoplasm.
The gametocytes of H. daviesensis sp. nov. were bean-shaped and were present in three forms: single, paired (double gametocyte), or more than two, inside the erythrocytes (Fig. 1). They are enclosed in a capsule and usually broad or could be slender, and were never curved. The parasite cytoplasm was pale blue, with granules surrounding the host cell extremities. In some cases, the gametocytes had a broad pole and, opposite this, a bent tail. The gametocyte nucleus presented diffuse or strongly condensed chromatin strands.
Maximum likelihood phylogenetic tree based on the partial sequences (1020 bp) of the 18S rRNA gene of Haemogregarinadaviesensis from Lepidosiren paradoxa, obtained in this study, and sequences available in GenBank. The branch length scale represents 0.04 substitutions per site. Cryptosporidium serpentis, Adelina dimidiata, and Adelina grylli were used as outgroup. The black dots are sequences identified in this study
Remarks
The gametocytes described here were monomorphic, with several stages of maturity, and its nucleus presented diffuse or strongly condensed chromatin strands. These differ from gametocytes of C. lignieresi which were remarkably dimorphic, named microgametocyte and macrogametocyte (Lainson 1981). Eiras et al. (1995) reported gametocytes of D. mugili from Mugil platanus that presented two morphotypes. The first morphotype (8.3–10.4 μm × 2.6–3.1 μm) had compact nucleus with well-defined margins and was curved in extremities, whereas the second (10.4–18.3 μm × 2.1–3.6 μm) was longer than the first and had C shape with extremities curved. Davies et al. (2008) described H. platessae in Paralichthys orbignyanus, reporting elongate and curved gametocytes (9.4 ± 0.9 μm × 3.1 ± 0.4 μm) with large vacuoles in cytoplasm. The morphology and morphometry recorded to species in abovementioned studies disagree with the presented here. Jepps (1929) analyzed morphological features of H. lepidosirenis from L. paradoxa and reported sausage-shaped parasites, with cytoplasmatic inclusions, produced through the process of schizogony. In this study, schizonts were not observed and gametocytes did not resemble those from H. lepidosirenis. It is important to highlight that few data about morphology and morphometry has been reported to fish haemogregarines.
Discussion
Based on morphology and morphometry of gamonts in blood and molecular phylogenetic analysis, we described the haemogregarine reported in this study as H. daviesensis sp. nov. The scarcity of molecular data on fish isolates makes genus identification difficult. Many haemogregarine species have been inconsistently described or have been reported as new records in hosts, because studies have been based only on morphological characteristics (Desser 1993; Alhaboubi et al. 2017; Úngari et al. 2018). Consequently, many descriptions made in the past are subject to new classifications and recombination (Mathew et al. 2000). Therefore, it is important to assemble data on morphology, life cycles, and genetics, so as to avoid misconceptions regarding taxonomy conclusions (Sloboda et al. 2007).
It is known that the genus Haemogregarina produces eight sporozoites, while Cyrilia produces more than 18 sporozoites in leeches (Siddall 1995). However, in the present study, we were unable to determine the genus of the intraerythrocytic haemogregarines in L. paradoxa using only morphological features because no invertebrate vector was found. Moreover, no merozoite stages were found, which made it impossible to compare the haemogregarines of this study with H. lepidosirenis in L. paradoxa (Jepps 1929) and Cyrilia lignieresi in Synbranchus marmoratus (Lainson 1981; Diniz et al. 2002). This absence of intraerythrocytic merozoites might suggest that these haemogregarines of L. paradoxa belong to the genus Desseria (Siddall 1995).
The gametocytes of H. daviesensis sp. nov. found in L. paradoxa were compared with the same stages found in other species: H. lepidosirenis, D. mugili, H. platessae, and C. lignieresi. In general, the morphological features observed in the gametocytes of H. daviesensis sp. nov. differed from the gametocytes found in the abovementioned species. The bean-shaped capsule, vacuole in cytoplasm, straight shape, undefined position of nuclei and presence of granules were different from these species (Jepps 1929; Lainson 1981; Eiras et al. 1995; Davies et al. 2008). With respect to C. lignieresi, Lainson (1981) reports gamonts (11.0 × 5.0 μm) that were larger than D. mugili (10.4 × 3.1 μm) and H. platessae (9.4 × 3.1 μm) (Lainson 1981; Eiras et al. 1995; Davies et al. 2008). These morphometric measures do not resemble the gametocytes of H. daviesensis from the present study. However, the morphometry of the gametocytes (15 × 3.2 μm) of Haemogregarina bertonii (Schouten 1941) was similar with H. daviesensis sp. nov. In sum, H. daviesensis presented morphological and morphometric differences with other species.
The molecular characterization of Haemogregarina species in the present study was based on analysis of the 18S rRNA gene sequences. In the phylogenetic analysis, H. daviesensis sp. nov. from L. paradoxa form a sister clade with a Haemogregarina sp. from one specimen of snapping turtle Macrochelys temminckii from Texas, USA (Alhaboubi et al. 2017). Macrochelys temminckii is a piscivorous aquatic chelonian that lives in swamps (Elsey 2006). This may suggest a host switch from turtles to fish, i.e., a host switch from turtle-infecting ancestral condition to fish-infecting condition. Moreover, this may indicate that a definitive host is a putative Glossiphoniidae leech (freshwater leech) in which sexual reproduction occurs. In other studies, lineages of hemosporidian parasites (Apicomplexa) perform host switch in avian, bats, primates, lizards, and turtles (Ricklefs et al. 2004; Duval et al. 2007; Úngari et al. 2018).
There are no sequences of Haemogregarina species in freshwater fish deposited in GenBank. The unique sequences of fish haemogregarine available in GenBank are from Haemogregarina (sensu latu) bigemina in marine fish (Hayes and Smit 2019). The scarcity of molecular and morphological data on haemogregarine developmental stages make difficult identification of Haemogregarina isolates in fish of the Neotropical region. However, the morphological and morphometric data were not identical to those observed for H. lepidosirenis (Jepps 1929), D. mugili (Eiras et al. 1995), H. platessae (Davies et al. 2008), and C. lignieresi (Lainson 1981).
To conclude, we describe a new species, H. daviesensis, in L. paradoxa from Brazil. In addition, this study on the morphological and molecular characterization in lungfish from the Amazon region compared isolates with the available morphological data on haemogregarine developmental stages that had previously been reported and provided the first data on the molecular phylogeny of an intraerythrocytic haemogregarine of freshwater fish in this Neotropical region. Lastly, our findings regarding the genetic sequences of haemogregarines of fish emphasize the importance of obtaining additional information on aspects of the general biology of these hemoparasites in fish populations, in order to achieve correct taxonomic classification.
References
Alhaboubi AR, Pollard DA, Holman PJ (2017) Molecular and morphological characterization of a haemogregarine in the alligator snapping turtle, Macrochelys temminckii (Testudines: Chelydridae). J Parasitol 116:207–215. https://doi.org/10.1007/s00436-016-5280-2
Almeida-Val VMF, Nozawa SR, Lopes NP, Rocha APH, Mesquita-Saad LS, Silva MNP, Val AL (2010) Biology of the South American lungfish, Lepidosiren paradoxa. In: Jorgensen JM, Joss J (eds) The biology of lungfishes, 1th edn. CRC Press, New York, pp 129–138
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth.2109
Davies AJ (1995) The biology of fish haemogregarines. Adv Parasitol 36:117–203. https://doi.org/10.1016/S0065-308X(08)60491-1
Davies AJ, Johnston MRL (2000) The biology of some intraerythrocytic parasites of fishes, amphibia and reptiles. Adv Parasitol 45:1–107. https://doi.org/10.1016/S0065-308X(00)45003-7
Davies AJ, Smit NJ (2001) The life cycle of Haemogregarina bigemina (Adeleina: Haemogregarinidae) in South African hosts. Folia Parasitol 48:169–177. https://doi.org/10.14411/fp.2001.029
Davies A, Amado LL, Cook R, Bianchini A, Eiras JC (2008) Potential environmental and host gender influences on prevalence of Haemogregarina platessae (Adeleorina: Haemogregarinidae) and suspected Haemohormidium terraenovae (incertae sedis) in Brazilian flounder from the Patos Lagoon Estuary, Southern Brazil. Folia Parasitol 55:161–170. https://doi.org/10.14411/fp.2008.023
Desser SS (1993) The Haemogregarinidae and Lankesterellidae. In: Kreier JP (ed) Parasitic Protozoa. Academic Press, London, pp 247–272
Diniz J, Silva E, De Souza W, Lainson R (2002) Some observations on the fine structure of trophozoites of the haemogregarine Cyrilia lignieresi (Adeleina: Haemogregarinidae) in erythrocytes of the fish Synbranchus marmoratus (Synbranchidae). Parasitol Res 88:593–597. https://doi.org/10.1007/s00436-002-0603-x
Duval L, Robert V, Csorba G, Hassanin A, Randrianarivelojosia M, Walston J, Nhim T, Goodman SM, Ariey F (2007) Multiple host-switching of Haemosporidia parasites in bats. Malar J 6:157. https://doi.org/10.1186/1475-2875-6-157
Eiras JC, Ranzani-Paiva MJT, Davies AJ (1995) Observations on Haemogregarina mugili (Apicomplexa) and Trypanosoma froesi (Sarcomastigophora) from the blood of Mugil platanus Günther, 1880 (Pisces: Mugilidae) in Brazil. Res Rev Parasitol 55:173–176
Eiras JC, Takemoto RM, Pavanelli GC, Luque JL (2012) Checklist of protozoan parasites of fishes from Brazil. Zootaxa 3221:1–25
Elsey RM (2006) Food habits of Macrochelys temminckii (alligator snapping turtle) from Arkansas and Louisiana. Southeast Nat 5:443–452. https://doi.org/10.1656/1528-7092(2006)5[443:FHOMTA]2.0.CO;2
Godfrey RD Jr, Fedynich AM, Pence DB (1987) Quantification of hematozoa in blood smear. J Wildl Dis 23:558–565. https://doi.org/10.7589/0090-3558-23.4.558
Godfrey RD Jr, Pence DB, Fedynich AM (1990) Effects of host and spatial factors on a haemoproteid community in mourning doves from western Texas. J Wildl Dis 26:435–441. https://doi.org/10.7589/0090-3558-26.4.435
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. https://doi.org/10.1093/sysbio/syq010
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hayes PM, Smit NJ (2019) Molecular insights into the identification and phylogenetics of the cosmopolitan marine fish blood parasite, Haemogregarina bigemina (Adeleorina: Haemogregarinidae). Int J Parasitol - Par 8:216–220. https://doi.org/10.1016/j.ijppaw.2019.01.006
Jepps MW (1927) Note on a haemogregarine in Lepidosiren paradoxa. Parasitology 19:285–287. https://doi.org/10.1017/S0031182000005722
Jepps MW (1929) Further note on Haemogregarina Lepidosirenis. Parasitology 21:282–287. https://doi.org/10.1017/S0031182000022976
Lainson R (1981) On Cyrilia gomesi (Neiva & Pinto, 1926) gen. nov. (Haemogregarinidae) and Trypanosoma bourouli Neiva & Pinto, in the fish Synbranchus marmoratus: simultaneous transmission by the leech Haementeria lutzi in: canning EU (ed) parasitological topics. (Society of Protozoologists special publication 1) Society of Protozoologists, London 150-158
Machado LP, Wellendorf H, Brito PM (2010) On the type material of Lepidosiren paradoxa Fitzinger, 1837 (Sarcopterygii, Dipnoi). C R Biol 333:56–60. https://doi.org/10.1016/j.crvi.2009.10.005
Magro NM, de-Oliveira AT, O'Dwyer LH (2016) First report and description of a Cyrilia sp.(Apicomplexa: Haemogregarinidae) from a freshwater Cururu Stingray Potamotrygon cf. histrix (Elasmobranchii: Potamotrygonidae), from the Amazon Region, Brazil. J Fish Dis 39: 907–911. https://doi.org/10.1111/jfd.12425
Mathew JS, Bussche RAVD, Ewing SA, Malayer JR, Latha BR, Panciera RJ (2000) Phylogenetic relationships of Hepatozoon (Apicomplexa: Adeleorina) based on molecular, morphologic, and life-cycle characters. J Parasitol 86:366–372. https://doi.org/10.1645/0022-3395(2000)086[0366:PROHAA]2.0.CO;2
Nelson JS, Grande TC, Wilson MVH (2016) Fishes of the world. John Wiley & Sons, New Jersey
Oliveira AT, Araújo MLG, Pantoja-Lima J, Aride PHR, Tavares-Dias M, Brinn RP, Marcon JL (2017) Cyrilia sp. (Apicomplexa: Haemogregarinidae) in the Amazonian freshwater stingray Potamotrygon wallacei (cururu stingray) in different hydrological phases of the Rio Negro. Braz J Biol 77: 413–416. https://doi.org/10.1590/1519-6984.00416
Ricklefs RE, Fallon SM, Bermingham E (2004) Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Syst Biol 53:111–119. https://doi.org/10.1080/10635150490264987
Schouten GB (1941) Haemogregarina bertonii n. sp. hematozoario de Lepidosiren paradoxa Fitinger. Rev Soc Cien Paraguay 5:113–115
Siddall ME (1995) Phylogeny of adeleid blood parasites with a partial systematic revision of the haemogregarine complex. J Eukaryot Microbiol 42:116–125. https://doi.org/10.1111/j.1550-7408.1995.tb01551.x
Siddall ME, Desser SS (1993) Ultrastructure of merogonic development of Haemogregarina (sensu lato) myoxocephali (Apicomplexa: Adeleina) in the marine leech Malmiana scorpii and localization of infective stages in the salivary cells. Eur J Protistol 29:191–201. https://doi.org/10.1016/S0932-4739(11)80273-7
Sloboda M, Kamler M, Bulantová J, Votýpka J, Modrý D (2007) A new species of Hepatozoon (Apicomplexa: Adeleorina) from Python regius (Serpentes: Pythonidae) and its experimental transmission by a mosquito vector. J Parasitol 93:1189–1198. https://doi.org/10.1645/GE-1200R.1
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Ujvari B, Madsen T, Olsson M (2004) High prevalence of Hepatozoon spp. (Apicomplexa, Hepatozoidae) infection in water pythons (Liasis fuscus) from tropical Australia. J Parasitol 90:670–672. https://doi.org/10.1645/GE-204R
Úngari LP, Santos ALQ, O’Dwyer LH, da Silva MRL, de Melo Fava NN, Paiva GCM, Cury MC (2018) Haemogregarina podocnemis sp. nov.: description of a new species of Haemogregarina Danilewsky 1885 (Adeleina: Haemogregarinaidae) in free-living and captive yellow-spotted river turtles Podocnemis unifilis (Testudines: Podocnemididae) from Brazil. J Parasitol Res 117:1535–1548. https://doi.org/10.1007/s00436-018-5817-7
Acknowledgements
The authors thank the National Council for Scientific and Technological Development (Conselho Nacional de Desenvolvimento Científico e Tecnológico, CNPq), Brazil for the research productivity grant awarded to Dr. M. Tavares-Dias (# 303013/2015-0). To Jackson Cleiton de Souza for help in fieldwork.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Astrid Holzer
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Esteves-Silva, P.H., da Silva, M.R.L., O’Dwyer, L.H. et al. Haemogregarina daviesensis sp. nov. (Apicomplexa: Haemogregarinidae) from South American lungfish Lepidosiren paradoxa (Sarcopterygii: Lepidosirenidae) in the eastern Amazon region. Parasitol Res 118, 2773–2779 (2019). https://doi.org/10.1007/s00436-019-06430-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-019-06430-7