Abstract
Main conclusion
OsPGIP4 overexpression enhances resistance to bacterial leaf streak in rice.
Polygalacturonase-inhibiting proteins are thought to play important roles in the innate immunity of rice against fungi. Here, we show that the chromosomal location of OsPGIP4 coincides with the major bacterial leaf streak resistance quantitative trait locus qBlsr5a on the short arm of chromosome 5. OsPGIP4 expression was up-regulated upon inoculation with the pathogen Xanthomonas oryzae pv. oryzicola strain RS105. OsPGIP4 overexpression enhanced the resistance of the susceptible rice variety Zhonghua 11 to RS105. In contrast, repressing OsPGIP4 expression resulted in an increase in disease lesions caused by RS105 in Zhonghua 11 and in Acc8558, a qBlsr5a resistance donor. More interestingly, upon inoculation, the activated expression of pathogenesis-related genes was attenuated for those genes involved in the salicylic acid pathway, while the activated expression of jasmonic acid pathway markers was increased in the overexpression lines. Our results not only provide the first report that rice PGIP could enhance resistant against a bacterial pathogen but also indicate that OsPGIP4 is a potential component of the qBlsr5a locus for bacterial leaf streak in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice (Oryza sativa) is the most important crop in the world, as more than half of the global population is dependent on it as the primary food source. The occurrence of rice diseases, such as blast and sheath blight caused by the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani, respectively, as well as bacterial leaf streak (BLS) and bacterial blight (BB) caused by the bacterial pathogens Xanthomonas oryzae pv. oryzicola (Xoc) and Xanthomonas oryzae pv. oryzae (Xoo), respectively, are major factors that limit rice yield and quality (Nino-Liu et al. 2006; Skaminioti and Gurr 2009). Breeding resistant rice varieties is an important strategy to control these diseases. Genetically, rice disease resistance can be classified into two types: qualitative resistance conferred by a single resistance (R) gene and quantitative resistance conferred by multiple genes or quantitative trait loci (Poland et al. 2009; Kou and Wang 2010). Qualitative resistance is primarily mediated by the recognition of avirulence (Avr) proteins in pathogens and guarded R proteins in the host. Qualitative resistance has been widely used to improve rice varieties because of its high-level resistance and easy manipulation. To date, more than 37 and 80 R genes have been identified that provide resistance against Xoo and M. oryzae, respectively (Kou and Wang 2010; Wang et al. 2012). Many of these R genes have been introduced as singletons or pyramids into rice varieties. However, no R gene has been identified in rice that provides resistance to BLS. However, a nonhost R gene, Rxo1, has been isolated from some maize varieties (Zhao et al. 2005). Additionally, R gene-mediated resistance is prone to easy breakdown due to high selection pressure, which could cause Avr gene mutations (Dai et al. 2010).
Quantitative resistance is considered a non-race-specific and more durable method. Quantitative resistance is usually controlled by multiple genes and is more favored in the genetic improvement of crops. However, due to the small effect of each QTL and the influence of the environment, isolating QTL genes and identifying their biochemical functions are difficult. To date, only a few resistance QTLs with large effects have been cloned through positional cloning. These resistance QTLs include pi21, which encodes a proline-rich protein containing a putative heavy metal-binding domain and putative protein–protein interaction motif (Fukuoka et al. 2009); Yr36, which encodes a kinase with a putative START lipid-binding domain that confers resistance to stripe rust in wheat (Fu et al. 2009); and Lr34, which encodes a putative ATP-binding cassette (ABC) transporter that confers durable resistance to multiple fungal pathogens in wheat (Krattinger et al. 2009). At least 13 QTLs involved in BLS quantitative resistance have been mapped from indica rice variety Acc8558 and Dular. The major QTL, qBlsr5a on the short arm of chromosome 5, has the largest effect, explaining ~14 % of the observed phenotypic variation in the population (Tang et al. 2000; Chen et al. 2006).
In recent years, some defense-related (DR) genes that co-localize with QTLs have been identified that may explain the small effect of QTLs (Kou and Wang 2010). OsDR10 (Os08g05960) coincided with BB resistance QTLs on chromosome 8; OsDR10-suppressed lines showed broad resistance to all tested Xoo strains (Xiao et al. 2009). A few cases of DR genes that confer resistance to Xoc have been reported. For instance, suppression of GH3-2, which encodes an indole-3-acetic-amino synthetase, and OsMPK6 mediated broad-spectrum resistance to both Xoo and Xoc (Fu et al. 2011; Shen et al. 2010). Suppression of NRRB, a receptor-like cytoplasmic kinase gene, and OsWRKY45-1 also enhanced resistance against Xoc (Tao et al. 2009; Guo et al. 2014). Additionally, these DRs and four stress-activated protein kinase genes were differentially expressed in the Rxo1-mediated resistance signaling pathway (Zhou et al. 2010; Xu et al. 2013).
Polygalacturonase-inhibiting proteins (PGIPs) inhibit the hydrolytic activity of polygalacturonase (PG), therefore delaying the hydrolysis of oligogalacturonides, which are components of the plant cell wall. PGIPs are members of DR gene family, which can be up-regulated by pathogen attack (Lu et al. 2012; Kalunke et al. 2015). PGIPs are typically leucine-rich repeat (LRR) proteins with ten imperfect LRRs, which are organized to form two β-sheets that interact with PG at the N-terminus or C-terminus of the enzyme and that include the loops surrounding the active site cleft (di Matteo et al. 2003; Benedetti et al. 2011; Gutierrez-Sanchez et al. 2012). In addition to identifying increasingly specific inhibitory effects between plant PGIPs and fungi PGs, dozens of plant PGIPs have been introduced into tomato, tobacco, potato, Brassica rapa, rapeseed, pea, grapevine, wheat, rice and Arabidopsis (Kalunke et al. 2015). PGIPs have also been identified that efficiently confer resistant against various necrotrophic and hemibiotrophic fungal pathogens (Kalunke et al. 2015; Wang et al. 2015). To inhibit PG directly, the interaction between PGs and PGIPs promotes the accumulation of oligogalacturonides (OGs), which could elicit a variety of defense responses in plants (Ferrari et al. 2013).
Compared with the many studies that have explored the interactions between plant PGIPs and fungal PGs, only a few studies examining resistance against PGs of bacteria have been performed (Kalunke et al. 2015). Constitutively expressing the pear PcPGIP gene was accidentally demonstrated to confer resistant against the bacterial pathogen Xylella fastidiosa and the fungal pathogen Botrytis cinerea in grapevine (Agüero et al. 2005). Later, this resistance was found to be caused by a specific interaction between PG and PGIP. The second case, transgenic tobacco and Chinese cabbage plants expressing B. rapa BrPGIP2 were resistant against the bacterial pathogen Pectobacterium carotovorum, the causal agent of soft rot disease (Hwang et al. 2010). The third study only indicated that the PG activity of Ralstonia solanacearum was inhibited by PGIP from a crude tomato stem extract (Schacht et al. 2011). Thus far, no evidence that PGIPs can protect rice against bacterial pathogens has been found.
Seven PGIP genes have been identified in rice; four of these genes are located on chromosome 5 within the region of qBlsr5a (Han et al. 2008; Lu et al. 2012). Only OsPGIP1 has been functionally analyzed regarding its ability to confer resistance against the fungal pathogen R. solani in rice (Wang et al. 2015). In this study, we performed a functional analysis of the role of OsPGIP4 in promoting resistance against Xoc. Gene expression patterns and transgenic rice were analyzed, and the results suggested that OsPGIP4 overexpression could enhanced the resistance capacity against Xoc.
Materials and methods
Plant materials and growth conditions
The BLS-susceptible rice variety Zhonghua 11 (ZH11, Oryzae sativa L. ssp. japonica) and moderate resistance variety Acc8558 (O. sativa L. ssp. indica) were used in this study. Seeds of the rice variety ZH11 were provided by Dr. Yuan Meng of the National Key Laboratory of Crop Genetic Improvement (Huazhong Agricultural University, Wuhan, China). Acc8558 seeds were obtained from Dr. Yang Long of Shandong Agricultural University. All rice plants were grown in a greenhouse at a temperature of 28 ± 2 °C, relative humidity of 85–100 %, and photoperiod of 16 h.
Vector construction
Genomic DNA was isolated from the leaves of Acc8558 rice plants using a Plant DNA Extraction Kit (CWBio, Jiangsu, China). The OsPGIP4 gene was amplified by PCR using the forward and reverse primers of OsPGIP4-OV (Table 1). The 50 μl PCR mixture contained 1× HF buffer with 1.5 mM MgCl2, 0.2 mM each dNTP, 0.8 μM each primer, 1 μl DNA (~100 ng) and 2 U Phusion HF DNA polymerase (New England Biolabs, Ipswich, MA, USA). The following PCR program was used: 98 °C for 30 s followed by 30 cycles of 98 °C for 10 s, 60 °C for 20 s, and 72 °C for 30 s with a final 5 min extension at 72 °C. The expected 1390 bp DNA fragment was gel-purified using a Gel Extraction Kit (Omega Bio-Tek, Norcoss, GA, USA) and then digested with Kpn I (New England Biolabs). The digestion product was purified using a PCR Purification Kit (Omega Bio-Tek). The DNA fragment was cloned into the binary vector pUbi1301-Kpn I-Cut (Li et al. 2013) and sequenced to confirm the authenticity of the sequence as the allele of OsPGIP4. Then, the plasmid construct pUbi1301-OsPGIP4 was transferred into the Agrobacterium tumefaciens strain EHA105.
To construct an RNAi vector, the expected 427 bp DNA fragment containing a portion of the encoding region and the 3′-UTR was amplified by PCR using the primers OsPGIP4-RI-F and OsPGIP4-RI-R (Table 1). To suppress OsPGIP4 expression, appropriate restriction sites were introduced into the PCR-amplified cDNA suitable for cloning steps (Spe I and Kpn I at the 5′ end and Sac I and BamH I at the 3′ end). The PCR-amplified portion of OsPGIP4 cDNA was first cloned into the KpnI-/BamHI-digested ds1301 expression vector (Li et al. 2013) and then into the SpeI-/SacI-digested ds1301 expression vector to obtain the ds1301::OsPGIP4 construct. They are spacing out by about 1.1 kb DNA fragment which is the intron of rice alcohol dehydrogenase (Adh) gene.
Rice transformation
For rice transformation, embryonic callus derived from mature embryos was infected by the Agrobacterium tumefaciens strain EHA105 containing the target genes. Callus culture and transformation for japonica variety ZH11 and indica variety Acc8558 were performed according to the published protocols (Lin and Zhang 2005; Ge et al. 2006).
PCR analysis of T0 generation transgenic plants
For transgenic lines, positive lines were validated by PCR. The PCR mixture consisted of 1 µl DNA template (approximately 20 ng), 0.5 µl each forward and reverse primers, 10 µl 2× Taq Master Mix (CWBio), and 8 µl ddH2O. The PCR reaction was performed at 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min, with a final 5 min extension at 72 °C. The forward primer (Ubi-F), which was designed based on the Ubi promoter sequence, and the reverse primer (JC-R), which was derived from the OsPGIP4 sequence, were used to select positive overexpression lines. The length of the PCR amplification product was approximately 250 bp. The primers ds1301-F2 and ds1301-R2 (Table 1) were used to amplify a 650 bp DNA fragment for positive selection with RNAi lines.
Pathogen inoculation and disease assessment
The X. oryzae pv oryzicola (Xoc) strain RS105 was grown on polypeptone-sucrose-agar medium (10 g l−1 polypeptone, 1 g l−1 glutamic acid, 10 g l−1 sucrose and 15 g l−1 agar) at 28 °C for 2 days and then suspended in sterile 10 mM MgCl2 to OD600 = 0.5. The more than five fully expanded leaves were infiltrated at three positions by inoculation with a non-needle syringe at the seedling stage or at the booting stage (Liu et al. 2014). The lengths of lesions on the transgenic and wild-type plants were scored at 14 days post-inoculation (dpi).
RNA extraction and cDNA synthesis
Infected and non-infected control plant leaves were harvested at 12, 48 and 72 h post-inoculation for total RNA isolation. Total RNA was extracted from 100 mg tissue using TRI Reagent (Sigma Aldrich) according to the manufacturer’s protocol. RNA pellets were resuspended in deionized water pretreated with 0.05 % (v/v) diethylpyrocarbonate (DEPC). First-strand cDNA synthesis was performed for each RNA sample using a PrimeScript™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
Quantitative polymerase chain reaction
Quantitative PCR was performed using SYBR® Premix Ex Taq™ (Tli RNase H Plus) and an IQ5 Real-Time PCR System (Bio-Rad) as previously reported (Li et al. 2013). The following PCR program was used: 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s, 55 °C for 20 s, and 72 °C for 30 s. A heat dissociation curve (55–95 °C) was checked after the final PCR cycle to determine the specificity of the PCR amplification. The OsActin (LOC_Os03g50890) gene of rice was used as an internal control to standardize the results. Expression levels of the OsPR1a (LOC_Os07g03710), OsPR1b (LOC_Os01g28450), OsPR10 (LOC_Os12g36880), OsAOS (LOC_Os03g12500) and OsAOC (LOC_Os03g32314) genes were analyzed by qRT-PCR assays, which were repeated at least twice with triplicate runs. Relative expression levels were measured using the 2−ΔΔCt analysis method. The primer sequences for each detected gene are listed in Table 1.
PGIP inhibition activity
About 1 g rice leaves were ground with liquid nitrogen and then flip cracked by 2 ml vegetable protein extract (CWBio) supplemented with 1 % (v/v) protease inhibitor cocktail at 4 °C for 1 h. The lysates were centrifuged at 3824g at 4 °C for 15 min, and supernatant were transferred to prechilled centrifuge tube for the follow-up experiment. The equivalent total plant proteins quantified by bradford assay were mixed with 0.1 mg/ml PG Pectinase from Aspergillus niger (>20 U/mg, BBI, Shanghai, China) at room temperature for 1 h. The mixtures and 0.25 % (m/v) polygalacturonic acid (Sigma) in sodium acetate buffer solution (pH 5.0) were incubated at 50 °C for 30 min. Then the reducing sugar products were quantified by DNS assay (McCleary and McGeough 2015). The relative activity of PGIP was calculated for the three overexpression rice lines compared with the ZH11.
Statistical analysis
The qRT-PCR estimates of certain gene transcript accumulation and the lesion lengths from BLS pathogenicity experiments were subjected to analysis of variance. Standard deviations were checked visually by error bars, and statistical significance was determined by analysis of variance. The data were subjected to one-way analysis of variance, the mean differences were compared by paired t test, and the P values <0.05 were considered significant. Correlation analysis was performed using SPSS software.
Results
Sequence analysis of OsPGIP4 in rice varieties Acc8558 and Zhonghua 11
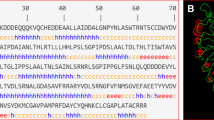
According to the gene annotation, OsPGIP4 (LOC_Os05g01444) in the Nipponbare genome contains a 1654 bp coding region without any intron and contains 1206 bp coding sequence (CDS) flanked by a 279 bp 5′ untranslated region (UTR) and a 169 bp 3′ UTR. The CDS of OsPGIP4 encodes a 401 residue polypeptide (Fig. 1a). After the alleles of OsPGIP4 in japonica rice ZH11 and in indica rice Acc8558 were sequenced, the allele of ZH11 was found to be identical to the annotation in Nipponbare, while 7 nucleotide differences that resulted in two amino acid substitutions were found for the allele of Acc8558 (Fig. 1b). The first substitution occurred at position 356, which is a cysteine in Nipponbare and ZH11 but a serine in Acc8558, and the second substitution, methionine to threonine, occurred at position 368.
The structure of OsPGIP4. a The DNA structure of OsPGIP4. The black boxes indicate the exons, and the gray boxes represent the 5′ and 3′ untranslated regions. b Protein alignment of OsPGIP4 homologs in Acc8558, Nipponbare (Nip) and Zhonghua 11 (ZH11). The substitution amino acids are highlighted in bold
Expression pattern of OsPGIP4
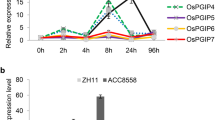
The expression profile of OsPGIP4 in different tissues and organs in ZH11 was detected by quantitative RT-PCR. Compared with the relative expression of OsPGIP4 in young root, the relative expression of OsPGIP4 was nearly two times higher in culm and leaf at the mature stage while less than half in anther, pistil and endosperm. In general, the relative expression of OsPGIP4 was similar in all tissues and organs (Fig. 2).
Pathogen challenge activated the expression of many DR genes; therefore, we measured the expression of OsPGIP4 post-inoculation with the Xoc strain RS105 in both the susceptible rice variety ZH11 and the moderate resistance rice variety Acc8558. The transcript level of OsPGIP4 increased quickly within 8 h after RS105 inoculation in both rice varieties. Then, this transcript level initially decreased from 24 h and nearly reached the same level as the control at 48 h. The tendency of transcription was similar in ZH11 and Acc8558. Notably, the expression level at 8 h post-inoculation was higher in the resistance variety Acc8558 than in ZH11, and the decrease in expression at 24 h post-inoculation was attenuated in Acc8558 (Fig. 3a, b). These observations suggest that OsPGIP4 expression could be quickly activated upon Xoc challenge. Upon inoculation with the Xoo strain PXO99, the transcript level of OsPGIP4 increased slightly in the susceptible variety IR24 (Fig. 3c) but decreased excessively in the resistance variety IRBB21 (Fig. 3d), which carries the resistance gene Xa21.
Expression analysis of OsPGIP4 in rice leaves upon Xoc and Xoo infection. Expression level of OsPGIP4 in response to RS105 in ZH11 (a susceptible), RS105 in Acc8558 (b moderate resistance), PXO99 in IR24 (c susceptible), and PXO99 in IRBB21 (d resistance) at 0, 8, 24 and 24 h post-inoculation. Bars represent the means (three replicates for gene expression) ± SD
OsPGIP4 overexpression enhanced the resistance against Xoc in ZH11
To access whether RS105-up-regulated OsPGIP4 expression could enhance resistance against Xoc, Agrobacterium-mediated transformation of the pUbi1301-OsPGIP4 construct was used to obtain 15 independent T0 lines. In the T0 generation, 9 of 15 individuals were identified as positive transgenic lines by PCR. These transgenic lines were inoculated with the Xoc strain RS105 at the booting stage. Disease severity was assessed by counting the lesion length per infiltration spot. All nine positive individuals exhibited enhanced resistance to RS105, with lesion lengths ranging from 1.0 to 1.8 cm (average of 1.3 cm) compared with 2.3 cm for wild-type ZH11 (Fig. 4a). Three individuals (OV7, OV13 and OV27) were chosen for detection of the OsPGIP4 expression level and for further analysis at the T1 generation based on the high-level resistance against RS105. All three lines showed over 200-fold increases in OsPGIP4 expression than did wild-type plants (Fig. 4b). OV23 showed the strongest expression of OsPGIP4, which was consistent with the shortest lesion length.
Resistance of the OsPGIP4 overexpression plants against the Xoc strain RS105. a Lesion lengths of the 12 individual T0 transgenic plants with pU1301::OsPGIP. b Relative expression levels of OsPGIP4 in three selected T0 transgenic plants. The qRT-PCR data for each sample were normalized to the amount of OsActin transcripts. Bars represent the means (three replicates for gene expression) ± SD
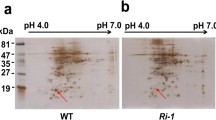
To evaluate the disease resistance of transgenic rice lines in the T1 generation, OsPGIP4-OXV lines and wild-type ZH11 were inoculated with RS105. In all three independent T1 lines, the progeny showed significant reductions in lesion length, as they were identified as positive transgenic individuals that carried the construct pUbi::OsPGIP4 (Fig. 5). After tested in T2 generation, all three lines were identified to significantly enhance the inhibition activity to PG (Suppl. Fig. S1), compared with wild type ZH11. These results further suggested that OsPGIP4 overexpression could improve resistance against Xoc.
Resistance of three OsPGIP4 overexpression lines to Xoc in the T1 generation. The average of lesion length for each plant was measured at more than ten inoculation sites. The gel figure indicates the plants carrying the pU1301::OsPGIP4 that were validated by PCR. Bars represent the mean ± SD. “*” and “**” indicate significant (t test, P < 0.05) and highly significant (t test, P < 0.01) differences detected in the lesion lengths between wild-type and transgenic plants, respectively
Repressing OsPGIP4 expression enhanced susceptibility to RS105
To further confirm that OsPGIP4 is involved in defense against Xoc, OsPGIP4 RNAi lines were generated in ZH11 by Agrobacterium-mediated transformation. Twelve independent T0 lines were obtained, eight of which were identified as positive lines by PCR. After the plants were inoculated with RS105, the eight positive lines exhibited significantly increased lesion lengths compared with wild-type ZH11 (Fig. 6). Three individuals (DS12, DS29 and DS48) were chosen from the T1 generation for further analysis based on their high-level susceptibility to RS105. These three lines also showed 50–80 % reduction of OsPGIP4 expression compared with ZH11 (Fig. 6b). In all three T1 lines, the positive transgenic plants exhibited significantly increased lesion lengths compared with wild-type ZH11 or negative individuals segregated from the progeny (Fig. 6c). These results suggested that OsPGIP4 acts as a positive regulator of resistance against Xoc in rice.
Repressing OsPGIP4 expression attenuated the resistance to RS105 in ZH11. a Lesion lengths of the 12 individual T0 transgenic plants with ds1301::OsPGIP4. b Relative expression levels of OsPGIP4 in three selected T0 transgenic plants. The qRT-PCR data for each sample were normalized to the amount of OsActin transcripts. Bars represent the means (three replicates for gene expression) ± SD. c The average lesion length for each individual of three T1 transgenic plants and the wild-type plant. The positive transgenic plants were identified by PCR using the primers ds1301-F2 and ds1301-R2
Resistance to RS105 was attenuated by repressing OsPGIP4 expression in Acc8558
Because OsPGIP4 is located on the same genomic region of the qBlsr5a locus, which is also responsible for bacterial leaf streak resistance in rice and because OsPGIP4 expression positively regulated Xoc resistance in this study, we questioned whether OsPGIP4 is one of the components of qBlsr5a. To answer this question, we generated OsPGIP4 RNAi lines in Acc8558, the resistance donor of qBlsr5a. Six individuals were generated, and five of these individuals were identified as positive transgenic plants. These five positive lines also showed 40–90 % reduction of OsPGIP4 expression, and 0.23–0.53 cm increased lesion length in response to RS105 infection compared with Acc8558 (Fig. 7a, b). At the T1 generation, all plants derived from two positive T0 individuals (ds1301::OsPGIP4-1 and ds1301::OsPGIP4-8) were investigated regarding their susceptibility to RS105. Consisted with the PCR identifications, all positive individuals exhibited significantly increased lesion lengths compared with wild-type Acc8558 (Fig. 7c). These results suggested that OsPGIP4 is also involved in resistance to Xoc in Acc8558.
Increased susceptibility to the Xoc strain RS105 was associated with the repressed expression of OsPGIP4 in Acc8558. a Lesion lengths of the 6 individual T0 transgenic plants with ds1301::OsPGIP4 in ACC8558 background. b OsPGIP4 expression was repressed in RNAi transgenic plants at the T0 generation. c Increased susceptibility to RS105 was identified in OsPGIP4-RNAi T1 families ds1301::OsPGIP4-1 and ds1301::OsPGIP4-8. The positive transgenic plants were identified by PCR using the primers ds1301-F2 and ds1301-R2
Increased expression of JA-related PR genes in OsPGIP4 overexpression lines
As the SA signaling pathway was reported to be involved in BLS resistance (Guo et al. 2014), quantitative real-time PCR was used to measure the basal levels of transcripts for several pathogenesis-related genes in three OsPGIP4 overexpression lines (OV17, OV23 and OV37) at the T2 generation. To our surprise, the expression levels of OsPR1a, OsPR1b and OsPR10 were significantly reduced in all three overexpression lines at 12 hpi compared with the wild-type plants (Fig. 8a–c). In contrast, all three OsPGIP4 overexpression lines showed significantly higher expression levels of OsAOC and OsAOS (Fig. 8d, e). Consistent with the highest levels of resistance to RS105, the activated expression levels of OsAOC and OsAOS were highest in the OV23 line.
Quantitative PCR amplification of the pathogenesis-related (PR) genes in wild-type and OsPGIP4 overexpression lines. Expression analysis was performed at 12 h post-infection in Xoc-infected leaves and non-infected leaves as a control. The qRT-PCR data for each sample were normalized to the amount of OsActin transcripts. Bars represent the means (three replicates for gene expression) ± SD
Discussion
PGIPs are conserved proteins that have been identified from various plant species; these proteins clearly function in inhibiting different fungal PGs (Kalunke et al. 2015). Additionally, PGIPs are members of plant DR gene families and, in most cases, are up-regulated following stress stimuli. Seven PGIP genes have been identified in rice (Janni et al. 2006; Lu et al. 2012), most of which could be activated to elicit up-regulated expression by various treatments with phytohormones or fungal infection (Lu et al. 2012). Two of these PGIP genes have been shown to functionally inhibit fungal PGs (Jang et al. 2003; Janni et al. 2006; Wang et al. 2015), and only one gene has been shown to enhance resistance against R. solani after being constitutively expressed in rice (Wang et al. 2015). In this study, we functional analyzed the role of OsPGIP4 in resistance against the bacterial pathogen Xoc in rice. Similar to most characterizations of plant PGIPs, OsPGIP4 expression was activated after Xoc inoculation (Fig. 3). Modulation of OsPGIP4 expression resulted in a resistance phenotype against RS105 in the susceptible rice variety ZH11 (Figs. 4, 5). These results strongly indicated that OsPGIP4 positively regulates the defense response to Xoc and provided the first evidence of a role for PGIP in resistance against a bacterial pathogen in rice. Despite the existence of a putative PG in Xoc, unfortunately, we failed to purify the fusion OsPGIP4 protein via prokaryotic expression in E. coli or transient expression in Nicotiana benthamiana (data not shown). Thus, we could not determine whether the enhanced resistance was directly caused by the interaction of PG-OsPGIP4.
Based on our data, we strongly suggested that this resistance was primarily dependent on the interaction of PG-OsPGIP4. Another rice bacterial pathogen, X. oryzae pv. oryzae, which shares approximately 91 % similarity with Xoc, causes bacterial blight in rice (Feng et al. 2015). This pathogen also contains only one putative PG, which is 95 % identical to Xoc-PG (Suppl. Fig. S2). Most substitutions occurred on the C-terminus, which is the primary active site of the enzyme. Regardless of compatible or incompatible interactions between Xoc and rice, OsPGIP4 expression was quickly activated and reached its highest level at 8 hpi (Fig. 3). However, OsPGIP4 expression was slightly up-regulated to threefold at 48 hpi with Xoo in the susceptible rice near isogenic line (NIL) IR24. In contrast, OsPGIP4 expression was reduced approximately ten folds at 8 hpi in the resistance NIL IRBB21 compared with the mock treatment (Fig. 3c, d). Notably, OsPGIP4 overexpression could enhanced resistance against the Xoc strain RS105 (Fig. 4) but had no effect on resistance against PXO99 (Suppl. Fig. S3). The OsPGIP4 RNAi lines also showed no difference regarding resistance against PXO99 compared with wild-type ZH11 (Suppl. Fig. S3). The specific resistance to Xoc, but not to Xoo, is hard to explain if the concept of specific recognition between Xoc-PG and OsPGIP4 is excluded. Additionally, direct interactions between Xoc-PG and OsPGIP4 proteins need to be confirmed in future studies.
Recently, defense-related (DR) genes that co-localize with QTLs have been identified that may explain the small effect of QTLs (Kou and Wang, 2010). Four clustering PGIPs (OsPGIP1 ~ OSPGIP4) located on chromosome 5 co-coincide with qBlsr5a, which has the largest effect of known QTLs for BLS resistance (Tang et al. 2000; Han et al. 2008). Our results show that OsPGIP4 overexpression enhanced resistance to Xoc and that reduction of OsPGIP4 expression resulted in greater susceptibility in ZH11. Moreover, the reduction of OsPGIP4 expression enhanced susceptibility to RS105 in Acc8558, the resistance donor of qBlsr5a. These results suggested that OsPGIP4 was possibly involved in qBlsr5a-mediated resistance against Xoc. Xie et al. (2014) further mapped qBlsr5a to a narrow region of approximately 30 kb with a set of overlapping sub-chromosome segment substitution lines (sub-CSSLs). The most possible candidate was the mutation Xa5 from the listed three candidates (Xie et al. 2014), and OsPGIP4 was excluded as a candidate for qBlsr5a. However, after deep analysis of the disease severity for the set of sub-CSSLs, we found that all sub-CSSLs showed resistance phenotypes with longer lesion lengths than the resistance parent H359-BLSR5A, including H91, P30, A33, B76 and D-101, which carried xa5 but were homozygous or heterozygous at the OsPGIP4 position in the susceptible parent. Additionally, comparison of susceptible parent H359 with those susceptible sub-CSSLs, including C-122, R-77, M35, F-9, H-59, G123, O-26 and B-102, we found that the lesion lengths were shorter in the susceptible sub-CSSLs than in H359. Thus, qBlsr5a had a genetic resistance residue effect in addition to the characterized region of the 30 kb DNA sequence (from the data of Xie et al. 2014). This finding should explain the small effect of qBlsr5a, which is located upstream of Xa5. Taken together, these findings and our results suggest that OsPGIP4 is a potential component of the qBlsr5a locus.
Defense response activation is always accompanied by induced expression of pathogenesis-related genes (Kou and Wang 2010). These genes are biomarkers that can be used to determine which phytohormone-mediated signaling pathway, such as SA, JA, ET or IAA, is involved in the defense response. Among the limited studies of rice-Xoc interactions, most studies have demonstrated that the expression of SA-related PR genes, such as OsPR1a, OsPR1b and OsPR10, is activated in the modulated expression transgenic lines upon inoculation (Xiao et al. 2009; Li et al. 2012; Guo et al. 2014). Even in Rxo1-mediated resistance against Xoc, stronger activation of OsPR1a and OsPR10 expression was observed in transgenic plants compared with the wild-type plants (Zhou et al. 2010). Additionally, in DEPG1-overexpressing lines, the expression of SA-related genes decreased simultaneously with increased susceptibility to Xoc (Guo et al. 2012). In this study, we found that the expression of SA-related PR genes, which were represented by OsPR1a, OsPR1b and OsPR10, was attenuated in all three OV lines post-inoculation with RS105. However, the expression of the JA-related PR genes OsAOC and OsAOS (Fig. 8) were highly activated, which suggested that JA might be in involved in the OsPGIP4-mediated resistance pathway. This finding is distinguished from the above-described Xoc-DR genes. As JA was reported to antagonize with SA in plants (Thaler et al. 2012), the opposite expression of OsPR1a and OsPR1b was supported by this hypothesis. A few studies have reported that PGIPs also favor the accumulation of elicitor-active OGs, which could induce JA synthesis in tomato (Ferrari et al. 2013). In turn, many studies have demonstrated that Xoo-mediated resistance was also dependent on the SA pathway; however, the OsPGIP4-RNAi lines did not visibly affect resistance against the Xoo strain PXO99. Whether OV lines inoculated with PXO99 could activate the expression of OsAOC, OsAOS and those SA-related genes remains to be determined and would help to identify the specific interaction between Xoc and OsPGIP4.
Author contribution statement
X.D. and Z.C. designed the experiments. C.F., X.Z. and T.W. performed the molecular experiments. B.Y. performed the pathogen inoculation. F.Y. performed the field management. X.D. analyzed the data. Z.C. wrote the manuscript.
Abbreviations
- BB:
-
Bacterial blight
- BLS:
-
Bacterial leaf streak
- DR:
-
Defense-related
- JA:
-
Jasmonic acid
- OGs:
-
Oligogalacturonides
- PG:
-
Polygalacturonase
- PGIP:
-
Polygalacturonase-inhibiting protein
- R:
-
Resistance
- SA:
-
Salicylic acid
- Xoc:
-
Xanthomonas oryzae pv. oryzicola
- Xoo:
-
Xanthomonas oryzae pv. oryzae
References
Agüero CB, Uratsu SL, Greve C, Powell AT, Labavitch JM, Meredith CP, Dandekar AM (2005) Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol Plant Pathol 6:43–51
Benedetti M, Leggio C, Federici L, de Lorenzo G, Viorel Pavel N, Cervone F (2011) Structural resolution of the complex between a fungal polygalacturonase and a plant polygalacturonase-inhibiting protein by small-angel X-ray scattering. Plant Physiol 157:599–607
Chen C, Zheng W, Huang X, Zhang D, Lin X (2006) Major QTL conferring resistance to rice bacterial leaf streak. Agric Sci China 5(3):216–220
Dai Y, Jia Y, Correll J, Wang X, Wang Y (2010) Diversification and evolution of the avirulence gene AVR-pita 1 in field isolates of Magnaporthe oryzae. Fungal Genet Biol 47:974–980
Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, de Lorenzo G, Tsernoglou D, Cervone F (2003) The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA 100:10124–10128
Feng W, Wang Y, Huang L, Feng C, Chu Z, Ding X, Yang L (2015) Genomic-associated markers and comparative genome maps of Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. World J Microbiol Biotechnol 31:1353–1359
Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, De Lorenzo G (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4:49
Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360
Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S (2011) Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol 155:589–602
Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325:998–1001
Ge X, Chu Z, Lin Y, Wang S (2006) A tissue culture system for different germplasms of indica rice. Plant Cell Rep 25:392–402
Guo L, Li M, Wang W, Wang L, Hao G, Guo C, Chen L (2012) Over-expression in the nucleotide-binding site-leucine rich repeat gene DEPG1 increase susceptibility to bacterial leaf streak disease in transgenic rice plants. Mol Biol Rep 39:3491–3504
Guo L, Guo C, Li M, Wang W, Luo C, Zhang Y, Chen L (2014) Suppression of expression of the putative receptor-like kinase gene NRRB enhances resistance to bacterial leaf streak in rice. Mol Biol Rep 41:2177–2187
Gutierrez-Sanchez G, King D, Kemp G, Bergmann C (2012) SPR and differential proteolysis/MS provide further insight into the interaction between PGIP2 and EPGs. Fungal Biol 116:737–746
Han QD, Chen ZW, Deng Y, Lan T, Guan HZ, Duan YL, Zhou YC, Lin MC, Wu WR (2008) Fine mapping of qBlsr5a, a QTL controlling resistance to bacterial leaf streak in rice (in Chinese). Acta Agron Sin 34(4):587–590
Hwang BH, Bae H, Lim HS, Kim KB, Kim SJ, Im MH, Park BS, Kim DS, Kim J (2010) Overexpression of polygalacturonase-inhibiting protein 2 (PGIP2) of Chinese cabbage (Brassica rapa ssp. pekinensis) increased resistance to the bacterial pathogen Pectobacterium carotovorum ssp. carotovorum. Plant Cell Tiss Organ Cult 103:293–305
Jang S, Lee B, Kim C, Kim SJ, Yim J, Han JJ, Lee S, Kim SR, An G (2003) The OsFOR1 gene encodes a polygalacturase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Mol Biol 53:357–369
Janni M, Di Giovanni M, Roberti S, Capodicasa C, D’Ovidio R (2006) Characterization of expressed Pgip genes in rice and wheat reveals similar extent of sequence variation to dicot PGIPs and identifies an active PGIP lacking an entire LRR repeat. Theor Appl Genet 113:1233–1245
Kalunke RM, Tundo S, Benedetti M, Cervone F, De Lorenzo G, D’Ovidio R (2015) An undate on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front Plant Sci 6:146
Kou Y, Wang S (2010) Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol 13:181–185
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFaddern H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Li D, Wang L, Teng S, Zhang G, Guo L, Mao Q, Wang W, Li M, Chen L (2012) Proteomics analysis of rice proteins up-regulated in response to bacterial leaf streak disease. J Plant Biol 55:316–324
Li N, Kong L, Zhou W, Zhang X, Wei S, Ding X, Chu Z (2013) Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant Cell Tiss Organ Cult 115:429–441
Lin Y, Zhang Q (2005) Optimizing the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep 23:540–547
Liu H, Chang Q, Feng W, Zhang B, Wu T, Li N, Yao F, Ding X, Chu Z (2014) Domain dissection of AvrRxo1 for suppressor, avirulence and cytotoxic functions. PLoS One 9(12):e113875
Lu L, Zhou F, Zhou Y, Fan X, Ye S, Wang L, Chen H, Lin Y (2012) Expression profile analysis of polygalacturonase-inhibiting protein genes in rice and their responses to phytohormones and fungal infection. Plant Cell Rep 31:1173–1187
McCleary BV, McGeough P (2015) A comparison of polysaccharide substrates and reducing sugar methods for the measurement of endo-1,4-β-xylanase. Appl Biochem Biotechnol 177:1152–1163
Nino-Liu DO, Ronald PC, Bogdanove AJ (2006) Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol 7:303–324
Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ (2009) Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 14:21–29
Schacht T, Unger C, Pich A, Wydra K (2011) Endo- and exopolygalactuonases of Rslstonia solanacearum are inhibited by polygalactuonase-inhibiting protein (PGIP) activity in tomato stem extracts. Plant Physiol Biochem 49:377–387
Shen X, Yuan B, Liu H, Li X, Xu C, Wang S (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 4:86–99
Skaminioti P, Gurr SJ (2009) Against the grain: safeguarding rice from blast disease. Trends Biotechnol 27:141–150
Tang D, Wu W, Li W, Lu H, Worland AJ (2000) Mapping of QTLs conferring resistance to bacterial leaf streak in rice. Theor Appl Genet 101:286–291
Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151:936–948
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270
Wang Y, Wang D, Deng X, Liu J, Sun P, Liu Y, Huang H, Jiang N, Kang H, Ning Y, Wang Z, Xiao J, Liu X, Liu E, Dai L, Wang GL (2012) Molecular mapping of the blast resistance genes Pi2-1 and Pi51 (t) in the durably resistant rice ‘Tianjingyeshengdao’. Phytopathology 102:779–786
Wang R, Lu L, Pan X, Hu Z, Ling F, Yan Y, Liu Y, Lin Y (2015) Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol Biol 87:181–191
Xiao W, Liu H, Li Y, Li X, Xu C, Long M, Wang S (2009) A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 4:e4603
Xie X, Chen Z, Cao J, Guan H, Lin D, Li C, Lan T, Duan Y, Mao D, Wu W (2014) Toward the positional cloning of qBlsr5a, a QTL underlying resistance to bacterial leaf streak, using overlapping sub-CSSLs in rice. PLoS One 9:e95751
Xu MR, Huang LY, Zhang F, Zhu LH, Zhou YL, Li ZK (2013) Genome-wide phylogenetic analysis of stress-activated protein kinase genes in rice (OsSAPKs) and expression profiling in response to Xanthomonas oryzae pv. oryzicola infection. Plant Mol Biol Rep 31:877–885
Zhao B, Lin X, Poland J, Trick H, Leach J, Hulbert S (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102:15383–15388
Zhou YL, Xu MR, Zhao MF, Xie XW, Zhu LH, Fu BY, Li ZK (2010) Genome-wide gene responses in a transgenic rice line carrying the maize resistance gene Rxo1 to the rice bacterial streak pathogen, Xanthomonas oryzae pv. oryzicola. BMC Genomics 11:78
Acknowledgments
This study was supported by the National Program of Transgenic Variety Development of China (2013ZX08009-004, 2014ZX08001-002), Foundation for the Author of National Excellent Doctoral Dissertation of PR China (Grant No. 201132), Shandong Provincial Natural Science Foundation, China (ZR2015CM004). Z.C. was funded by the Taishan Scholar Program of Shandong Province and by the Shandong Modern Agricultural Technology & Industry System.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The authors declare that the experiments comply with the laws of China.
Additional information
C. Feng and X. Zhang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2016_2480_MOESM1_ESM.pdf
Suppl. Fig. S1 The inhibition of PG activity for OsPGIP4 overexpression rice. Three individual OsPGIP4 overexpression lines were performed for the inhibition of PG activity that compared with the wild type ZH11. Suppl. Fig. S2 The PG homologs of Xanthomonas oryzae. Alignment of PG homologs in the Xoo strains PXO99A and PXO86 and in the Xoc strains BLS256 and RS105. The substitution amino acids are highlighted in bold. Suppl. Fig. S3 The lesion areas to PXO99 for OsPGIP4 overexpression and RNAi plants. Lesion areas were counted by the ratio between the lesion length and leaf length after inoculation with the Xoo strain PXO99 in T1 generation. Each line included at least 5 positive individuals for the experiment (PDF 74 kb)
Rights and permissions
About this article
Cite this article
Feng, C., Zhang, X., Wu, T. et al. The polygalacturonase-inhibiting protein 4 (OsPGIP4), a potential component of the qBlsr5a locus, confers resistance to bacterial leaf streak in rice. Planta 243, 1297–1308 (2016). https://doi.org/10.1007/s00425-016-2480-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2480-z