Abstract
Main conclusion
BrSOC1b may promote early flowering of Chinese cabbage by acting on BrAGL9 a, BrAGL9 b, BrAGL2 and BrAGL8 proteins.
Abstract
SOC1 is a flowering signal integrator that acts as a key regulator in controlling plant flowering time. This study focuses on the cloning of the open reading frame of SOC1b (BrSOC1b, Gene ID: Bra000393) gene, and analyzes its structure and phylogenetic relationships. Additionally, various techniques such as vector construction, transgenic technology, virus-induced gene silencing technology, and protein interaction technology were employed to investigate the function of the BrSOC1b gene and its interactions with other proteins. The results indicate that BrSOC1b consists of 642 bp and encodes 213 amino acids. It contains conserved domains such as the MADS domain, K (keratin-like) domain, and SOC1 box. The phylogenetic analysis reveals that BrSOC1b shares the closest homology with BjSOC1 from Brassica juncea. Tissue localization analysis demonstrates that BrSOC1b exhibits the highest expression in the stem during the seedling stage and the highest expression in flowers during the early stage of pod formation. Sub-cellular localization analysis reveals that BrSOC1b is localized in the nucleus and plasma membrane. Furthermore, through genetic transformation of the BrSOC1b gene, it was observed that Arabidopsis thaliana plants expressing BrSOC1b flowered earlier and bolted earlier than wild-type plants. Conversely, Chinese cabbage plants with silenced BrSOC1b exhibited delayed bolting and flowering compared to the control plants. These findings indicate that BrSOC1b promotes early flowering in Chinese cabbage. Yeast two-hybrid and quantitative real-time PCR (qRT-PCR) analyses suggest that BrSOC1b may participate in the regulation of flowering by interacting with BrAGL9a, BrAGL9b, BrAGL2, and BrAGL8 proteins. Overall, this research holds significant implications for the analysis of key genes involved in regulating bolting and flowering in Chinese cabbage, as well as for enhancing germplasm innovation in Chinese cabbage breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chinese cabbage (Brassica rapa ssp. pekinensis), belonging to the Cruciferae Brassica genus and native to China, is a popular vegetable species. It is often referred to as the “head vegetable” and holds a prominent position in the “vegetable basket.” Chinese cabbage has the largest cultivation area and is the most consumed vegetable in China (Liu 2016). However, during the cultivation process, Chinese cabbage is prone to early bolting and flowering due to variations in climate. Early bolting greatly affects both the yield and quality of the plants. Therefore, it is crucial to analyze the mechanisms behind the key genes regulating bolting and flowering in Chinese cabbage to improve its yield and germplasm innovation.
Six flowering signaling pathways have been identified, including the photoperiod pathway (Fernández et al. 2016), vernalization pathway (Zhu et al. 2015), gibberellin pathway (Galvao et al. 2015), autonomous pathway (Simpson 2004), thermosensitive pathway (Lievens et al. 2017), and age pathway (Vishal and Kumar 2018). These pathways sense and transmit flowering signals, and they converge on two key flowering integration factors, namely FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Yang et al. 2021). The SOC1 gene and its homologous genes belong to the class of transcription factors that encode MADS-box and play a crucial role in regulating plant flowering.
The MADS protein is characterized by a conserved 58–60 amino acid domain called the MADS domain, located at the N-terminus, which serves as the DNA binding domain. This domain binds to the CArG box (Yanofsky et al. 1990). Phylogenetic analysis divides the plant MADS gene family into Type I and Type II. Type II MADS genes consist of 95 identified genes, while Type I MADS genes consist of 65 confirmed genes, with Type II genes being the predominant group (Bemer 2008). Type I genes are further categorized into M-α, M-β, and M-γ, while Type II genes are divided into 13 subgroups: A, B, CD, E, AGL6, AGL12, TM3-like (SOC1), AGL17, SVP, AGL18/15, BS(TT16), FLC/MAF, and MIKC. Among these subgroups, TM3-like (SOC1) has the highest number of proteins (16), whereas AGL12, AGL6, and BS (TT16) have the fewest number of proteins (only 3 each) (Zhang 2021). MIKC genes are further divided into two subgroups: MIKCC and MIKC* (Henschel et al. 2002).
SOC1 is a flowering signal integrator that participates in the photoperiod pathway, autonomous pathway, vernalization pathway, and age pathway. It acts as a key regulator in controlling the flowering time of plants (Moon et al. 2003). Extensive research on the SOC1 gene has revealed its functions in different species. For instance, the GhSOC1 gene in Gerbera hybrida is expressed in leaves, leaf buds, inflorescences, stems, and roots of wild-type Arabidopsis thaliana in Colombia. Its expression level increases with plant growth, particularly in mature leaves where it is the highest (Lee et al. 2000). Additionally, the GhSOC1 gene influences flower development (Ruokolainen et al. 2011). Overexpression of MtSOC1a promotes flowering and main stem elongation in Medicago truncatula, while its mutants exhibit delayed flowering time and shortened main stem (Jaudal et al. 2018). Petunia hybrida Fbp20/UNS (FLORAL BINDING PROTEIN20/UNSHAVEN), a SOC1 homologous gene, when overexpressed, stimulates plant flowering (Ferrario et al. 2004). However, overexpression of Fbp20/UNS with missing fragments (specifically the MADS domain) results in delayed flowering of truncated plants (Ferrario et al. 2004). ZmMADS1, a SOC1/TM3-like gene, was cloned from Zea mays by Heuer et al. (2001). Its high expression during maize flower development suggests an important role in maize flowering. In Dendrobium officinale, overexpression of the DOSOC1 gene promotes early flowering (Ding et al. 2013). These findings highlight the crucial role of the SOC1 gene in regulating flowering.
Bolting and flowering are not only important agronomic traits in Chinese cabbage crop production but also crucial for the yield and quality of the plant organs, which directly impact human life. Based on the classification of Arabidopsis thaliana, the Chinese cabbage MADS family was categorized, and within a specific classification, there are 16 genes that share similarity with the Arabidopsis thaliana SOC1 gene (Duan et al. 2015). However, it has been discovered that the SOC1a (BrSOC1a, Gene ID: Bra000393) gene from Brassica rapa promotes flowering in Brassica oleracea (Hong et al. 2013). Whether the homologous gene BrSOC1b (gene ID: Bra004928) is involved in flowering regulation remains unclear. Therefore, this study aims to analyze the function of BrSOC1b in flowering and further explore its molecular mechanism. The results of this research carry significant implications for understanding the mechanisms of key genes involved in regulating bolting and flowering in Chinese cabbage and for enhancing germplasm innovation in Chinese cabbage.
Materials and methods
Cloning of BrSOC1b gene
To clone the nucleotide sequence of BrSOC1b, total RNA was extracted from the leaves of 'HK-8' using the RNA simple total RNA kit (Tiangen, Beijing). The RNA was then reverse transcribed into cDNA using the PrimeScriptRT reagent kit (TaKaRa, Dalian, China). Specific primers (Table S1) were designed based on the nucleotide sequence of BrSOC1b in the Chinese cabbage database (http://brassicadb.cn/#/), and PCR amplification was conducted using cDNA as the template. The amplified product was ligated to the pEASY-Blunt Cloning vector and transformed into E. coli (DH5α). Monoclonal, shaking, bacterial PCR detection, and sequencing were performed to confirm the correct cloning recombinant vector, which was named pEASY-Blunt-BrSOC1b.
Analysis of physical and chemical characteristics of BrSOC1b
The number of amino acids, theoretical isoelectric point (pI), relative molecular mass, protein stability, and hydrophobicity of BrSOC1b were analyzed using the ExPASy online software (https://web.expasy.org/cgi-bin/protparam/protparam). The secondary structure and transmembrane structure of the BraSOC1 protein were analyzed using SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) and DeepTMHMM Server v.2.0 (https://dtu.biolib.com/DeepTMHMM) (Hallgren et al. 2022), respectively. DNAMAN software (http://www.lynnon.com/) was utilized to align the cloned DNA fragment with the BrSOC1b fragment in the database. The conserved domain and motif of BrSOC1b were analyzed using SMART software (http://smart.embl-heidelberg.de/) (Letunic et al. 2012) and MEME online software (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) (Bailey et al. 2006), respectively. To determine the subcellular localization of BrSOC1b, we employed Plant-mPLoc (Chou and Shen 2010) for protein prediction.
Phylogenetic analysis of BrSOC1b
The phylogenetic relationship between BrSOC1b and SOC1 in other species was analyzed using the Maximum Likelihood method with MEGA5.0 software (Tamura et al. 2011). The protein sequences of OsSOC1, TaSOC1, and GmSOC1 were obtained from previous reports (Lee et al. 2004; Shitsukawa et al. 2007; Zhong et al. 2012). The sequences of AtSOC1, BjSOC1, and BnSOC1 were retrieved from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). The abbreviations and accession numbers of the protein sequences used are provided in Table S2.
Analysis of tissue expression pattern of BrSOC1b
To analyze the tissue expression pattern of BrSOC1b, the root, stem and leaf of Chinese cabbage 'HK-8' cultured normally were taken as samples. In the early stage of podding, the root, stem, stem old leaves, stem young leaves, flowers, pods and buds were taken as samples to analyze the tissue expression pattern of BrSOC1b. Each tissue selected three biological replicates.
Construction of BrSOC1b-heterologous expression vector and its genetic transformation in Arabidopsis thaliana
To construct the BrSOC1b-heterologous expression vector, specific primers containing Kpn I and Spe I restriction sites were designed (Table S1). Firstly, the cDNA fragment containing Kpn I and Spe I restriction sites was cloned from pEASY-Blunt-BrSOC1b. Secondly, the amplified fragment was ligated to the PTCK303 vector digested by Kpn I-Spe I using T4 ligase and then transformed into E. coli (DH5α). Monoclonal shaking, bacterial PCR detection, and sequencing were performed. The correct recombinant vector was named pTCK303-BrSOC1b (Fig. S4).
The vector pTCK303-BrSCO1b was transformed into Agrobacterium GV3101 using the freeze–thaw method (Yu et al. 2003). It was then plated on LB plates containing Kanamycin and incubated in an inverted position in a 28℃ dark incubator for 2–3 days. Randomly selected single colonies were subjected to PCR, and the colonies with correct bands were selected for further use. Agrobacterium containing pTCK303-BrSCO1b was inoculated into LB liquid medium containing Kanamycin and Rifampicin antibiotics, and cultured at 28 °C and 250 rpm until the OD600 reached approximately 1.0. After centrifugation at 6,000 rpm for 5 min at 20 °C, the cells were collected. The cells were then evenly resuspended in a solution containing 5% sucrose (with 0.02% silwet L-77) to achieve an OD600 of approximately 1.0. The well-grown and flowering Arabidopsis thaliana plants were watered one day in advance. The small pots containing Arabidopsis thaliana plants were inverted the next day, ensuring that all inflorescences were immersed in the suspension buffer for approximately 50 s. After 7 days, the transformation process was repeated using the same method. After 2–3 weeks, watering with nutrient solution was minimized, and the plants were subjected to accelerated aging until mature seeds were obtained. Subsequently, the T3 homozygous lines were obtained through multi-generation selfing and single plant seed collection.
Positive identification of BrSOC1b-heterologous expression Arabidopsis thaliana plants
Considering that the pTCK303 vector carries hygromycin resistance genes, we performed DNA detection of the hygromycin resistance genes in BrSOC1b-infected Arabidopsis thaliana plants. Furthermore, since the pTCK303 vector also contains a GUS reporter gene, GUS histochemical staining was employed to identify BrSOC1b-heterologous expression Arabidopsis thaliana plants. Additionally, RNA was extracted from the leaves of transgenic and wild-type Arabidopsis thaliana plants, and the relative expression of BrSOC1b was determined using reverse transcription and quantitative real-time PCR (qRT-PCR) technology.
Construction of Brsoc1b-silenced vector and its genetic transformation in Chinese cabbage
For the virus-induced gene silencing (VIGS) experiments, a suitable 40 bp sequence and its reverse complementary sequence were selected from BrSOC1b: 5'-ATAGGATCATGTTCGATAGAGGAGCTGCAGCAAATTGAGCGCTCAATTTGCTGCAGCTCCTCTATCGAACATGATCCTAT-3'. These sequences were synthesized, and the resulting 80 bp sequence was phosphorylated. The vector pTY was digested with the SnaB I restriction enzyme and subsequently dephosphorylated. The phosphorylated DNA fragment and the dephosphorylated pTY vector were ligated using T4 ligase. The ligated construct was then transformed into DH5α, followed by screening and sequencing, leading to the identification of positive clones. Following the principles of virus-induced gene silencing, Brsoc1b-silenced Chinese cabbage plants were obtained through friction inoculation (Yu et al. 2018) and designated as pTY-Brsoc1b (Fig. S5).
Statistics of flowering related indexes of transgenic plants
According to the method of Chen et al. (2012), the days required for bolting and initial flowering of transgenic plants and control plants were counted.
Construction and transient transformation of BrSOC1b-subcellular localization vector
To investigate the subcellular localization of the BrSOC1b protein, an open reading frame fragment of BrSOC1b (without the stop codon) containing the SalI-KpnI restriction site was cloned using pEASY-Blunt-BrSOC1b as a template. The amplified fragment was then ligated to the PRI101-GFP vector, which had been digested with SalI-KpnI, using T4 ligase. The resulting construct was transformed into E. coli (DH5α) for monoclonal selection, followed by bacterial PCR detection and sequencing. The correctly recombined vector was designated as PRI101-GFP-BrSOC1b.
The generated recombinant vector PRI101-GFP-BrSOC1b (Fig. S6) and the empty PRI101-GFP vector were separately transformed into Agrobacterium GV3101 using the freeze–thaw method (Dupadahalli 2007). Subsequently, the Agrobacterium strains were infiltrated into tobacco leaves using the injection infiltration method (Li and Zhang 2010). The PRI101-GFP vector was used as a control in the experiment. After 40 h of dark incubation, the subcellular localization of the BrSOC1b protein was analyzed using laser confocal microscopy.
Verification of the interaction between BrSOC1b and the target proteins
To clarify the network relationship of BrSOC1b in flowering regulation, we utilized the STRING online tool (https://www.string-db.org) to predict the protein interaction network of BrSOC1b. To validate the interaction between BrSOC1b and the target proteins, we constructed the pGBKT7-BrSOC1b bait vector and the pGADT7-target protein vectors. Firstly, we verified the self-activation of the BrSOC1b protein, and subsequently confirmed the interaction between BrSOC1b and the target proteins.
-
a.
Construction of yeast two-hybrid vector
To construct pGBKT7-BrSOC1b bait vector, the cDNA fragmenof of BrSOC1b containing Nco I / BamH I restriction sites (without the stop codon) was cloned using pEASY-Blunt-BrSOC1b as template and specific primers containing Nco I / BamH I restriction sites. The amplified fragment was ligated to the pGBKT7 vector digested by Nco I / BamH I using T4 ligase. The target protein gene was inserted into the EcoR I / Xho I restriction site of pGADT7 using the same method, and the vector was named pGADT7-target protein vector. The information of proteins interacting with BrSOC1b in this experiment is list in Table S3.
-
b.
Co-transformation and interaction verification of bait vector and target protein
The pGBKT7-BrSOC1b and pGADT7-target proteins were co-transformed into yeast strain Y2HGold by Yeastmaker yeast transformation system 2 (Cat. No.630439), and then cultured on SD / -Trp / -Leu medium. Monoclonal cells normally grown on SD / -Trp / -Leu solid medium were inoculated into 5 mL SD / -Trp / -Leu liquid medium and cultured at 30 ℃ until OD600 was 1.0. The growth of yeast was observed after 3–5 days on SD / -Trp / -Leu / -His / -Ade / + X-α-Gal solid medium.
RNA extraction, reverse transcription and qRT-PCR analysis
Total RNA was extracted from leaf samples using RNA simple Total RNA Kit (Tiangen, Beijing), and RNA was reverse transcribed into cDNA using Prime Script RT reagent kit (TaKaRa, Dalian, China). Finally, qRT-PCR was performed using the SYBR Premix ExTaq kit (TaKaRa, Dalian, Beijing). The total PCR system was 20 μL, including 10 μL 2 × SYBR® Premix Ex Taq™ II (TaKaRa), 6.8 μL ddH2O, 0.4 μL ROX II, 0.8 μL primers and 2.0 μL cDNA template. PCR conditions were as follows: stage 1, 95 °C 20 s; stage 2, 95 °C 3 s; 60 °C 30 s (40 cycles); stage 3, 95 °C 15 s; 60 °C 1 min; 95 °C 15 s. Stage 3 was used to create a melting curve. The results were calculated according to the 2−ΔΔCt method (Livak and Schmittgen 2001) and plotted using Origin.
Statistical analysis
Data were collected from a minimum of three biological replicates and analyzed using Student's t-test or one-way ANOVA followed by post hoc Duncan's test using SPSS software (version 20, Chicago, IL, USA). The presented data represent the mean of three replicates, and the error bars indicate the standard deviation. Different letters above the columns indicate significant differences between different tissues or lines (P < 0.05).
Results and analysis
Cloning and bioinformatics analysis of the BrSOC1b genes
By cloning the nucleotide sequence of BrSOC1b, we found that BrSOC1b contains 642 bp (Fig. S1). The nucleotide sequence of the cloned BrSOC1b was aligned with the nucleotide sequence of the gene in the Chinese cabbage database, and the sequence identity was 100.00%. BrSOC1b was analyzed by ExPASy online software (https://web.expasy.org/cgi-bin/protparam/protparam). The results showed that BrSOC1b encoded 213 amino acids (Fig. 1a), the theoretical isoelectric point (pI) was 9.16, and the relative molecular mass was 24 399.92. The protein belongs to unstable protein (instability index is 59.72) and hydrophobic protein (hydrophilic average index is − 0.814). SOPMA analysis showed that BrSOC1b protein was composed of 56.34% α-helix, 26.76% random coil, 11.27% extended strand and 5.63% β-turn (Fig. 1b). In addition, the transmembrane structure of BrSOC1b protein was predicted by DeepTMHMM Server v.2.0, and it was found that the protein belonged to the non-trans-membrane proteins protein (Fig. 1c). The conserved domain of BrSOC1b was analyzed by SMART online software. The results showed that BrSOC1b protein contained conserved MADS domain and K domain (Fig. 1a). Finally, the subcellular localization of BrSOC1b protein was predicted by Plant-mPLoc, and it was speculated that BrSOC1b may be localized in the nucleus.
Bioinformatics analysis of BrSOC1b. a The predicted amino acid sequence and conserved domain prediction of BrSOC1b. MADS, MADS domain, K-box, K domain. b The secondary structure of the BrSOC1b. Helix, represents α-helix; Sheet, represents extended strand; Turn, represents β-turn; Coil, represents random coil. c The transmembrane structure prediction of BrSOC1b
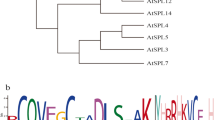
Amino acid sequence alignment and phylogenetic tree analysis of BrSOC1b and SOC1 of other species
By comparing the amino acid sequence of BrSOC1b with SOC1 protein sequences from other species, it was observed that BrSOC1b and SOC1 proteins of other species shared the same domains, namely the MADS domain, K (keratin-like) domain, and SOC1 box at the C-terminus (Fig. 2a). To further investigate the evolutionary relationship between BrSOC1b and SOC1 proteins from Oryza sativa, Triticum aestivum, Hordeum vulgare, Arabidopsis thaliana, Brassica juncea, and Brassica napus, phylogenetic analysis was performed using MEGA5.0 software. The results revealed that BrSOC1b exhibited the closest homology with BjSOC1b from Brassica juncea, followed by BnSOC1b from Brassica napus. Additionally, BrSOC1b displayed relatively close homology with BrSOC1a (Fig. 2b). The evolutionary branch indicates that BrSOC1b is relatively distant from OsSOC1, TaSOC1, and GmSOC1.
Amino acid sequence alignment and phylogenetic relationship between BrSOC1b and SOC1 proteins of other species. a Amino acid sequence and conserved domain analysis of BrSOC1b protein. Identical and conserved residues are highlighted with the same color background color. The corresponding conserved domains are marked by thick black lines. b A phylogenetic tree of BrSOC1b and SOC1 proteins from other species. The phylogenetic tree was constructed by MEGA5.0 software with the method of Maximum Likelihood. Bootstrap values higher than 89% are shown on the nodes
Tissue expression pattern analysis of BrSOC1b gene
To explore the expression of BrSOC1b gene in various tissues, we explored it at seedling stage and early podding stage. Firstly, the relative expression of BrSOC1b in roots, stems and leaves was determined at seedling stage. The results showed that BrSOC1b had the highest expression in stem, which was 77.12 times higher than that in root, followed by the expression in leaf, which was 45.21 times higher than that in root (Fig. 3a).
Expression profiles of BrSOC1b in various tissues. a Expression profile of BrSOC1b in root, stem and leaf at seedling stage. b Expression profiles of BrSOC1b in the root, stem, stem born old leave, stem born young leave, flower, pod, and bud. The data represent the mean of three biological replicates and three technical replicates, and the error line represents the standard deviation of the mean. The gene expression was carried out by 2−ΔΔCt method, and then the expression of BrSOC1b in the root was corrected to 1. BrActin (Bra028615) was as reference gene. Different letters above the column indicated significant differences between different tissues (P < 0.05)
Furthermore, we studied the relative expression of BrSOC1b in roots, stems, old leaves, young leaves, flowers, pods and buds at the early stage of pod formation. The results showed that BrSOC1b had the highest expression level in flowers, which was 11.56 times that of roots (Fig. 3b). Secondly, it is also expressed in stems, stem born old leaves and stem born young leaves. The lowest expression level was in the bud, which was 0.27 times that of the root (Fig. 3b). Based on the above experimental results, we speculate that BrSOC1b may be related to the growth and development of stems and leaves at the seedling stage, and is mainly expressed in flowers at the early stage of pod formation. Therefore, we speculate that BrSOC1b may play a key role in flower development.
Sub-cellular localization analysis of BrSOC1b
To explore the subcellular localization of BrSOC1b protein, we transiently expressed BrSOC1b gene in tobacco. The results showed that PRI101-GFP, as a control, emitted green light in the whole cell of tobacco, while GFP, which formed a fusion protein with BrSOC1b, emitted light in the nucleus and cell membrane, and the green fluorescence on the membrane coincided with the red fluorescence of the membrane tag mCherry (Fig. 4). Therefore, we speculated that BrSOC1b was localized in the nucleus and plasma membrane.
Sub-cellular localization analysis of BrSOC1b in Nicotiana benthamiana leaf cells. mCherry, membrane marker; pm-rbCD3-1008; GFP, dark field image of tobacco leaf cells; Bright field, bright field image of tobacco leaf cells; Merged, the superposition of red fluorescence image, green fluorescence image and bright field; Scale: 20 μm
Positive identification of BrSOC1b-heterologous expression Arabidopsis thaliana plants
To investigate the function of BrSOC1b, we conducted heterologous expression of BrSOC1b to generate transgenic Arabidopsis thaliana plants. To identify positive seedlings of BrSOC1b-heterologous expression Arabidopsis thaliana plants, we initially performed hygromycin resistance gene detection using DNA extracted from transgenic Arabidopsis thaliana leaves. The results indicated the presence of hygromycin resistance genes in 11 T1 plants (Fig. 5a). Through successive selfing and individual plant selection, we obtained T3 generation plants.
Positive identification of BrSOC1b-heterologous expression Arabidopsis thaliana plants. a DNA analysis of hygromycin resistance genes in BrSOC1b-heterologous expression Arabidopsis thaliana. Note: Column 1 is shown as a positive control. Column 2 is 1000 marker, 100, 250, 500, 750 and 1000 bp from bottom to top. Column 3 is negative control. Column 4 and 5 are transgenic seedling detection samples. b β-glucuronidase (GUS) histochemical staining of BrSOC1b-heterologous expression Arabidopsis thaliana lines. c The qRT-PCR analysis of BrSOC1b in the leaves of BrSOC1b-heterologous expression Arabidopsis thaliana lines. The gene expression was carried out by 2−ΔΔCt method, and then the expression of BrSOC1b in the root was corrected to 1. The eIF4Actin gene was used as an internal control. The data represents the average of three replicates, and the error line represents the standard deviation of the average. Different letters above the column indicated significant differences between different tissues (P < 0.05)
In the T3 generation, we employed the GUS (β-D-glucuronidase) histochemical staining method to verify partial staining in the leaves of transgenic Arabidopsis thaliana under a stereomicroscope, whereas no staining was observed in the leaves of wild-type Arabidopsis thaliana (Fig. 5b). Subsequently, we randomly examined the relative expression levels of BrSOC1b in two transgenic lines (OE1 and OE4) as well as wild-type plants using qRT-PCR. The results revealed that the relative expression level of BrSOC1b in wild-type plants was very low, while in transgenic plants, it was 10.09–12.57 times higher than that in wild-type plants (Fig. 5c). Based on this analysis, we selected the transgenic lines OE1 and OE4 for further phenotypic observation and statistical analysis.
Phenotypic observation and flowering time statistics of BrSOC1b-heterologous expression Arabidopsis thaliana plants
To investigate the impact of BrSOC1b on plant flowering, we utilized two lines of transgenic plants (OE1 and OE4) along with wild-type plants as the experimental materials. We observed the phenotypes of these plants and recorded the bolting time and flowering time. The results revealed that the bolting time and flowering time of the transgenic plants were significantly earlier compared to those of the wild-type plants (Fig. 6a, b). Specifically, the bolting time of BrSOC1b-overexpressing Arabidopsis thaliana plants occurred 7 to 11 days earlier than that of the wild type (Fig. 6c). Moreover, the flowering time of BrSOC1b-heterologous expression Arabidopsis thaliana plants was 6 to 7 days earlier than that of the wild type (Fig. 6d).
Phenotypic observation and related data statistics of BrSOC1b-heterologous expression Arabidopsis thaliana plants and wild-type plants. a The phenotype of BrSOC1b-heterologous expression Arabidopsis thaliana plants and wild-type plants at bolting stage. b Phenotypes of BrSOC1b-heterologous expression Arabidopsis thaliana plants and wild-type plants at flowering stage. c Bolting time statistics of BrSOC1b-heterologous expression Arabidopsis thaliana plants and wild-type plants. d Flowering time statistics of BrSOC1b-heterologous expression Arabidopsis thaliana plants and wild-type plants. We take the initial flowering period as the flowering time. Each line and wild-type plants were counted 8 plants. The data represents the average, and the error line represents the standard deviation of the average. Different letters above the column indicated that there were significant differences between different lines (P < 0.05)
BrSOC1b protein interaction network prediction and the self-activation detection of BrSOC1b
To clarify the interaction relationship of BrSOC1b protein, we used STRING online tool to predict the interaction network diagram of BrSOC1b protein (Fig. S2). The interaction diagram showed that BrSOC1b interacted with BrAP1 (Bra038326), BrAGL9a / SEP3 (Bra010955), BrAGL9b / SEP3 (Bra030032), BrAGL2 / SEP1 (Bra008674), FUL / BrAGL8 (Bra029347) (Fig. S2). Therefore, I speculate that BrSOC1b may interact with the above proteins.
To verify the interaction between BrSOC1b and the above proteins, the self-activation of BrSOC1b protein was first verified. The pGBKT7-BrSOC1b and pGADT7 were co-transformed into the yeast strain Y2HGold as the experimental group. The negative control was the co-transformation of pGBKT7 and pGADT7 in the yeast strain, and the positive control was the co-transformation of pGBKT7-53 and pGADT7-T in the yeast strain. The positive control, negative control and experimental groups were cultured on SD / -Trp / -Leu and SD / -Trp / -Leu / -His / -Ade / + X-α-Gal solid medium, respectively. As shown in Fig. S3, these three groups can grow normally on SD / -Trp / -Leu, and the positive control can grow normally and show blue on SD / -Trp / -Leu / -His / -Ade / + X-α-Gal solid medium. However, negative control and experimental groups in SD / -Trp / -Leu / -His / -Ade / + X-α-Gal could not grow normally (Fig. S3). Based on the above analysis, we speculate that there is no self-activation of BrSOC1b protein in yeast.
Verification of the interaction between BrSOC1b and predicted interacting proteins
To explore the interaction between BrSOC1b and BrAP1, BrAGL9a, BrAGL9b, BrAGL2, BrAGL8 proteins, we used BrSOC1b as a bait protein and verified by yeast two-hybrid technology. PGBKT7-BrSOC1b + pGADT7-BrAP1, pGBKT7-BrSOC1b + pGADT7-BrAGL9a, pGBKT7-BrSOC1b + pGADT7-BrAGL9b, pGBKT7-BrSOC1b + pGADT7-BrAGL8, pGBKT7-BrSOC1b + pGADT7-BrAGL2 were coated on SD / -Trp / -Leu and SD / -Trp / -Leu / -His / -Ade / + X-α-Gal plates, respectively. After 3 days of culture, it was found that all combinations could grow on SD / -Trp / -Leu medium (Fig. 7a). In addition, the experimental groups pGBKT7-BrSOC1b + pGADT7-BrAGL9a, pGBKT7-BrSOC1b + pGADT7-BrAGL9b, pGBKT7-BrSOC1b + pGADT7-BrAGL8 and pGBKT7-BrSOC1b + pGADT7-BrAGL2 could grow and turn blue on SD / -Trp / -Leu / -His / -Ade / + X-α-Gal medium. However, the experimental group pGBKT7-BrSOC1b + pGADT7-BrAP1 could not grow normally. Based on the above experimental results, we concluded that BrSOC1b interacts with BrAGL9a, BrAGL9b, BrAGL2, BrAGL8 proteins, but not with BrAP1 (Fig. 7b). Considering that BrAGL9a, BrAGL9b, BrAGL2 and BrAGL8 proteins all belong to MADS transcription factors, this family plays a key role in regulating plant flowering pathways, so we speculate that BrSOC1b protein may interact with the above proteins to regulate flowering.
Verification of the interaction between BrSOC1b and BrAP1, BrAGL9a, BrAGL9b, BrAGL2, BrAGL8 proteins. The pGBKT7-BrSOC1b bait vector and the target vector were co-transformed into the yeast strain Y2HGold, and then detected on SD / -Trp / -Leu (a) and SD / -Trp / -Leu / -His / -Ade / + X-α-Gal (b) plates
Phenotypic observation of Brsoc1b-silenced Chinese cabbage plants
The leaves of Brsoc1b-silenced Chinese cabbage plants exhibited significant chlorosis compared to the wild-type (WT) plants, 20 days after virus inoculation. They displayed typical symptoms of turnip yellow mosaic virus (Fig. 8a). A total of 20 pTY-Brsoc1b plants and 10 pTY control plants were obtained. The Brsoc1b-silenced Chinese cabbage plants, as well as the control (pTY) plants, were further cultivated, and it was observed that the Brsoc1b-silenced Chinese cabbage plants exhibited delayed bolting and flowering compared to the control plants (Fig. 8b, c). The expression of the BrSOC1b gene was assessed by qRT-PCR when Chinese cabbage plants were close to bolting. The results demonstrated that the relative expression of BrSOC1b in pTY-Brsoc1b plants was approximately 0.65–0.78 times that of pTY plants (Fig. 9a). Consequently, we hypothesized that BrSOC1b was partially silenced in Chinese cabbage, and this gene played a positive role in regulating the bolting time and flowering time of Chinese cabbage.
Phenotypic analysis of Brsoc1b-silenced Chinese cabbage plants and control plants. a Local phenotypes of Brsoc1b-silenced Chinese cabbage pTY-Brsoc1b and control pTY leaves. b Phenotype of Brsoc1b-silenced Chinese cabbage plants and control plants at bolting stage. c The phenotype of Brsoc1b-silenced Chinese cabbage plants and control plants at the initial flowering stage
Expression analysis of genes interacting with BrSOC1b in pTY-BrSOC1b plants and pTY plants. BrActin (Bra028615) gene was used as an internal control. The gene expression was carried out by 2−ΔΔCt method. The data represents the average of three replicates, and the error line represents the standard deviation of the average. Different letters above the column indicated significant differences between different lines (P < 0.05)
Expression analysis of genes interacting with BrSOC1b in pTY-Brsoc1b plants and pTY plants
Three pTY-Brsoc1b plants and three pTY control plants were selected for further analysis. The relative expression profiles of BrAGL9a, BrAGL9b, BrAGL2, BrAGL8, and BrAP1 genes were measured in the pTY-Brsoc1b plants and control plants. It was observed that when the BrSOC1b gene was partially silenced, the expression of BrAGL9a, BrAGL9b, BrAGL2, and BrAGL8 genes was significantly down-regulated, except for the BrAP1 gene (Fig. 9). For instance, in pTY-Brsoc1b plants, the expression of BrAGL9a was approximately 0.38–0.45 times that of pTY plants (Fig. 9c), and the expression of BrAGL2 was around 0.65–0.76 times that of pTY plants (Fig. 9f). Taking into account the results of the protein interaction experiments, it is speculated that BrAGL9a, BrAGL9b, BrAGL2, and BrAGL8 genes are down-regulated genes of the BrSOC1b gene, which may play a pivotal role in flowering regulation.
Discussion
Flowering time is a critical period for plant reproduction, and the regulation of flowering holds great significance for plant production and breeding. The SOC1 gene acts as an integrator that combines multiple flowering signals. Through the analysis of the structure of BrSOC1b in Chinese cabbage, it was observed that the BrSOC1b protein contains a SOC1 box, which is unique to the TM3-like branch of the MADS family gene. This suggests that BrSOC1b belongs to the TM3-like branch gene (Fig. 2a). Furthermore, the transmembrane structure prediction of BrSOC1b protein revealed that it is a non-transmembrane protein (Fig. 1c). Additionally, the subcellular localization prediction of BrSOC1b protein indicated that it localizes to the nucleus. However, when BrSOC1b was transiently expressed in tobacco cells, it was observed to localize to both the nucleus and plasma membrane (Fig. 4). Based on these findings, it can be speculated that BrSOC1b may exhibit spatiotemporal differential expression characteristics.
BrSOC1b promotes early flowering in Arabidopsis thaliana, and the phylogenetic relationship suggests that BrSOC1b has the closest homology with BjSOC1b. This indicates that BjSOC1b may also be involved in flowering regulation in mustard. However, further experiments are required to confirm this speculation. Moreover, considering that Oryza sativa, Triticum aestivum, and Hordeum vulgare are monocots, while Arabidopsis thaliana, Brassica juncea, and Brassica napus are dicots, it can be observed that the homologous relationship between BrSOC1b and SOC1 proteins in dicots is closer than that in monocots. This finding is consistent with the evolutionary characteristics of organisms.
Currently, researchers have conducted extensive investigations into the function of the SOC1 gene. The Arabidopsis thaliana SOC1 (AtSOC1) gene has been found to regulate flowering time and floral tube development (Zhao et al. 2014). Overexpression of the SOC1 gene promotes early flowering in alfalfa (Mauren et al. 2018). In Bambusa oldhamii, SOC1 exhibits the highest expression in flower buds and the lowest expression in stems (Onouchi et al. 2000). Additionally, CsSOC1a was found to accumulate the most in mature leaves and also promoted flowering induction in Gannan early navel orange trees (He et al. 2022). BrSOC1b displays high expression levels in stems and leaves during the seedling stage. Furthermore, BrSOC1b is highly expressed in flowers during the early podding stage (Fig. 3), suggesting a potential key role for BrSOC1b in flower development.
Through phenotypic analysis of BrSOC1b-heterologous expression in Arabidopsis thaliana plants and wild-type Arabidopsis thaliana plants, it was discovered that heterologous expression of BrSOC1b promotes flowering in Arabidopsis thaliana. Conversely, partial silencing of BrSOC1b results in delayed flowering in Chinese cabbage. These results indicate that BrSOC1b plays a crucial role in the regulation of flowering in Chinese cabbage. While this study reveals that BrSOC1b regulates the flowering time of plants, the molecular mechanism underlying SOC1's regulation of plant flowering remains unclear (Xian et al. 2013). Current research has mainly focused on the regulation of SOC1 in the shoot apical meristem to control the process of flowering induction in plants. However, it is still necessary to verify whether SOC1 in leaves can translocate to the shoot apex, warranting further exploration of the molecular mechanism of SOC1.
Proteins serve as the executors of function, and protein interactions are the primary means through which functions are performed. The yeast two-hybrid experiment revealed interactions between BrSOC1b and BrAGL9a, BrAGL9b, BrAGL8b, and BrAGL2b. The AtAGL9 gene was initially cloned in Arabidopsis thaliana, and its close association with flower development was observed (Mandel and Yanofsky 1998; Yang et al. 2019). Based on this, we speculate that BrSOC1b may regulate plant flower development by interacting with BrAGL9a and BrAGL9b, potentially affecting flowering time regulation in plants. Additionally, several studies have reported on the AP1 gene. It was discovered that the apple AP1 gene (MdAP1) and petunia AP1 gene promote early flowering in transgenic Arabidopsis thaliana plants (Kotoda et al. 2002; An et al. 2001). Heterologous expression of Arabidopsis thaliana AP1 in tomato shortens the vegetative cycle without affecting plant production (Ellul et al. 2004). In this study, yeast two-hybrid assays did not indicate an interaction between BrSOC1b and BrAP1. However, qRT-PCR analysis demonstrated an increase in BrAP1 expression after partial silencing of BrSOC1b (Fig. 9b), suggesting that the BrSOC1b gene may indirectly influence the expression of BrAP1. Further verification is necessary to determine if BrAP1 is involved in flowering regulation. These results provide a foundation for further investigation into the mechanisms underlying BrSOC1b's involvement in flowering regulation.
Conclusion
Based on the aforementioned studies, BrSOC1b consists of 642 base pairs, encoding 213 amino acids. It contains a conserved MADS domain, K (keratin-like) domain, and SOC1 box. The BrSOC1b protein is localized in the nucleus and plasma membrane, with specific expression primarily observed in Chinese cabbage flowers. BrSOC1b promotes early flowering in plants and may regulate the flowering time of Chinese cabbage by interacting with BrAGL9a, BrAGL9b, BrAGL2, and BrAGL8 proteins. This study lays the groundwork for the comprehensive analysis of the molecular mechanisms underlying the regulation of Chinese cabbage flowering by the BrSOC1b gene. Furthermore, it holds significant implications for the genetic enhancement of Chinese cabbage germplasm resources in future breeding efforts.
Author contribution statement
JY (Jingping Yuan) and CS conceived and designed the experiments, BS, RC and DL performed the experiments. XG, CW, NK and BC performed the qRT-PCR. JY, CS and XL wrote the manuscript. All authors read and approved the final manuscript.
Data availability
No data was used for the research described in the article.
References
An L, Liu R, Chen Z, Li Y (2001) Studies on Petunia hybirda transformed with flower-meristem-identity gene AP1. Acta Bot Sin 43(1):63–66. https://doi.org/10.3321/j.issn:1672-9072.2001.01.012(inchinese)
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:369–373. https://doi.org/10.1093/nar/gkl198
Bemer M (2008) Functional and evolutionary studies of type I MADS box genes in Petunia hybrida and Arabidopsis thaliana. Nucl Phys B-proc Sup
Chen L, Qu X, Hou B, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Sci 335(6065):207–211. https://doi.org/10.1126/science.1213351
Chou K, Shen H (2010) Plant-mploc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE. https://doi.org/10.1371/journal.pone.0011335
Ding L, Wang Y, Yu H (2013) Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid dendrobium chao parya smile. Plant Cell Physiol 54(4):595–608. https://doi.org/10.1093/pcp/pct026
Duan W, Song X, Liu T, Huang Z, Li Y (2015) Genome-wide analysis of the mads-box gene family in Brassica rapa (Chinese cabbage). Mol Genet Genom 290(1):239–255. https://doi.org/10.1007/s00438-014-0912-7
Dupadahalli K (2007) A modified freeze-thaw method for the efficient transformation of Agrobacterium tumefaciens. Curr Sci 93(6):770. http://www.jstor.org/stable/24099118
Ellul P, Angosto T, Garcia-Sogo B, Garcia-Hurtado N, Martin-Trillo M, Salinas M, Moreno V, Lozano R, Martinez-Zapater M (2004) Expression of Arabidopsis APETALA1 in tomato reduces its vegetative cycle without affecting plant production. Mol Breed 13(2):155–163. https://doi.org/10.1023/B:MOLB.0000018763.64585.6b
Fernández V, Takahashi Y, Le Gourrierec J, Coupland G (2016) Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J 86(5):426–440. https://doi.org/10.1111/tpj.13183
Ferrario S, Busscher J, Franken J, Gerats T, Vandenbussche M, Angenent GC, Immink RGH (2004) Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 16(6):1490–1505. https://doi.org/10.1105/tpc.019679
Galvao VC, Collani S, Horrer D, Schmid M (2015) Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J 84(5):949–962. https://doi.org/10.1111/tpj.13051
Hallgren J, Tsirigos KD, Pedersen MD, Almagro Armenteros JJ, Marcatili P, Nielsen H, Krogh A, Winther O (2022) DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv. https://doi.org/10.1101/2022.04.08.487609
He X, Yang J, Gao Q, Tong X, Zhang X, Li X (2022) Cloning of navel orange SOC1 gene and its expression analysis during flower development. Mol Plant Breeding 20(15):4948–4957. https://doi.org/10.13271/j.mpb.020.004948 (in Chinese)
Henschel K, Kofuji R, Hasebe M, Saedler H, Munster T, Theissen G (2002) Two ancient classes of MIKC-type MADS-box genes are Present in the Moss Physcomitrella patens. Mol Biol Evol 19(6):801–814. https://doi.org/10.1093/oxfordjournals.molbev.a004137
Heuer S, Hansen S, Bantin J, Brettschneider R, Kranz E, Lorz H, Dresselhaus T (2001) The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiol 127(1):33–45. https://doi.org/10.1104/pp.127.1.33
Hong JK, Kim SY, Kim KS, Kwon SJ, Kim JS, Kim JA, Lee SI, Lee YH (2013) Overexpression of a Brassica rapa mads-box gene, BrAGL20, induces early flowering time phenotypes in Brassica napus. Plant Biotechnol Rep 7(3):231–237. https://doi.org/10.1007/s11816-012-0254-z
Jaudal M, Zhang L, Che C, Li G, Tang Y, Wen J, Mysore KS, Putterill J (2018) A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J Exp Bot 69(20):4867–4880. https://doi.org/10.1093/jxb/ery284
Kotoda N, Wada M, Kusaba S, Kano-Murakami Y, Masuda T, Soejima J (2002) Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci 162(5):679–687. https://doi.org/10.1016/S0168-9452(02)00024-9
Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I (2000) The AGAMOUS-like 20 MADS domain protein integrates floral inductive path ways in Arabidopsis. Gene Dev 14(18):2366–2376. https://doi.org/10.1101/gad.813600
Lee S, Kim J, Han J, Han M, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO1/ AGAMOUS-LIKE 20 (SOC1/ AGL20) ortholog in rice. Plant J 38(5):754–764. https://doi.org/10.1111/j.1365-313X.2004.02082.x
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40(1):302–305. https://doi.org/10.1093/nar/gkr931
Li Y, Zhang Y (2010) Experimental study on transformation of tobacco by agrobacterium injection infiltration. Exp Technol Manag 27(11):3. https://doi.org/10.3969/j.issn.1002-4956.2010.11.015(inChinese)
Lievens L, Pollier J, Goossens A, Beyaert R, Staal J (2017) Abscisic acidas pathogen effector and immune regulator. Front Plant Sci 8:587. https://doi.org/10.3389/fpls.2017.00587.eCollection2017
Liu T (2016) A pioneer leader of Chinese cabbage seed industry - interview with Zhang Fenglan, scientist of Chinese cabbage breeding post of national bulk vegetable industry technology system. Yangtze River Vegetables 22:3. CNKI:SUN:CJSC.0.2016-22-003 (in Chinese)
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Mandel MA, Yanofsky MF (1998) The Arabidopsis AGL9 MADS box gene is expressed in young flower primordia. Sex Plant Reprod 11(1):22–28. https://doi.org/10.1007/s004970050116
Mauren J, Zhang L, Che C, Li G, Tang Y, Wen KSM, Joanna P (2018) A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J Exp Bot 69(20):4867–4880. https://doi.org/10.1093/jxb/ery284
Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, Kim SG, Lee I (2003) The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J 35(5):613–623. https://doi.org/10.1046/j.1365-313X.2003.01833.x
Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12(6):885–900. https://doi.org/10.1105/tpc.12.6.885
Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH (2011) Overexpression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann Bot 107(9):1491–1499. https://doi.org/10.1093/aob/mcr112
Shitsukawa N, Ikari C, Mitsuya T, Sakiyama T, Ishikawa A, Takumi S, Murai K (2007) Wheat BraSOC1 functions independently of WAP1/VRN1, an integrator of vernalization and photoperiod flowering promotion pathways. Physiol Plantarum 130(4):627–636. https://doi.org/10.1111/j.1399-3054.2007.00927.x
Simpson GG (2004) The autonomous pathway: epigenetic and posttranscriptional gene regulation in the control of Arabidopsis flowering time. Curr Opin Plant Biol 7(5):570–574. https://doi.org/10.1016/j.pbi.2004.07.002
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Vishal B, Kumar PP (2018) Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00838
Xian D, Jiang W, Zhao X, Tang Q, Song M, Wang Z (2013) Molecular mechanism of flowering integrator SOC1 flowering regulation. Chinese Vegetables 6:1–8. https://doi.org/10.3969/j.issn.1000-6346.2013.06.001(inChinese)
Yang F, Xia L, Chen L, Tian Y, Song W, Liang M (2019) Cloning and expression analysis on family gene AGL9 of mads-box in tea plant. Xinan Nongye Xuebao 32(10):2299–2303. https://doi.org/10.16213/j.cnki.scjas.2019.10.007 (in Chinese)
Yang X, Li X, Liao W (2021) Research progress on genetic regulation pathways of flowering time in plants. Biodivers 29(6):825–842. https://doi.org/10.17520/biods.2020370 (in Chinese)
Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346(6279):35–39. https://doi.org/10.1038/346035a0
Yu Y, Du J, Wang G, Ji J (2003) Studies on the freeze-thaw method of transforming recombinant plasmid DNA into agrobacterium tumefaciens. J Jilin Agr Univ 25(3):257–259. https://doi.org/10.3969/j.issn.1000-5684.2003.03.006
Yu J, Yang XD, Wang Q, Gao LW, Yang Y, Xiao D, Liu TK, Li Y, Hou XL, Zhang CW (2018) Efficient virus-induced gene silencing in Brassica rapa, using a turnip yellow mosaic virus vector. Biol Plantarum 62(4):826–834. https://doi.org/10.1007/s10535-018-0803-6
Zhang A (2021) Identification and functional analysis of key flowering genes in cotton. GanSu Agr Univ https://doi.org/10.27025/d.cnki.ggsnu.2021.000217 (in Chinese)
Zhao S, Lou Y, Zhang Z, Xu M, Wang W, Zhao Y, Zhang L, Fan Y, Wang L (2014) ZmSOC1, a MADS-Box transcription factor from Zea mays, promotes flowering in Arabidopsis. Int J Mol Sci 15(11):19987–20003. https://doi.org/10.3390/ijms151119987
Zhong X, Dai X, Xv JH, Wu HY, Liu B, Li H (2012) Cloning and expression analysis of GmGAL1, BraSOC1 homolog gene in soybean. Mol Biol Rep 39(6):6967–6974. https://doi.org/10.1007/s11033-012-1524-0
Zhu D, Rosa S, Dean C (2015) Nuclear organization changes and theepigenetic silencing of FLC during vernalization. J Mol Biol 427(3):659–669. https://doi.org/10.1016/j.jmb.2014.08.025
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32102393), the Science and Technology Program of Henan Province (222102110042), the Scientific Research Foundation for High-level Talent (103010620001/015 and 2017034). Funding body have no role in the study design, data collection, analysis and manuscript writing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, X., Shen, C., Chen, R. et al. Function of BrSOC1b gene in flowering regulation of Chinese cabbage and its protein interaction. Planta 258, 21 (2023). https://doi.org/10.1007/s00425-023-04173-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04173-5