Abstract
Procamallanus (Spirocamallanus) huacraensis infecting the catfish Trichomycterus spegazzinii from Escoipe River, Salta province (Argentina), is redescribed and genetically characterised for the first time, based on three genetic markers (nuclear 18S and 28S rRNA; cytochrome c oxidase subunit I [cox1] mtDNA). The phylogeny of Camallanidae was also discussed. Morphological evaluation of P. (S.) huacraensis using light and scanning electron microscopy revealed the previously undescribed features: location of deirids, accurate morphology of larvae (L1) and ovijector in females, as well as phasmids in males. Differences were found comparing the newly collected material and the type specimens, probably because the original description lacked detailing. Unfortunately, type specimens of P. (S.) huacraensis were no available for loan. The results of morphological and genetic analyses supported the validity of P. (S.) huacraensis. Inconsistencies regarding the taxonomic identification of species of Camallanidae in GenBank database were noted. Based on the current genetic database of Camallanidae, phylogenetic reconstructions using the 18S rRNA sequences were most consistent, due to the inclusion of higher number of taxa. Procamallanus (S.) huacraensis appeared as sister group of P. (S.) rarus, also isolated from a catfish in a neighbouring region. The order and habitat of hosts were also similar within some well-supported parasite lineages, but without common geographic origin. However, it is still premature to make definitive affirmations regarding the role of such features in the phylogenetic patterns of Camallanidae, given the scarcity of genetic data. The phylogenetic reconstructions also confirmed the artificiality of the morphology-based systematics of the family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Camallanidae Railliet & Henry, 1915 is a monophyletic family of nematodes that infect marine and freshwater fishes, exhibiting broad global distribution (Anderson et al. 2009; Černotíková et al. 2011). Despite its species richness, proportionally few taxa within the Camallanidae has been genetically characterised (Wu et al. 2008, 2009; Kuzmin et al. 2011; Gaither et al. 2013), and their phylogenetic relationships are poorly understood (Sardella et al. 2017).
The morphological taxonomy of camallanids is problematic, especially regarding species of Procamallanus Baylis, 1923 (Sardella et al. 2017). This genus has been subdivided into several subgenera based on the structure of the buccal capsule, a fact that generates debate (see Moravec and Sey 1988; Moravec and Thatcher 1997; Rigby and Adamson 1997; Anderson et al. 2009; Moravec and Van As 2015). The current genetic database does not support the resolution of such systematic problems (Černotíková et al. 2011; Sardella et al. 2017).
Procamallanus (Spirocamallanus) Olsen, 1952, is the most specious subgenera of Procamallanus; however, several species remain poorly described and only a few representatives were genetically characterised, also resulting in taxonomic problems (Moravec and Justine 2017; Sardella et al. 2017). In this sense, improvement of the morphological and genetic knowledge pertaining to these parasites is necessary.
During parasitological examinations in Trichomycterus spegazzinii (Berg) (Siluriformes: Trichomycteridae) from River Escoipe, province of Salta, Argentina, some specimens of nematodes were recovered from the anterior intestine of the fish. Detailed observation revealed that the specimens were similar to P. (S.) huacraensis Ramallo 2008 (a species with poorly detailed morphological description). Here, we redescribed P. (S.) huacraensis, using light and scanning electron microscopy and provided its first genetic characterisation based on nuclear and mitochondrial genetic markers. Additionally, phylogenetic reconstructions were used to evaluate the phylogenetic position of P. (S.) huacraensis within Camallanidae and its relationships with other taxa. Some aspects of the phylogeny of Camallanidae were also discussed.
Materials and methods
Collection and examination of nematodes
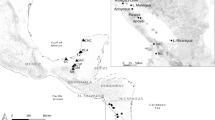
During February 2017 to March 2019, 111 specimens of T. spegazzinii (standard length 33.00–162.79 mm) were caught using fishing rods in Escoipe River (25°06′49″S; 65°36′0.4″W), municipality of Chicoana, Salta province, Argentina. Fish were kept alive in small water tanks and brought to the laboratory prior to helminthological examinations. Host nomenclature and classification were updated according to Froese and Pauly (2019). Nematodes were found alive, washed in saline, fixed in hot 4% formaldehyde solution and preserved in 70% ethanol. For morphological observations, nematodes were cleared in glycerine. Two males had the mid body excised and fixed in molecular-grade 96–99% ethanol for genetic studies; anterior and posterior parts were fixed from morphological identification.
Drawings were made using a drawing tube attached to a microscope Olympus BX51. Measurements are given in micrometres, unless otherwise stated. Specimens used for SEM (2 males and 2 females) were dehydrated through a graded ethanol series, dried by evaporation with hexamethyldisilazane, coated with gold and observed in a JEOL JSM 6460-LV, at an accelerating voltage of 15 kV. The systematic classification of nematodes follows the proposal of Moravec and Thatcher (1997). Voucher specimens were deposited in the Parasitological Collection of the Museo Argentino de Ciencias Naturales ‘Bernardino Rivadavia’, Buenos Aires, Argentina (MACN-Pa).
Genetic procedures
Genomic DNA was isolated using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. Three genetic regions were amplified: the 5′ end of the 18S nuclear rRNA, the D2 and D3 domains of the nuclear 28S rRNA and the cox1 of the mtDNA. All PCR reactions, cycling conditions and primers are detailed in Online Resource 1. PCR products were purified through an enzymatic treatment with ExoProStar™ (GE Helathcare) and sent for sequencing at ACTGene (Ludwig Biotec, Rio Grande do Sul, Brazil) with the same primers used in PCR reactions.
Contiguous sequences were assembled in Geneious (Geneious ver. 9.1.5 created by Biomatters, available from http://www.geneious.com/) and deposited in the GenBank (see taxonomic summary for accession numbers). Preliminary BLAST search on GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) was performed to confirm the genetic proximity between the present sequences and those from representatives of Camallanidae.
Phylogenetic analyses of molecular data
The phylogenetic reconstructions were based on two different datasets, one containing sequences of the 18S rRNA and one containing those of cox1 mtDNA. Due to the small number of sequences of the 28S rRNA, phylogenies based on this genetic region were provided as Online Resource 2 and were not discussed here. Sequences were chosen according to the following criteria: genetic regions congruent with those obtained in the present study and minimum length of 744 bp and 355 bp for the 18S and the cox1, respectively (for details see Table 1). Sequences from the same isolate, clones or with 100% of genetic similarity were mostly not considered. The outgroup was chosen according to previous phylogenies of Camallanidae (see Černotíková et al. 2011). Sequences were aligned using M-Cofee (Notredame et al. 2000), then evaluated by the transitive consistency score, to verify the reliability of aligned positions and, based on score values, ambiguous aligned positions were trimmed (Chang et al. 2014). Datasets were subjected to maximum likelihood (ML) and Bayesian inference (BI) analyses, using PHYML and MrBayes, respectively (Huelsenbeck and Ronquist 2001; Guindon and Gascuel 2003). The model of evolution (nucleotide substitution) and its fixed parameters for each dataset were chosen and estimated under the Akaike information criterion with jModelTest 2 (Guindon and Gascuel 2003; Darriba et al. 2012) and are detailed in Online Resource 3. Nodal supports for ML were based on 1000 bootstrap non-parametric replications. The same, but for Bayesian posterior probability, was determined after running the Markov chain Monte Carlo (2 runs 4 chains) for 4 × 106 generations, with sampling frequency every 4 × 103 generations and discarding the initial one fourth of sampled trees (1 × 106) as bur-in. In order to check chain convergence, analyses were run in duplicates and inspected using Tracer.
Results
The description was based on 19 nematodes collected in the intestine of T. spegazzinii from River Escoipe, Northwestern Argentina. Detailed morphometric data of the newly collected and of the type specimens from P. (S.) huacraensis is presented in Table 2.
Camallanidae Railliet & Henry, 1915;
Procamallanus (Spirocamallanus) Olsen, 1952;
Procamallanus (Spirocamallanus) huacraensis Ramallo, 2008.
Redescription
General
Medium-sized to large nematodes, reddish when alive. Cuticle finely transversely striated (Fig. 2d–g). Oral opening circular, surrounded by eight submedian cephalic papillae, arranged in two circles: outer circle with distinctly larger papillae than those of inner circle (Figs. 1e and 2a, c). Pair of small lateral amphids present (Figs. 1e and 2a, c). Small pore-like cephalic structures absent. Buccal capsule orange-brown, thick-walled, barrel-shaped, longer than wide, with well-developed basal ring (Fig. 1c, d). Inner surface of buccal capsule showing slight sexual dimorphism; two–three blade-like sclerotized structures arising between mid-length and bottom of buccal capsule, present in both sexes (Figs. 1c, d and 2b). Muscular oesophagus club-shaped, shorter than glandular portion (Fig. 1a, b). Glandular oesophagus cylindrical, with a smooth constriction at posterior end (Fig. 1a, b). Nerve ring encircling muscular oesophagus somewhat anterior to its mid-length (Fig. 1a, b). Excretory pore posterior to muscular/glandular oesophagus junction (Fig. 1b). Deirids very small, simple, anterior to muscular/glandular oesophagus junction (Fig. 1a, b). Intestine brown and narrow. Tail conical, with finger-like constriction rounded at end, longer in males (Figs. 1f–h and 2d–g).
Procamallanus (Spirocamallanus) huacraensis. a, b Anterior end of female and male, respectively, dorsoventral and lateral views, respectively. c, d Cephalic end of female and male, respectively, lateral view. e Cephalic end, apical view. f, g Posterior end of male, lateral and ventral views, respectively. h Posterior end of female, lateral view. i Region of vulva, lateral view; j First-stage larva dissected from uterus
SEM micrographs of Procamallanus (Spirocamallanus) huacraensis.a, b Cephalic end of a larvigerous female, apical view (arrowheads indicate blade-like structures of buccal capsule). c Cephalic end of male, sublateral view. d Posterior end of male, lateral view. e Precloacal papillae, lateral view. f Tail of female, subapical view. g Tail of male, lateral view. Abbreviations: a, amphid; c, cephalic papilla; p, phasmid
Male (based on eight adult specimens)
Length of body 5.2–7.6 mm, maximum width 115–230. Buccal capsule including basal ring 49–63 long and 37–49 wide, with four inner spiral ridges (two anterior incomplete); basal ring 5–6 long, 20–32 wide (Fig. 1d). Maximum width/length ratio of buccal capsule 1:1.1–1.5. Muscular oesophagus 221–275 long, 79–98 wide; glandular oesophagus 295–393 long, 56–98 wide; length ratio of muscular/glandular parts 1:1.3–1.5. Length of entire oesophagus and buccal capsule representing 9.3–12.4% of total body length. Nerve ring, deirids and excretory pore 135–159, 233–334 and 330–387, respectively, from anterior end (Fig. 1b). Spicules short, similar and equal, with straight proximal end and sharply pointed at distal extremity (Fig. 1f), 78–91 long, representing 1.1–1.6% of total body length. Gubernaculum and caudal alae absent. Precloacal papillae: four pairs of subventral papillae, first and second pairs larger and laterally displaced, third and fourth pairs smaller and ventrally directed; papillae pairs may be slightly asymmetric (Figs. 1f, g and 2d, e). Postcloacal papillae: two subventral pairs, somewhat asymmetrically arranged (Figs. 1f, g and 2d, g). Pair of lateral, small and inconspicuous phasmids, posterior to last pair of postcloacal papillae (Figs. 1f, g and 2d, g). Tail 208–275 long; its constricted portion 98–118 long (Figs. 1f, g and 2d, g).
Female (based on two specimens containing larvae and nine containing eggs [measurements in parentheses])
Length of body 25.5–28.2 mm (15.3–24.3), maximum width 547–576 (297–574). Buccal capsule including basal ring 64 (59–71) long and 49 (49–66) wide, with two inner complete spiral ridges; basal ring 5–7 (5–8) long, 27–29 (28–37) wide (Fig. 1c). Maximum width/length ratio of buccal capsule 1:1.3 (1:1.0–1.2). Muscular oesophagus 344–354 (324–393) long, 118–128 (98–177) wide; glandular oesophagus 491–540 (472–560) long, 108–137 (98–157) wide; length ratio of muscular/glandular parts 1:1.4–1.6 (1:1.4–1.7). Length of entire oesophagus and buccal capsule representing 3.2–3.7% (3.0–3.5) of body length. Nerve ring, deirids and excretory pore 187–196 (157–255), 294–368 (362–429) and 392–480 (363–502), respectively, from anterior end (Fig. 1a). Vulva ranging from equatorial to postequatorial, 9.7–12.2 mm (8.4–12.3) from posterior end, at 57–62 (49–63) % of body length. Vulval lips not elevated (Fig. 1i). Vagina with striated musculature, followed by muscular ovijector bearing anterior chamber; both posteriorly directed from vulva (Fig. 1i). Phasmids not observed. Tail 285–275 long (255–403); its constricted portion 79–118 long (88–118) (Figs. 1h and 2f). Uterus of larvigerous females filled with first-stage larvae (n = 4), 283–320 long, 16–17 wide; with 2-min labia and rudimentary oesophagus 64–84 long, nerve ring 24–27 from anterior end and tail 62–70 long, ending in sharp point (Fig. 1j).
Taxonomic summary
Host. Trichomycterus spegazzinii (Berg) (Siluriformes: Trichomycteridae).
Site of infection. Intestine.
Locality. River Escoipe (25° 06′49″S; 65° 36′0.4″W), municipality of Chicoana, province of Salta, Argentina.
Prevalence and mean intensity. 17% (19 fish infected/111 fish examined); 1.5 ± 0.9 (range 1–5) nematodes per infected fish.
Voucher specimens deposited. Six males and nine females (MACN-Pa 662).
Genetic data (GenBank accession numbers). 18S rRNA (MK794615), 28S (MK793794) rRNA, cox1 mtDNA (MK780067).
Remarks
Procamallanus (S.) huacraensis was originally described from Trichomycterus corduvensis Weyenbergh in River Huacra, Catamarca province, Argentina (Ramallo 2008). Ramallo and Padilla Bortayro (2011) reported the species infecting a conspecific to the type host but collected from a different river (Vis-Vis) in the same province; the authors provided no taxonomic description. The newly collected material showed the following differences in contrast to the original description of P. (S.) huacraensis: buccal capsule with four spiral ridges, two of which incomplete, in males (vs. all complete) and only two complete spiral ridges in females (vs. 3–4), more posterior position of deirids and excretore pore, and smaller spicules (78–91 long, representing 1.1–1.6% of total body length vs. 180–190 long, representing 2.7–3.7% of total body length) (Ramallo 2008). However, in addition to the congeneric hosts and to the neighbouring geographic origins, the present material and the types of P. (S.) huacraensis share the same number and arrangement of caudal papillae in males as well as most of the morphometric features (see Table 2). Since the types of P. (S.) huacraensis were not available for comparison, we believed that the most prudent was to tentatively retain the present parasites in the referred species.
Morphological observations revealed difficulties for observing the accurate structure of cephalic papillae and buccal capsule, as well as the location of deirids (they are very small) and excretory pore, especially in large females that have thick muscular and cuticular layers. These features were not represented in details by (Ramallo 2008), and it cannot be discarded that they might have been inaccurately reported. In this sense and contrary to the original description, the basal ring of buccal capsule is clearly distinct (vs. reported as indistinct) and the sclerotized blade-like structures arising from its inner walls are present in both male and female (vs. described only in females) (Ramallo 2008).
The present results also confirmed the morphology of cephalic papillae in P. (S.) huacraensis, as well as showed the absence of pore-like structures surrounding mouth, which are difficult to observe within species of Procamallanus (Spirocamallanus).
Furthermore, the following features were described for the first time in P. (S.) huacraensis: deirids, accurate morphology of ovijector and of larvae (L1) in females, and phasmids in males.
According to the conception of Moravec et al. (2004), P. (S.) huacraensis belongs to the group of Procamallanus (S.) spp. in which males lack caudal alae and have two small equal spicules. The following congeneric parasites of freshwater fishes in the Neotropical region also belong to this group: P. (S.) belenensis Giese et al. 2009, P. (S.) chimusensis (Freitas & Ibáñez, 1968), P. (S.) hilarii Vaz & Pereira, 1934, P. (S.) inopinatus Travassos, Artigas & Pereira, 1928, P. (S.) krameri (Petter, 1974), P. (S.) neocaballeroi (Caballero-Deloya, 1977), P. (S.) paraensis Pinto & Noronha, 1976, P. (S.) pintoi (Kohn & Fernandes, 1988) and P. (S.) saofranciscencis (Moreira et al. 1944) (Moravec 1998; Moravec et al. 2004; Ramallo 2008; Giese et al. 2009). However, P. (S.) huacraensis clearly differs from the abovementioned species based on the fewer spiral ridges in buccal capsule (2–4 vs. 8–18 as overall range), the small number of postcloacal pairs of papillae (2 vs. 3 at minimum) and by the morphology of tail in females (other than with a long finger-like constriction) (see Moravec 1998; Giese et al. 2009). The structure of tail in females seems to be constant within species of Procamallanus (Spirocamallanus) (Moravec et al. 2006).
Procamallanus (S.) chimusensis, a parasite of Trichomycterus spp. from Peru and Colombia (Freitas and Ibáñez 1968; Tantaleán et al. 1985; Moravec et al. 2004), somewhat resembles P. (S.) huacraensis. However males of P. (S.) chimusensis have 9–10 spiral ridges in the buccal capsule and eight pairs of caudal papillae (with different arrangement) vs. 4 spiral ridges and six pairs of caudal papillae in those of P. (S.) huacraensis (Moravec et al. 2004; Ramallo 2008; present study). Therefore, based on the previous differential diagnosis, the validity of P. (S.) huacraensis was confirmed.
Molecular characterisation and phylogenetic analyses
The partial sequences of the 18S (826 bp) and 28S (780 bp) rRNA and cox1 (387 bp) mtDNA were obtained for P. (S.) huacraensis; sequences generated from the two males were identical and only one isolate was used for analyses. In the alignment of 18S sequences, P. (S.) huacraensis was most genetically similar (sequence identity 96.5%) to P. (S.) rarus Travassos, Artigas & Pereira, 1928, ex. Calophysus macropterus (Lichtenstein) (Siluriformes: Pimelodidae) from Peru, followed by (sequence identity 95.4%) Cammallanus lacustris (Zoega, 1776), ex. Sander lucioperca (Linnaeus) (Perciformes: Percidae) from Czech Republic. The alignment of 28S sequences was poorly informative, since it included only few representatives of Camallanus Railliet & Henry, 1915 and one of Serpinema Yeh, 1960 (see Table 1 and Online Resource 1); therefore, these results were omitted. Regarding the alignment from sequences of cox1, P. (S.) huacraensis was most genetically similar (sequence identity 80.4%) to P. (P.) spiculogubernaculus Agarwal, 1958, ex. Heteropneustes fossilis (Bloch) (Siluriformes: Heteropneustidae) from India, followed by (sequence identity 80.4%) P. (S.) istiblenni (Noble, 1966) ex. Lutjanus kasmira (Forsskål) (Perciformes: Lutjanidae) from Hawaii.
The general topology of the phylogenetic reconstructions generated from BI and ML were not different (i.e. well and moderately supported assemblages were the same) (see Fig. 3, Online Resource 1 and 4). The phylogenetic reconstruction using the 18S sequences contained more taxa and was consequently more informative than that of the cox1 (Fig. 3a, b). Procamallanus (Procamallanus) Baylis, 1923 and Procamallanus (Spirocamallanus) were not monophyletic and, except for Camallanus sp. (GenBank accession number DQ442664), Camallanus was monophyletic (Fig. 3a, b). Procamallanus (S.) huacraensis was sister group of P. (S.) rarus forming a well-supported sister assemblage to that formed by Camallanus spp. from freshwater fishes (Fig. 3a). In the absence of P. (S.) rarus (i.e. in the phylogeny of cox1), P. (S.) huacraensis remained as a sister lineage to Camallanus, albeit with low support (Fig. 3b). Camallanus cotti Fujita, 1927, C. oxycephalus Ward & Magath, 1916, P. (S.) istiblenni and P. (S.) rarus appeared as polyphyletic (Fig. 3a); however, some of these species identification along with those of Procamallanus sp. (FJ172980, GU082486, GQ265672, KJ566935) are doubtful (Fig. 3a, b). Moderate to well-supported clades were formed sharing the same order of host and habitat; the clades seldom shared similar geographic origins (Fig. 3, Table 1). Important information pertaining to the genetic sequences was occasionally missing (see Table 1), which complicated the interpretation of some results.
Phylogenetic trees generated using maximum likelihood (ML) from sequences of the 18S rDNA (a) and Bayesian inference from those of the cox1 mtDNA (b). Nodal supports of Bayesian posterior probability (BPP) were estimated after running the Markov chain Monte Carlo (2 runs 4 chains, 4 × 106 generations, sampling frequency = 4 × 103, bur-in = 1 × 106); those of ML were generated after 1000 non-parametric bootstrap replications. Full circles = BPP > 0.96 and ML > 80%; empty circles = 0.90 < BPP < 0.96 and 50% < ML < 80%. Low nodal supports were not represented and considered. Shaded lineages have dubious taxonomic diagnosis. Sequences in bold are from the present study. Orders of hosts are depicted. *Considered taxon inquirendum; **named as Procamallanus slomei in GenBank
Discussion
Here is presented the third report of Procamallanus (S.) huacraensis, in which T. spegazzinii and Salta province represent new host and locality records, respectively. It should be mentioned that the original differential diagnosis for the species was restrict, regarding the comparisons among congeners (Ramallo 2008). In the present study, we provided detailed remarks for Procamallanus (S.) huacraensis, confirming its validity. Despite the differences noted between the present specimens and the types of Procamallanus (S.) huacraensis, we strongly believe that they belong to a same species, based on the closely related hosts and geographic origins, as well as other similarities (see remarks). It was not possible to conclude if these differences may be accounted by intraspecific variations or if they resulted from misinterpretations, since type specimens were inaccessible. The genetic characterisation of Procamallanus (S.) huacraensis also differentiated it from the congeners characterised so far. In all the phylogenetic reconstructions, this species formed an independent lineage supporting its validity.
The phylogenetic reconstruction using sequences of the 18S was the most informative, because it included more taxa. Therefore, much of the discussion was based on these results. It could be noted that the order of host and habitat (freshwater/marine) showed relationship with the assembling of certain clades, e.g., formed by P. (P.) annulatus, P. (P.) sigani, P. (S.) istiblenni (EF180076, KC505629, KC505630), P. (S.) macanesis, P. (S.) monotaxis, P. (S.) philippinensis and P. (S.) rebecae, which are all isolated from marine perciforms (except by P. (S.) rebecae). Alike, the assemblage of P. (P.) laeviconchus, P. (P.) spiculogubernaculus, P. (S.) fulvidraconis and P. (S.) rarus (DQ494195), with exception of P. (P.) pacificus, included parasites of freshwater catfishes (even though nodal supports were generally low). Procamallanus (S.) huacraensis was sister group to P. (S.) rarus (FJ803921), both parasites of catfishes from neighbouring regions (Argentina and Peru, respectively). Even though host taxa, habitat and geographic origin appeared to be related with some phylogenetic patterns within Camallanidae, it is not prudent to make affirmations until the genetic database is improved and the systematic impasses within the family are resolved.
The sequences of C. cotti (GU082507), C. oxycephalus (GU082496) and P. (S.) istiblenni (GU082491) made these species polyphyletic. Most likely, this is a case of wrong species determination. The same could be related to some Procamallanus sp. (FJ172980, GQ265672, GU082486, KJ566935) (see Fig. 3). None of these sequences were published in scientific papers, and information pertaining to them is scarce. Černotíková et al. (2011) highlighted that one of the sequences assigned to P. (S.) rarus (i.e. DQ494195, JF803912) may be also a case of species misidentification.
Regarding the genus Camallanus, the sequence Camallanus sp. (DQ442664) probably was isolated from C. carangis (Olsen, 1954), ex. the marine perciform Upeneus vittatus (Forsskål), from New Caledonia (see Wijová et al. 2006; Černotíková et al. 2011). This sequence appeared as a basal lineage to the in group, far from the congeners parasitic in freshwater fishes; consequently, Camallanus appeared as paraphyletic (Fig. 3a). In fact, there was a complete lack of monophyly of genera and subgenera within Camallanidae, suggesting artificially on the current morphology-based systematics, as previously asserted (Wijová et al. 2006; Černotíková et al. 2011).
It is assumed that the systematics of Camallanidae is complicated and unresolved. As stated above, the current genetic knowledge and the morphological diagnosis based on buccal capsule structures are not enough to solve these problems. Therefore, the genetic database should be substantially improved. In this sense, the first genetic characterisation of P. (S.) huacraensis represents a contribution. Moreover, the morphological knowledge of the species was improved, and some insights on the phylogeny of Camallanidae came to light.
References
Anderson RC, Chabaud AG, Willmott S (2009) Keys to the nematode parasites of vertebrates: archival volume. CABI Publishing, Wallingford
Černotíková E, Horák A, Moravec F (2011) Phylogenetic relationships of some spirurine nematodes (Nematoda: Chromadorea: Rhabditida: Spirurina) parasitic in fishes inferred from SSU rRNA gene sequences. Folia Parasitol 58:135–148. https://doi.org/10.14411/fp.2011.013
Chang J-M, Di Tommaso P, Notredame C (2014) TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol Biol Evol 31:1625–1637. https://doi.org/10.1093/molbev/msu117
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth
Freitas JFT, Ibáñez HN (1968) Fauna helmintológica del Perú: nueva especie del género Spirocamallanus Olsen, 1952 (Nematoda, Camallanoidea). Bol Chil Parasitol 23:146–148
Froese R, Pauly D (Eds) (2019) FishBase. World Wide Web electronic publication. http://www.fishbase.org, version 02/2019. Accessed 22 Feb 2019
Gaither MR, Aeby G, Vignon M, Meguro Y, Rigby M, Runyon C, Toonen RJ, Wood CL, Bowen BW (2013) An invasive fish and the time-lagged spread of its parasite across the Hawaiian archipelago. PLoS One 8:e56940. https://doi.org/10.1371/journal.pone.0056940
Giese EG, Santos JN, Lanfredi RM (2009) A new species of Camallanidae from Ageneiosus ucayalensis (Pisces: Siluriformes) from Pará state, Brazil. J Parasitol 95:407–412. https://doi.org/10.1645/GE-1680.1
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. https://doi.org/10.1080/10635150390235520
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Kuzmin Y, Tkach VV, Snyder SD, Bell JA (2011) Camallanus Railliet et Henry, 1915 (Nematoda, Camallanidae) from Australian freshwater turtles with descriptions of two new species and molecular differentiation of known. Acta Parasitol 56:213–226. https://doi.org/10.2478/s11686-011-0015-0
Moravec F (1998) Nematodes of freshwater fishes of the Neotropical region. Academia, Praha
Moravec F, Chara J, Shinn AP (2004) Two nematodes, Dentinema trichomycteri n. g., n. sp. (Cosmocercidae) and Procamallanus chimusensis Freitas & Ibáñez, 1968 (Camallanidae), from catfishes Trichomycterus spp. (Pisces) in Colombia. Syst Parasitol 59:189–197. https://doi.org/10.1023/B:SYPA.0000048098.80098.26
Moravec F, Justine J-L, Würtz J, Taraschewski H, Sasal P (2006) A new species of Procamallanus (Nematoda: Camallanidae) from Pacific eels (Anguilla spp.). J Parasitol 92:130–137. https://doi.org/10.1645/GE-3509.1
Moravec F, Justine J-L (2017) Two new species of nematode parasites, Cucullanus epinepheli sp. n. (Cucullanidae) and Procamallanus (Spirocamallanus) sinespinis sp. n. (Camallanidae), from marine serranid and haemulid fishes off New Caledonia. Folia Parasitol 64(011). https://doi.org/10.14411/fp.2017.011
Moravec F, Sey O (1988) Nematodes of freshwater fishes from North Vietnam. Part 1. Camallanoidea and Habronematoidea. Acta Soc Zool Bohemoslov 52:128–148
Moravec F, Thatcher VE (1997) Procamallanus (Denticamallanus subgen. n.) dentatus n. sp. (Nematoda: Camallanidae) from the characid fish, Bryconops alburnoides, in the Brazilian Amazon. Parasite 4:239–243
Moravec F, Van As LL (2015) Procamallanus (Spirocamallanus) spp. (Nematoda: Camallanidae) from fishes of the Okavango River, Botswana, including P. (S.) serranochromis n. sp. parasitic in Serranochromis spp. (Cichlidae). Syst Parasitol 90:151–164. https://doi.org/10.1007/s11230-014-9542-z
Notredame C, Higgins DG, Heringa J (2000) T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. https://doi.org/10.1006/jmbi.2000.4042
Ramallo G (2008) Nueva especie de Procamallanus (Spirocamallanus) (Nematoda, Camallanidae), parásito de Trichomycterus corduvensis (Siluriformes: Trichomycteridae), en el Norte de Argentina. Acta Zool Lilloana 52:25–29
Ramallo G, Padilla Bortayro G (2011) Nemátodos parásitos de peces dulceacuícolas de la Provincia de Catamarca, Argentina. Acta Zool Lilloana 55:261–263
Rigby MC, Adamson ML (1997) Spirocamallanus species of French Polynesian coral reef fishes. Can J Zool 75:1270–1279. https://doi.org/10.1139/z97-150
Sardella CJ, Pereira FB, Luque JL (2017) Redescription and first genetic characterisation of Procamallanus (Spirocamallanus) macaensis Vicente & Santos, 1972 (Nematoda: Camallanidae), including re-evaluation of the species of Procamallanus (Spirocamallanus) from marine fishes off Brazil. Syst Parasitol 94:657–668. https://doi.org/10.1007/s11230-017-9728-2
Tantaleán VM, Franco AH, Huayta EH (1985) Helmintos parásitos de peces de agua dulce del Perú. Instituto de Investigaciones para el Desarrollo Social del Altiplano, Puno
Wijová M, Moravec F, Horák A, Lukeš J (2006) Evolutionary relationships of Spirurina (Nematoda: Chromadorea: Rhabditida) with special emphasis on dracunculoid nematodes inferred from SSU rRNA gene sequences. Int J Parasitol 36:1067–1075. https://doi.org/10.1016/j.ijpara.2006.04.005
Wu SG, Wang GT, Xi BW, Gao D, Nie P (2008) Molecular characteristics of Camallanus spp. (Spirurida: Camallanidae) in fishes from China based on its rDNA sequences. J Parasitol 94:731–736. https://doi.org/10.1645/GE-1219.1
Wu SG, Wang GT, Xi BW, Xiong F, Liu T, Nie P (2009) Population genetic structure of the parasitic nematode Camallanus cotti inferred from DNA sequences of ITS1 rDNA and the mitochondrial COI gene. Vet Parasitol 164:248–256. https://doi.org/10.1016/j.vetpar.2009.04.030
Acknowledgements
The authors would like to thank Dr. Elias Nogueira de Aguiar from the Laboratório Multiusuário de Análises de Materiais do Instituto de Física (MULTILAM-INFI), from the Universidade Federal de Mato Grosso do Sul (UFMS), for the help with scanning electron microscopical procedures, and Dr. Carina Elisei from the Universidade Católica Dom Bosco, for providing the facilities for molecular studies. Thanks are also due to the Secretaría de Medio Ambiente del Gobierno de la Provincia de Salta for allowing us to make the collection of ichthyology, and Florencia Liquín, José Saravia, Roberto Sanchez, Federico Soria and Facundo Leguizamon for their assistance in the field work.
Funding
This work was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES)-Finance code 001 and by the Consejo de Investigación de la Universidad Nacional de Salta (CIUNSa) (grant number 2193/3). F. B. P. was supported by a Post-doctoral fellowship PNPD-CAPES (Programa Nacional de Pós-Doutorado-Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-CAPES, Brazil). L. A. C. was supported by a Doctoral fellowship from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina); L. E. R. T. was supported by a researcher’s fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) (313292/2018-3).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Guillermo Salgado-Maldonado
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ailán-Choke, L.G., Davies, D.A., Tavares, L.E. et al. An integrative taxonomic assessment of Procamallanus (Spirocamallanus) huacraensis (Nematoda: Camallanidae), infecting the freshwater catfish Trichomycterus spegazzinii (Siluriformes: Trichomycteridae) in Argentina. Parasitol Res 118, 2819–2829 (2019). https://doi.org/10.1007/s00436-019-06429-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-019-06429-0