Abstract
Polygalacturonase-inhibiting proteins (PGIPs) are leucine-rich repeat (LRR) proteins involved in plant defence. A number of PGIPs have been characterized from dicot species, whereas only a few data are available from monocots. Database searches and genome-specific cloning strategies allowed the identification of four rice (Oryza sativa L.) and two wheat (Triticum aestivum L.) Pgip genes. The rice Pgip genes (Ospgip1, Ospgip2, Ospgip3 and Ospgip4) are distributed over a 30 kbp region of the short arm of chromosome 5, whereas the wheat Pgip genes, Tapgip1 and Tapgip2, are localized on the short arm of chromosome 7B and 7D, respectively. Deduced amino acid sequences show the typical LRR modular organization and a conserved distribution of the eight cysteines at the N- and C-terminal regions. Sequence comparison suggests that monocot and dicot PGIPs form two separate clusters sharing about 40% identity and shows that this value is close to the extent of variability observed within each cluster. Gene-specific RT-PCR and biochemical analyses demonstrate that both Ospgips and Tapgips are expressed in the whole plant or in a tissue-specific manner, and that OsPGIP1, lacking an entire LRR repeat, is an active inhibitor of fungal polygalacturonases. This last finding can contribute to define the molecular features of PG–PGIP interactions and highlights that the genetic events that can generate variability at the Pgip locus are not only limited to substitutions or small insertions/deletions, as so far reported, but can also involve variation in the number of LRRs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polygalacturonase-inhibiting proteins (PGIPs) are plant cell wall glycoproteins that inhibit fungal endopolygalacturonases (PG) and modulate their activity favouring the accumulation of elicitor-active oligogalacturonides (De Lorenzo et al. 2001; Federici et al. 2006). PGIPs are induced by pathogen infection and a number of stress-related signals (De Lorenzo et al. 2001). In Arabidopsis it has been also shown that the up regulation of Atpgip1 and Atpgip2 occurs through separate signal transduction pathways (Ferrari et al. 2003). On the basis of their properties, PGIPs are considered typical defence proteins that are able to limit the growth of those fungal pathogens producing PG during tissue colonization. The involvement of PGIP in limiting plant disease has been demonstrated in transgenic tomato, arabidopsis, tobacco and grape plants, where over-expression of PGIP is correlated with a reduction of symptoms caused by the fungal pathogen Botrytis cinerea (Powell et al. 2000; Ferrari et al. 2003; Manfredini et al. 2005; Aguero et al. 2005;).
PGIPs, like the products of many resistance genes, belong to the subclass of proteins containing leucine-rich repeats (LRRs) of the extracytoplasmic type (Jones and Jones 1997). They typically contain 10 imperfect LRRs of 24 residues, each containing the consensus xxLxLxx motif composed of hydrophobic structural residues (L) and by non-conserved residues (x) responsible for PG recognition (Di Matteo et al. 2003).
Plant genomes contain a small number of Pgip genes and those of Arabidopsis thaliana (L.) Heynh. (Ferrari et al. 2003), Brassica napus (Li et al. 2003), Phaseolus vulgaris L. (D’Ovidio et al. 2004) and Glycine max (L.) Merr (D’Ovidio et al. 2006) have been characterized. These analyses demonstrated that the different members of a Pgip family can be quite divergent and can undergo a different transcript regulation during growth and development or following biotic and abiotic stresses. The encoded products can possess different inhibiting activities against fungal or insect PGs and this capability can be affected by single amino acid substitutions and small insertions/deletions (indels) within the xxLxLxx motifs (Leckie et al. 1999; D’Ovidio et al. 2004).
The role of PGIP in monocots has been poorly investigated and, until recently, PGIP activity has been characterised only in pectin-rich species such as Allium cepa (Favaron et al. 1993) and A. porrum (Favaron et al. 1997; Favaron 2001). Lately, PGIPs have been characterized in wheat, although their N-terminal sequences (Lin and Li 2002; Kemp et al. 2003) do not show any similarity with the typical PGIP sequences determined from purified proteins or deduced from gene sequences (De Lorenzo et al. 2001). Moreover, a rice Pgip gene has been reported to be involved in flower development (Jang et al. 2003). The recent demonstration that PG is a pathogenicity factor in Claviceps purpurea during the infection of rye (Oeser et al. 2002) has reinforced the interest for a defence role of PGIP also in monocots.
The objective of the present work was the identification of Pgip genes in the monocots wheat and rice and the analysis of their genome organization, expression features and relationships with PGIPs from dicot species. We determined also the inhibition activities of the product encoded by one of the rice Pgip genes lacking an entire LRR.
Materials and methods
Plant materials
Seeds were surface sterilized by immersion in sodium hypochlorite (0.5% v/v) for 30 min, and then rinsed thoroughly in sterile water. Rice and wheat plants were grown at 18–23°C with a 14 h light period with a light intensity of 300 μE m−2 s−1, while Nicotiana benthamiana plants were grown at 24°C with 16/8 h day/night period at 100 μE m−2 s−1 light intensity. Spike, leaf and root tissues were collected from rice (Oryza sativa L.) and wheat (Triticum aestivum L.) flowering plants, except wheat roots that were collected from 7-day-old seedlings.
Chromosome assignment was carried out by using nulli-tetrasomic and ditelosomic lines of T. aestivum cv. Chinese Spring (Sears 1966) and D-genome-chromosome intervarietal substitution lines of T. durum cv. Langdon (Joppa 1988).
DNA analyses and manipulation
Genomic DNA was extracted from 0.5 g of green material following the procedure reported by Tai and Tanksley (1991).
PCR reactions for cloning Pgip genes were carried out in a reaction volume of 50 μl with 50 ng of genomic DNA, 2.5 units of FastStart High Fidelity PCR system (Roche Diagnostics Monza, Italy), 1× Taq PCR buffer, 50 ng of each of the two primers and 100 μM of each deoxyribonucleotide. PCR conditions were: 1 cycle at 95°C for 2 min, 30 cycles at 95°C for 30 s, 60°C for 1 min, 72°C for 1 min, and a final step at 72°C for 5 min. Oligonucleotides used to isolate the complete coding region of Pgip genes were synthesized on the basis of nucleotide sequences available at data banks as specified in the Results section, and have the following sequences: (PGIPWH1F) 5′-ATGAGCACTCCACCCTCGGCACAT and (PGIPWH1R) 5′-TTATTTCTTGCATGGATCTGGGAG for both Tapgip1 and Tapgip2; (Ospgip1F) 5′-TGACTCGCTATTGCATGCG and (Ospgip2R) 5′-TGGGAGCTTAATTGCAGGGA for Ospgip1; (OsPGIP72F) 5′-ATACACGGCATTGCATGCAC and (OsPGIP72R) 5′-CTTACACTCGTTCTCCGTAC for Ospgip2; (OsPGIP63F) 5′-TAGAAGAGAGGAAGCACGCA and (OsPGIP63R) 5′-GTTGGTGGCCTGAGATAGGT for Ospgip3; (OsPGIP4_3F) 5′-TGTCGTGCACTTGTGTTCAA and (OsPGIP4_3R) 5′-GCATTAGCTGGTTGCTTC for Ospgip4.
Amplified products were fractionated on 1.2% agarose gel, recovered using the GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare, Cologno Monzese, Italy) and cloned into pGEM-T Easy Vector (Promega Italia Srl., Milano, Italy) or directly subjected to nucleotide sequencing. Plasmid DNA was purified from recombinant bacterial clones with the NucleoBond® Plasmid Purification Kit (Clontech, Mountain View, USA).
Sequencing reactions were performed using the “ABI PRISM dye terminator cycle sequencing ready reaction” kit and DNA sequences were determined with the semiautomatic ABI PRISM 310 sequencer (Applied Biosystem, Monza, Italy). Nucleotide sequences were also determined by the MWG-Biotech AG (Ebersberg, Germany) and Invitrogen (San Giuliano Milanese, Italy) Sequencing Services.
Sequence analyses were performed using the DNAMAN software (Lynnon Biosoft, Quebec, Canada) that uses ClustalW algorithm for multiple sequence alignment (Thompson et al. 1994). The parameters for sequence alignment were: a protein gap open penalty of 10 and a protein gap extension penalty of 0.1. However, the automatic multiple sequence alignments has been improved by manual editing based on the LRR modular organization of PvPGIP2 (Di Matteo et al. 2003). Trees were generated by DNAMAN program by using both the UPGMA (Sneath and Sokal 1973) and Neighbor-Joining (Saitou and Nei 1987) methods. Bootstrap values were based on 1,000 replications.
Blast searches (McGinnis and Madden, 2004) at TIGR (http://www.tigr.org/tdb/tgi/plant.shtml) and at the International Nucleotide Sequence Databases (http://www.ncbi.nlm.nih.gov/) databases were performed with programs tBlastn, tBlastx and Blastn using default parameters.
Genomic DNA (10 μg) was digested with EcoRI (Invitrogen) and fractionated on 1.2% agarose gel using the conditions specified by the manufacturer. Southern blot analyses were carried out using standard conditions for specific target sequences (Sambrook et al. 1989) by using as probe the complete coding region of Tapgip1 labelled with digoxigenin (Digoxigenin−11-uridine-5(-triphosphate, Roche Diagnostics) following the procedure reported by D’Ovidio and Anderson (1994).
PCR reactions for chromosomal assignment of wheat Pgip genes were performed using the RedTaqTM ReadyMix (Sigma-Aldrich, Milano, Italy) in a 50 μl final volume and following the manufacturer’s procedure. Amplification conditions were: 32 cycles at 94°C for 1 min, 65°C for 1 min, 72°C for 40 s and a final step at 72°C for 5 min. Oligonucleotides used were those specified for RT–PCR experiments (see below).
RNA extraction and RT–PCR
Total RNA was extracted using RNeasy Plant Mini kit (Qiagen SpA, Milano, Italy) following the manufacturer’s procedure. Contaminating DNA was removed using the DNA-free TM mix (Ambion Ltd., Huntingdon, United Kingdom) and RNA concentration was determined spectrophotometrically.
RT–PCR experiments were performed by using the QuantyTect® SYBR® Green RT–PCR (Qiagen) in a MyCycler™ thermal cycler (Bio-Rad Life Science, Segrate, Italy). Each RT–PCR was performed in a total volume of 25 μl using 250 ng of total RNA and 50 ng of each of the two primers. cDNA synthesis and amplification conditions were: 30 min at 50°C for first-strand cDNA synthesis; 15 min at 95°C to inactivate the reverse transcriptase; then, 40 cycles at 94°C for 15 s, 60°C for 30 s and 72°C for 30 s for cDNA amplification. Two negative controls missing total RNA template or reverse transcriptase were included in each experiment. Oligonucleotide primers specific for each Tapgip and Ospgip genes were:
(PGIPWH1F) 5′-ATGAGCACTCCACCCTCGGCACAT and (PGIPWH10R) 5′-GGCGTCGTTGCAAGTGAT for Tapgip1; (PGIPWH1F) 5′-ATGAGCACTCCACCCTCGGCACAT and (PGIPWH11R) 5′-GGCGTCGTTGCAAGTGTC for Tapgip2; (PGIPWH1F); (OSPGIP5F) 5′-AGAAGGAGTGCAACGCCGGC and (OSPGIP1_3R) 5′-GGTGGTGTCGTCGCAGGT for Ospgip1; (OSPGIP2_F5) 5′-AGAAGGTGCAATGCCATGAG and (OSPGIP73_R4) 5′-GATGGTGAGGTGGGTGAGGT for Ospgip2; (OSPGIP3F4) 5′-TCATCAACCTCGCCATCACA and (OSPGIP64R7) 5′-CCTTGAAGAAGGTGATGTAC for Ospgip3; (OSPGIP4_1F) 5′-TGCTCGTGTTCGTGGTGT and (OSPGIP4_1R) 5′-ACTGGAAGTAGGACGCGTTG for Ospgip4.
The specificity of the primers was assessed in separate PCR experiments using, as a template, recombinant plasmid DNA containing the appropriate Pgip clone. PCR assays using up to 200 ng of plasmid DNA confirmed that each oligonucleotide pair amplified specifically the correct Pgip gene (data not shown). The identity of each PCR and RT–PCR product was confirmed by direct nucleotide sequencing of the amplicon. Amplification of 18S rRNA was used as reference for transcript amplification. The 18S rRNA primers used were conserved in both wheat and rice and have the following sequence: (OS18SRNAF) 5′-ATGATAACTCGACGGATCGC and (OS18SRNAR) 5′-CTTGGATGTGGTAGCCGTTT.
Fungal polygalacturonases (PGs), PGIP preparations and enzymatic assays
The endo-PGs of Sclerotinia sclerotiorum isolate B-24 and F. graminearum isolate 3827 were kindly provided by F. Favaron (University of Padova, Italy) while those of Aspergillus niger, Fusarium moniliforme isolate FC10, and Botrytis cinerea strain B05–10 were kindly provided by G. De Lorenzo (University of Rome, Italy). Endo-PGs of S. sclerotiorum (Favaron et al. 1997), A. niger (Cervone et al. 1987), F. moniliforme (Caprari et al. 1996) and F. graminearum (F. Favaron, unpublished) were purified at homogeneity, while that of B. cinerea (D’Ovidio et al. 2004) was purified from culture filtrate.
Transient expression of OsPGIP1 and GmPGIP3 was performed on Nicotiana benthamiana plants by using the potato virus X. The complete coding region of both Ospgip1 and Gmpgip3 genes were amplified by PCR using sequence specific oligonucleotides including the restriction site for ClaI or SalI at the 5′ ends of the forward or reverse primers, respectively. The amplified fragments were double digested with ClaI and SalI and cloned into the pPVX201 expression vector. The plasmids obtained were used to inoculate N. benthamiana plants using 30 μg of DNA/plant as described by Baulcombe et al. (1995).
Transiently expressed PGIPs were extracted from leaves of N. benthamiana plants infected with a single PVX-Pgip construct or with the empty vector, as previously described (D’Ovidio et al. 2004). Protein concentration was determined with the Coomassie plus-the better Bradford assay kit (Pierce, Rockford, IL, USA). SDS-PAGE and immunoblotting were performed as previously described (Desiderio et al. 1997). Polyclonal antibodies raised against PGIP purified from P. vulgaris pods (kindly provided by Prof. G. De Lorenzo, University of Rome ‘La Sapienza’, Italy) were used for immunoblotting experiments.
Enzymatic activity of PGs was determined as an increase in reducing end-groups over time. Reducing end-groups were measured using the method of Milner and Avigad (1967), as slightly modified by Sella et al. (2004) using d-galacuronic acid as a standard. The incubation mixture contained 200 μl of 0.5% polygalacturonic acid (PGA, from orange, Sigma; estimated average degree of polymerization, DP = 37) in 100 mM sodium acetate, pH 4.7. After enzyme addition, the mixture was incubated at 30°C for 30 min. One enzyme unit was defined as the amount of enzyme required to release 1 nmol per second of reducing end groups (nkat). The same mixture containing 0.018 nkat was used to assess the inhibitory activity of PGIP. Relative inhibitory activities were calculated as the ratio between the amount of protein extract required to inhibit by 50% 0.018 nkat of the specific PG at pH 4.7 and that required to inhibit by 50% 0.018 nkat of PGb of S. sclerotiorum, taken as a standard. The assays were performed in triplicates and repeated three times.
Results
Isolation and characterization of rice and wheat Pgip genes
To identify rice Pgip sequences we performed Blast searches on the TIGR Rice EST database (http://www.tigrblast.tigr.org/tgi/) and on the International Nucleotide Sequence Databases (http://www.ncbi.nlm.nih.gov/) by using, in separate searches, the coding region of a representative set of Pgip genes so far reported (bean, soybean, pear, Arabidopsis, grape, tomato, Brassica napus and Citrus sinensis). We identified four different sequences on the BAC clone P0668H12 (accession number AC084818) and two additional sequences, OsFOR1 (Jang et al. 2003) and LOC_Os09g31450, located on different regions of the rice genome, showing a slightly divergent sequence from the other four clustered genes. In particular, the putative mature coding region of the LOC_Os09g31450 contains a higher number of cysteine codons (11 codons) with respect to the typical 8–9 codons so far reported for all PGIP genes (De Lorenzo et al. 2001).
By performing similar Blast searches on the TIGR Wheat EST database and on the International Nucleotide Sequence Databases using the above mentioned Pgip genes and the newly identified rice Pgip genes, we found two overlapping ESTs (BJ280223 and BJ285213, now included in the tentative contig TC30134), sharing more than 60% identity with the rice Pgip genes.
In a recent database search (April 2006), it was confirmed that the selected rice and wheat sequences are those showing the higher sequence similarity to Pgip genes so far reported.
In order to investigate the genomic organization and functional features of the selected Pgip sequences and their encoded products, we isolated and sequenced the corresponding genes from the rice cv. Roma and the hexaploid wheat cv. Chinese Spring. For the rice Pgip sequences we limited our analysis to the four genes clustered on the BAC clone P0668H12, because OsFOR1 has been already characterized (Jang et al. 2003) and the LOC_Os09g31450, as specified above, is a more divergent sequence.
Each rice and wheat Pgip gene was amplified by PCR using specific primers spanning the complete coding region.
The Pgip genes isolated from the rice cv. Roma, named Ospgip1, Ospgip2, Ospgip3 and Ospgip4 contained open reading frames (ORFs) of 927, 1,029, 1,020 and 1,050 bp, respectively. These genes were identical to the corresponding Pgip genes from which the primers were derived, except Ospgip2 that showed two non-synonymous substitutions.
The encoded products of the four sequences show the typical PGIP topology with a putative signal peptide (Fig. 1a) and an LRR domain (Fig. 1c) flanked by short N- and C- terminal regions (Fig. 1b, d, respectively). The signal peptides vary between 17 (OsPGIP1), 22 (OsPGIP2), 21 (OsPGIP3) and 25 (OsPGIP4) amino acids (predicted by SignalP, www.expasy.org) and they should direct the mature protein into the apoplast (predicted by PSORT, www.expasy.org). OsPGIP2, OsPGIP3 and OSPGIP4 contain ten LRR modules, whereas OsPGIP1 possesses only nine LRR because the seventh LRR is missing (Fig. 1). The consensus sequence of the LRR region is identical in all four genes (xxL(D/N)LSxNxLxGxIPxxLxxLxxL) and matches precisely the extracytoplasmic LRR consensus sequence of resistance genes (Jones and Jones 1997). Additional differences between the four mature OsPGIPs reside in the number of cysteine residues and predicted glycosylation sites. Besides the conserved eight cysteines residues, OsPGIP2, OsPGIP3 and OsPGIP4 contain an extra cysteine at corresponding positions in the C-terminal region (Fig. 1). The predicted glycosylation sites are 4, 10, 7 and 5 in OsPGIP1, OsPGIP2, OsPGIP3 and OSPGIP4, respectively (Fig. 1). Only three of these potential glycosylation sites are in corresponding positions in all four proteins and one of them, located in the last LRR, is conserved also in the bean PGIPs (D’Ovidio et al. 2004). OsPGIP1, OsPGIP2, OsPGIP3 and OsPGIP4 have a calculated pI of 6.6, 4.6, 5.7 and 7.8, respectively. These values, except that of OsPGIP4, are much lower than that observed in PGIPs of dicot species that have pI values around 8.0 and 9.0.
Sequence organization and variability of OsPGIPs. Deduced amino acid sequences of rice PGIPs (OsPGIP1-4) are divided in four regions (a, b, c and d) and aligned. Numbering is referred to the OsPGIP1 sequence and starts from the first residue of the mature protein as predicted by SignalP, www.expasy.org. Regions are on the basis of crystallographic analysis of the bean PvPGIP2 (Di Matteo et al. 2003) and represent the signal peptide (a), the N-terminal region (b), the modular LRR region (c) and the C-terminal region (d). The xxLxLxx region is boxed. Empty spaces indicate gaps. Cysteine residues are in bold; putative glycosylation sites are underlined and in italics. The consensus sequence of the LRR region is: xxL(D/N)LSxNxLxGxIPxxLxxLxxL, where L is an aliphatic residues L, I, M, V, F and x indicates any residue. Where single residues are shown these comprise more than 50% of the residues at this position. Where two residues are shown, the two residues together comprise more than 50% of the residues at that position, with the upper of the two being more frequent
The wheat Pgip gene was amplified from the genomic DNA of cv. Chinese Spring using the primers PGIPWH1F/PGIPWH1R and the single amplicon obtained, named Tapgip1, contained an ORF of 1,011 bp that showed only a single synonymous nucleotide substitution when compared with TC30134. The protein encoded by this gene possesses the typical PGIP structure including a putative signal peptide (Fig. 2a) and the characteristic 10 LRR modules (Fig. 2c) flanked by conserved N- and C- terminal regions (Fig. 2b and d, respectively). The most likely cleavage site of the signal peptide is between position 26 and 27 (predicted by SignalP, www.expasy.org) and the resulting mature protein may be directed into the vacuole (90%) or in the apoplast (82%) (predicted by PSORT, www.expasy.org). Similarly to the rice OsPGIPs, the consensus sequence of the LRR matches precisely the extracytoplasmic LRR consensus sequence of resistance genes (Jones and Jones 1997) and contains the typical eight cysteine residues at conserved positions: four at N-terminus, one in the tenth LRR and three at the C-terminus (Fig. 2). The protein should be also glycosylated since five Asn-Xaa-Ser/Thr sequons are present and that in the last LRR is also conserved in the rice PGIPs. Like the rice OsPGIPs the calculated pI of the mature TaPGIP1 differs greatly from those of dicot PGIPs, being 6.05.
Sequence organization and variability of TaPGIPs. Deduced amino acid sequence of wheat PGIPs are divided in regions and aligned. Numbering is referred to the TaPGIP1 sequence and starts from the first residue of the mature protein as predicted by SignalP, www.expasy.org. Regions are on the basis of crystallographic analysis of the bean PvPGIP2 (Di Matteo et al. 2003). See legend of Fig. 1
Chromosomal localization of rice and wheat Pgip genes
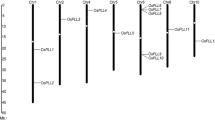
As described above, Ospgip1, Ospgip2, Ospgip3 and Ospgip4 are clustered on the same BAC clone P0668H12. This clone has been assigned to the short arm of chromosome 5 and the four Pgip genes are distributed over a 30 kbp region at distances varying from 2.8 kbp (Ospgip1 and Ospgip2) in one direction, and 16.6 kbp (Ospgip1 and Ospgip3) and 24.2 kbp (Ospgip1 and Ospgip4) in the opposite direction.
Since the large wheat genome has not been sequenced yet, the genomic organization of the wheat Pgip genes has been examined by Southern blot analysis using the Tapgip1 as probe. Two main hybridizing fragments of about 3.7 and 4.0 kbp were detected on genomic DNA of T. aestivum cv. Chinese Spring digested with EcoRI (Fig. 3a). The same analysis performed on genomic DNA of T. durum cultivar Langdon identified only a main EcoRI hybridizing fragment of 4.0 kbp (Fig. 3a).
Chromosomal localization of wheat Pgip genes. Southern blotting of genomic DNA (10 μg) digested with EcoRI and probed with Tapgip1. T. aestivum cv. Chinese spring (1, 5) and its ditelosomic lines lacking chromosome arms 7BS (6) and 7DS (7); T. durum cv. Langdon (2) and its D-substitution lines of chromosomes 7A (3) and 7B (4). The lack of hybridization fragment indicates the specific chromosomal assignment. The presence in both Langdon substitution lines of the 3.7 kbp fragment indicates its localization on chromosome 7D
In order to identify the chromosomal localization of these hybridizing fragments, we performed Southern blot analysis of genomic DNA of both nulli-tetrasomic lines of T. aestivum cv. Chinese Spring and of the D-genome substitution lines of T. durum cv. Langdon. These analyses demonstrated that the 3.7 kbp and 4.0 kbp EcoRI fragments were localized on chromosome 7D and 7B, respectively (Fig. 3b). The chromosomal localization of the Pgip hybridizing fragments was further defined by Southern blot analyses on genomic DNA of ditelosomic lines of T. aestivum cv. Chinese Spring. Based on these analyses the 3.7 and 4.0 kbp EcoRI fragments were assigned to the short arm of chromosome 7D and 7B, respectively (Fig. 3C).
Isolation of Pgip gene homeologs in wheat
In order to isolate wheat Pgip gene homeologs, we used a strategy based on PCR amplification of genomic DNA of nulli-tetrasomic (NT) lines of T. aestivum cv. Chinese Spring. On the basis of the chromosomal assignment described above, we amplified separately the genomic DNA of N7BT7A and N7DT7A by using PGIPWH1F/PGIPWH1R primers and the resulting amplicons were subjected to direct nucleotide sequencing. The nucleotide sequence of the amplicon obtained from N7DT7A was identical to Tapgip1, whereas that from the N7BT7A showed a few differences consisting of single substitutions and two short deletions in the N-terminal part of the coding region. This novel wheat Pgip sequence, named Tapgip2, contains an ORF of 999 bp and the deduced protein of 333 amino acids contains a signal peptide (23 residues) and the characteristic 10 LRRs flanked by conserved N- and C- terminal regions. Similarly to what predicted for TaPGIP1, the mature TaPGIP2 may be directed into the vacuole (90%) or, with a lower probability, into the apoplast (38%) (Predicted by PSORT, www.expasy.org). Like TaPGIP1, TaPGIP2 contains the typical eight-cysteine residues at conserved positions, five putative glycosylation sites (Fig. 2) and a calculated pI of 6.25.
To further verify the genome origin of Tapgip1 and Tapgip2, we developed the gene-specific primer pairs PGIPWH1F/PGIPWH10R, specific for Tapgip1, and PGIPWH1F/PGIPWH11R, specific for Tapgip2. PCR amplifications using these primers on genomic DNAs of nulli-tetrasomic lines of homeologous group 7 chromosomes of T. aestivum cv. Chinese Spring demonstrated that Tapgip1 and Tapgip2 are located on chromosome 7B and 7D, respectively (Fig. 4).
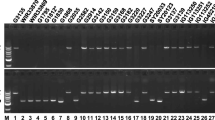
Chromosomal assignment of Tapgip1 and Tapgip2. Agarose gel of 1.6% PCR products obtained with the primer pair PGIPWH1F/PGIPWH10R specific for Tapgip1 (lanes 5–8) or with the primer pair PGIPWH1F/PGIPWH11R specific for Tapgip2 (lanes 1–4). The lack of amplification product indicates the specific chromosomal assignment. 1,5 T. aestivum cv. Chinese spring; 2, 6 N7AT7B; 3, 7 N7BT7A; 4, 8 N7DT7A
Transcript analysis of wheat and rice Pgip genes
Since only sequences corresponding to Ospgip1 and Tapgip1 were present in the EST databases, we verified by RT–PCR analysis whether the expression of the identified wheat and rice Pgip genes occurs in plant tissues during normal plant growth and development. In addition to the previously mentioned primers specific for Tapgip1 and Tapgip2, we developed gene-specific primers for each of the four Ospgips and used them to amplify total RNA extracted from roots, leaves and spikes. The amplification of wheat RNAs showed that Tapgip1 and Tapgip2 were expressed in roots, leaves and spikes. Conversely, when the analysis was performed on rice RNAs, only Ospgip1 and to a lesser extent Ospgip4 were clearly expressed in roots, leaves and flowering spikes. Ospgip3 was slightly expressed in all three tissues and Ospgip2 was expressed only in roots (Fig. 5).
Differential expression of Pgip genes in different tissues of rice and wheat plants. RT–PCR was performed on total RNA using gene-specific primers and the amplification products fractionated on 2% agarose gel. Amplicon sizes were: Ospgip1,137 bp; Ospgip2, 315 bp; Ospgip3, 102 bp; Ospgip4, 106 bp; Tapgip1, 219 bp; Tapgip2, 213 bp; 18SRNA (18S), 180 bp. S spikes, L leaves, R roots
Sequence variability of wheat and rice PGIPs
Sequence alignment of the deduced mature protein and subsequent cluster analyses showed that wheat PGIPs group separately from OsPGIPs, and share an average sequence identity of 55%. The wheat PGIPs share a very high sequence identity (88%) whereas the OsPGIPs are separated into two distinct sub-groups formed by the pairs OsPGIP1/OsPGIP2 (60% identity) and OsPGIP3/OsPGIP4 (79% identity), sharing only 56% of sequence identity (Fig. 6). Sequence analysis showed also that OsFOR1 and LOC_Os09g31450 form a cluster separated from OsPGIPs and wheat PGIPs and shares with them 43% sequence identity (Fig. 6). A similar clustering result has been obtained in Neighbor-Joining trees generated from the same multiple sequence alignment (data not shown), although the TaPGIP and OsPGIP clusters have bootstrap values ≤55.
UPGMA tree showing the relationships between the mature proteins of PGIPs from monocots and a representative number of PGIPs from dicots. Species origin and accession number in parenthesis. Numbers at branch points represent average identities. Dicots: AdPGIP1 (Actinidia deliciosa, Z49063); AtPGIP1, AtPGIP2 (Arabidopsis thaliana, AF229249, AF229250); BnPGIP1, BnPGIP2, (Brassica napus, AF529691, AF529693); CsPGIP1 (Citrus sinensis, Y08618); EgPGIP1 (Eucalyptus grandis, AF159167); GmPGIP1, GmPGIP2, GmPGIP3, GmPGIP4 (Glycine max, AJ972660 to AJ972663); LePGIP1 (Lycopersicon esculentum, L26529); PcPGIP1 (Pyrus communis, L09264); PvPGIP1, PvPGIP2, PvPGIP3, PvPGIP4 (Phaseolus vulgaris, AJ786408 to AJ786411); VvPGIP1 (Vitis vinifera, AF499451). Monocots: TaPGIP1, TaPGIP2 (Triticum aestivum, this paper, AM180656, AM180657); OsPGIP1, OsPGIP2, OsPGIP3, OsPGIP4 (Oryza sativa, this paper, AM180652 to AM180655); OsFOR1 (Oriza sativa, AF466357); Os09g31 (Oriza sativa, the LOC_Os09g31450 sequence is located on the clone OSJNBb0052C07 with accession number AC108762)
In order to ascertain the distribution of sequence variation along these proteins, sequence comparison was also performed between the different regions of wheat or rice PGIPs. Regions were based on the crystallographic analysis of the bean PvPGIP2 (Di Matteo et al. 2003) and included the N-terminal portion (region B), the LRR region with and without the xxLxLxx motifs (region C and region C-out, respectively), the portion corresponding to the xxLxLxx motifs with and without the conserved hydrophobic residues (xxLxLxx and xx(L)x(L)xx regions, respectively), and the C-terminal portion (region D). This analysis revealed an alternating pattern of variable and conserved sites among the different regions of TaPGIP1 and TaPGIP2 (S1). In particular, the xxLxLxx region showed a more pronounced variation as compared to the B, D and C regions. For example, the xx(L)x(L)xx regions share 70% of sequence identity in contrast with the 93% of identity of the C-out regions (S1). The same analysis performed between the rice PGIPs showed a more uniform variation between the different regions, except for the pair OsPGIP3/OsPGIP4 where the xx(L)x(L)xx region, similar to what observed between TaPGIP1 and TaPGIP2, showed a stronger variability (62.7%) as compared to the C-out region (81.9%)(S2).
Monocot and dicot PGIPs possess a similar extent of sequence variability
Sequence comparison, at both nucleotide and amino acid levels, between all known PGIP sequences showed that TaPGIPs and OsPGIP form a cluster separated from the dicot PGIPs (Fig. 6). The range of sequence identity between the two groups varies between 47 and 61% at nucleotide level, and 33 and 47% at protein level. For example, TaPGIP1 showed the lowest and highest protein sequence identity with the soybean GmPGIP1 (35%) and pear PcPGIP1 (42%), respectively, whereas OsPGIP1 shared 36% of identity with the soybean GmPGIP2 and 47% of identity with tomato LePGIP1. This range of sequence identity is close to the extent of variability observed within each of the two groups. For example, PGIPs from bean and soybean share a protein sequence identity of 42–49% with most PGIP sequences from other dicot species (Fig. 6), whereas wheat and rice PGIPs possess a sequence identity within the range 42–54%. The clustering results reported in Fig. 6 were also confirmed with the Neighbor Joining method using the same multiple sequence alignment (data not shown). In this analysis the bootstrap values of the major groups (monocot PGIPs, dicot PGIPs excluding legumes and legume PGIPs) have bootstrap values ≥99.
OsPGIP1 is an effective inhibitor of fungal PGs
Since the most striking variation observed in the deduced products of the rice and wheat Pgip genes was the lack of the entire seventh LRR in OsPGIP1 (Fig. 1), and since this type of variation has not been observed in the PGIPs so far reported, we analyzed the inhibitor activities of this protein against different fungal PGs. The expression of Ospgip1 was performed in Nicotiana benthamiana using a vector based on potato virus X (PVX; Baulcombe et al. 1995). Western blotting analyses using an antibody raised against the bean PGIP showed the presence of a strong hybridizing bands of the expected size in the total protein extract of N. benthamiana plants inoculated with PVX-Ospgip1, while no hybridising band was detected in extracts prepared from control plants (inoculated with the empty vector) (Fig. 7).
The same protein extract was then used to test the activity of OsPGIP1 against five PGs from fungal pathogens (Table 1). As positive control of the assays we have used the soybean GmPGIP3, known to have a wide spectrum of inhibitor activities against fungal PGs (D’Ovidio et al. 2006).
Either total protein extract (<1 μg) from N. benthamiana plants infected with PVX-Gmpgip3 or PVX-Ospgip1 was able to inhibit completely the different PGs used, except that of F. moniliforme that was not affected by the latter sample (Table 1). On the contrary, total protein extract (up to 1 μg) from N. benthamiana plants infected with the empty vector (PVX-201) did not affect the activity of the PGs used, except that of B. cinerea, that was inhibited to a maximum of 20% with 1 μg of protein extract. In this case, the inhibition value was subtracted from that obtained with the PVX-pgip samples. Control samples heated at 100°C for 10 min did not affect the activity of none of the PG tested.
As shown in Table 1, the inhibitor activity of OsPGIP1 varies between the analysed PGs and is mostly effective against the PGb of S. sclerotiorum. Comparison between OsPGIP1 and the very efficient GmPGIP3 showed that OsPGIP1 is less effective in inhibiting the PGs of F. moniliforme, A. niger and B. cinerea, but is more active against the purified PG of F. graminearum.
Discussion
Similarly to dicot plant species, wheat and rice genomes contain a few Pgip genes and, as in the majority of species so far analyzed, they are not interrupted by introns. The rice Pgip genes are located on chromosome 5, clustered on a short genomic region spanning 30 kbp. A clustered organization of Pgip genes has been reported also in Arabidopsis, where Atpgip1 and Atpgip2 are located 507 bp apart on chromosome 5 (Ferrari et al. 2003); in bean, where the four Pgip genes (Pvpgip1, Pvpgip2, Pvpgip3 and Pvpgip4) are distributed over a 50 kbp region (D’Ovidio et al. 2004) and in soybean where Gmpgip1 and Gmpgip2 are about 3 kbp apart (D’Ovidio et al. 2006). Moreover, two additional soybean members (Gmpgip3 and Gmpgip4) have been identified in a BAC clone harbouring a 60 kbp insert (D’Ovidio et al. 2006), and Bnpgip3 and Bnpgip4 are tandemly arrayed in Brassica napus cv. Westar (Li et al. 2003).
Despite their cluster organization, additional Pgip genes can be found at different genome locations. In rice, OsFOR1, a more divergent Pgip gene involved in flower development, is located on chromosome 7 (Jang et al. 2003) and in B. napus line DH12075, Bnpgip1 and Bnpgip2 are located on separate chromosomal regions (Li et al. 2003).
Genomic organization of Pgip genes in wheat has been analyzed by Southern blot analyses of genomic DNA of aneuploid lines and the two main EcoRI hybridizing fragments of about 4.0 and 3.7 kbp, have been assigned to the short arm of chromosome 7B and 7D, respectively. Accordingly, gene-specific PCR analyses assigned Tapgip1 and Tapgip2 to the short arm of chromosome 7B and 7D, respectively.
The lack of Pgip hybridizing fragments from the A genome of both T. durum cv. Langdon and T. aestivum cv. Chinese Spring can be due to gene loss or to a marked sequence rearrangement that could strongly reduce the capability of the probe to detect the target sequence. Several studies have shown that polyploidization can have many consequences on gene expression and genome organization (Adams and Wendel 2005). In wheat, a number of deletion events have been reported to occur at the polyploid level (Akhunov et al. 2003) and, by using newly synthesized wheat allotetraploids, it has been also shown that gene loss and gene silencing occur immediately upon allopolyploid formation (Kashkush et al. 2002). Activation of retro-transposons has been also reported to occur during allotetraploid formation in wheat (Kashkush et al. 2003) and the insertion of a Wis-2 retroelement has been identified within the coding region of the Ay high molecular weight glutenin subunit gene (Harberd et al. 1987). On this basis, the analysis of Pgip genes in the A-genome progenitor, Triticum urartu, and in other wild wheat relatives should shed light on the genomic events responsible for the lack of PGIP hybridizing fragments in the A-genome of durum and bread wheat.
Comparative mapping studies have shown that homoeologous group 7 chromosomes of wheat shares large blocks of synteny with the rice chromosomes 6 and 8 (Kurata et al. 1994; Sorrells et al. 2003; Hossain et al. 2004). These studies revealed also that these blocks of synteny are broken by a few sequences located on different rice chromosomes (for a map figure see http://www.tigr.org/tdb/synteny/wheat/figureview_desc.shtml). The finding that Pgip genes are located on the short arms of both rice chromosome 5 and wheat chromosomes 7 contributes to define better the extent of the region and the type of sequences interrupting the synteny between the wheat homoeologous group 7 chromosomes and the rice chromosomes 6 and 8.
The deduced proteins from both rice and wheat Pgip genes show the typical LRR modular organization flanked by non repetitive N- and C-terminal regions and a conserved number and distribution of cysteine residues, as reported for PGIPs from dicot species. Despite the conservation of these structural features, the overall amino acid sequence identity between monocot (wheat and rice) and dicot PGIPs can reach values as low as 34%, as between OsPGIP3 and the soybean GmPGIP2. This low value of sequence identity, however, is not surprising if we consider that legume and other dicot PGIPs share an average sequence identity of 45%.
As is observed in bean (D’Ovidio et al. 2004) and soybean (D’Ovidio et al. 2006) PGIPs, sequence variation between TaPGIP1 and TaPGIP2 is distributed along the entire sequence, but it is more pronounced within the xxLxLxx region. This observation is particularly evident when considering the high value of sequence identity between their C-out regions (93%) and the significant lower value shared between their xx(L)x(L)xx regions (70%). On the basis of the proposed role of ligand recognition for the xxLxLxx region, the higher sequence variation within this region might be indicative of a functional diversification of the two products. As proposed by Di Matteo et al. (2003), a stable interaction between PG and PGIP likely requires a network of multiple and relatively weak contacts and only one or very few strong contacts that “lock” the complex. These limited numbers of locking contacts may be different in different PGIP–PG interactions and involve non-conserved residues within or close the xxLxLxx region.
The same sequence comparison performed between the rice PGIPs showed a more uniform variation along the sequences. Only the comparison between OsPGIP3 and OsPGIP4 showed an increase of sequence variation within the xxLxLxx region. The C-out regions of these proteins share 82% of sequence identity in contrast with the 62% of identity between their xx(L)x(L)xx regions. Whether the OsPGIPs represent redundant or diversified functional products has to be verified experimentally; however, their low degree of overall identity (about 54%) suggests that functional diversification could have been achieved through multiple changes along the entire sequence. It is also possible that the high sequence variation between members reflects a different physiological role in planta, including the possibility to interact with a different ligand molecule. The recent reports on the capabilities of PvPGIP2 to interact with pectins (Mattei B. personal communication) and the effect of OsFOR1, a rice protein possessing PG inhibiting capabilities, on the formation and/or maintenance of floral organ primordia (Jang et al. 2003) support this possibility.
A first evidence of a functional diversification of Ospgip genes derives from their different regulation during plant growth and development. Our transcript analysis demonstrated that Ospgip1 and Ospgip4 are clearly expressed in spikes, leaves and roots, whereas Ospgip3 is weakly expressed in all three tissues and Ospgip2 is expressed only in roots. Similar to Ospgip1 and Ospgip4, both Tapgip genes accumulate at comparable levels in spikes, leaves and roots. A constitutive or a more specific expression of the different members of a Pgip family has been already reported in dicot species. In bean, Pvpgip2, Pvpgip3, Pvpgip4 are expressed in 5-day-old seedlings, whereas Pvpgip1 is expressed only following a wounding stress (D’Ovidio et al. 2004). Similarly, the soybean Gmpgip1, Gmpgip3, Gmpgip4 are expressed in 7-day-old seedlings, whereas Gmpgip2 is expressed only following Sclerotinia sclerotiorum infection (D’Ovidio et al. 2006). In Brassica napus, Bnpgip1 is more strongly expressed than Bnpgip2 in flower buds, but both of them are expressed at similar levels in roots, open flowers and stems, and are not expressed in leaf blade (Li et al. 2003). In Arabidopsis, Atpgip1 and Atpgip2 are up regulated coordinately in response to Botrytis cinerea infection, but through separate signal transduction pathways. Moreover, Atpgip1, but not Atpgip2 transcripts, accumulate in seedlings following a cold treatment (Ferrari et al. 2003).
PGIPs are glycosylated proteins (Mattei et al. 2001) and, accordingly, the deduced proteins encoded by both Tapgips and Ospgips possess a few putative glycosylation sites. These are variable in number and distribution between the different PGIP members and only the one within the tenth LRR was conserved in both wheat, rice and in the majority of PGIPs from dicot species. In spite of this evident conservation, however, this site has been found not occupied by any polysaccharide in the bean PGIP2 (Mattei et al. 2001).
A clear difference between monocot and dicot PGIPs is represented by the values of their calculated pIs. Whether this feature is maintained in the native proteins has to be proven experimentally, however, both wheat and rice PGIPs, except OsPGIP4, have calculated pIs ranging between 4.7 (OsPGIP2) and 6.6 (OsPGIP1) that are much lower than those observed in dicot PGIPs, showing pI values >8.0.
As previously mentioned, rice and wheat PGIPs show the characteristic LRR modular organization composed by 10 imperfect repeats of about 24 amino acids. This feature is conserved in all Pgip gene products so far characterized (De Lorenzo et al. 2001) but now we report, for the first time, the occurrence of a Pgip gene encoding a product with nine LRRs. We have already reported that gene duplication and subsequent diversification, by substitutions or short deletions, could have played an important role in the evolution of the bean Pgip family (D’Ovidio et al. 2004). The finding of nine LRRs in OsPGIP1 demonstrates that deletion events can be also responsible for generating variation in the LRR copy number of a Pgip gene. We do not know the molecular mechanism responsible for this variation, but unequal crossing-over or slippage during replication is a likely process. The repetitive structure of the LRR region (about 50% average nucleotide identity between all nine repeats of Ospgip1 and 63% nucleotide identity between repeats 3 and 4) should facilitate the occurrence of such processes leading to expansion and contraction of the LRR copy number, as already reported for some resistance genes, such as the RPP5 in Arabidopsis (Parker et al. 1997), the M in flax (Anderson et al. 1997) and the Cf-2/Cf5 gene family in tomato (Dixon et al. 1998).
By sequence comparison and inhibition assays we demonstrated that OsPGIP1 lacks the entire seventh LRR and is an active inhibitor of fungal PGs. Comparison between the inhibitory activities of OsPGIP1 and those of the very efficient soybean GmPGIP3 showed that OsPGIP1 is less effective to inhibit the PGs of A. niger and B. cinerea but is more active against a purified PG of F. graminearum. The finding that OsPGIP1 is inactive against the PG of F. moniliforme is not surprising since this PG is resistant to most PGIP inhibitors, except PvPGIP2 and GmPGIP3 (D’Ovidio et. al. 2004, 2006; Sella et al. 2004).
In addition to the specific contribution that OsPGIP1 can exert in controlling the activity of fungal PGs, these results are of particular interest to define the molecular features of PG–PGIP interaction because they demonstrate that ten LRRs are not essential for the inhibitory activity of PGIP and that the lack of the entire seventh LRR does not cause the loss of activity against fungal PGs.
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Aguero CB, Uratsu SL, Greve C, Powell AT, Labavitch JM, Meredith CP, Dandekar AM (2005) Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol Plant Pathol 6:43–51
Akhunov ED, Akhunova AR., Linkiewicz AM, Dubcovsky J, Hummel D et al (2003) Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. PNAS 100:10836–10841
Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9:641–651
Baulcombe D, Chapman S, Santa Cruz S (1995) Jellyfish green fluorescent protein as a reporter for virus infections. Plant J 7:1045–1053
Caprari C, Mattei B, Basile ML, Salvi G, Crescenzi V, De Lorenzo G, Cervone F (1996) Mutagenesis of endopolygalacturonase from Fusarium moniliforme: histidine residue 234 is critical for enzymatic and macerating activities and not for binding to polygalacturonase-inhibiting protein (PGIP). Mol Plant–Microbe Interact 9:617–624
Cervone F, De Lorenzo G, Degrà L, Salvi G (1987) Elicitation of necrosis in Vigna unguiculata Walp. by homogeneous Aspergillus niger endo-polygalacturonase and by a-d-galacturonate oligomers. Plant Physiol 85:626–630
D’Ovidio R, Anderson OD (1994) PCR analysis to distinguish between alleles of a member of a multigene family correlated with wheat quality. Theor Appl Genet 88:759–763
D’Ovidio R, Raiola A, Capodicasa C, Devoto A, Pontiggia D, Roberti S, Galletti R, Conti E, O’Sullivan D, De Lorenzo G (2004) Characterization of the complex locus of Phaseolus vulgaris encoding polygalacturonase-inhibiting proteins (PGIPs) reveals sub-functionalization for defense against fungi and insects. Plant Physiol 135:2424–2435
D’Ovidio R, Roberti S, Di Giovanni M, Capodicasa C, Melaragni M, Sella L, Tosi P, Favaron F (2006) The characterization of the soybean Pgip family reveals that a single members is responsible for the activity detected in soybean tissues. Planta, DOI 10.1007/s00425-006-0235-y
De Lorenzo G, D’Ovidio R, Cervone F (2001) The role of polygacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol 39:313–335
Desiderio A, Aracri B, Leckie F, Mattei B, Salvi G, Tigelaar H, Van Roekel JS, Baulcombe DC, Melchers LS, De Lorenzo G, Cervone F (1997) Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol Plant–Microbe Interact 10:852–860
Di Matteo A, Federici L, Mattei B, Salvi G, Johnson KA, Savino C, De Lorenzo G, Tsernoglou D, Cervone F (2003) The crystal structure of PGIP (polygalacturonase-inhibiting protein), a leucine-rich repeat protein involved in plant defense. Proc Natl Acad Sci USA 100:10124–10128
Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10:1915–1926
Favaron F (2001). Gel detection of allium porrum polygalacturonase-inhibiting protein reveals a high number of isoforms. Physiol Mol Plant Pathol 58:239–245
Favaron F, Castiglioni C, D’Ovidio R, Alghisi P (1997) Polygalacturonase inhibiting proteins from Allium porrum L. and protection of plant tissue from fungal endo-polygalacturonase degradation. Physiol Mol Plant Pathol 50:403–417
Favaron F, Castiglioni C, Di Lenna P (1993) Inhibition of some rot fungi polygalacturonases by Allium cepa L. and Allium porrum L. extracts. J Phytopathol 139:201–206
Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F (2006) Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci 11:65–70
Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Arabidopsis polygalacturonase-inhibiting proteins (PGIP) are regulated by different signal transduction pathways during fungal infection. Plant Cell 15:93–106
Harberd NP, Flavell RB, Thompson RD (1987) Identification of a transposon-like insertion in a Glu-1 allele of wheat. Mol Gen Genet 209:326–332
Hossain KG, Kalavacharla V, Lazo GR, Hegstad J, Wentz MJ et al. (2004) A chromosome bin map of 2,148 expressed sequence tag loci of wheat homoeologous group 7. Genetics 168:687–699
Jang S, Lee B, Kim C, Yim J, Han J-J, Lee S, Kim S-R, An G (2003) The OsFOR1 gene encodes a polygalacturonase-inhibiting protein (PGIP) that regulates floral organ number in rice. Plant Mol Biol 53:357–369
Jones DA, Jones JDG (1997) The roles of leucine rich repeats in plant defences. Adv Bot Res. 24:90–167
Joppa LR, Williams ND (1988) Langdon durum disomic substitution lines and aneuploid analysis in tetraploid wheat. Genome 30:222–228
Kashkush K, Feldman M, Levy A (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160:1651–1659
Kashkush K, Feldman M, Levy A (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33:102–106
Kemp G, Bergmann CW, Clay R, Van der Westhuizen AJ, Pretorius ZA (2003) Isolation of a polygalacturonase-inhibiting protein (PGIP) from wheat. Mol Plant–Microbe Interact 16:955–961
Kurata N, Moore G, Nagamura Y, Foote T, Yano M, Minobe Y, Gale M (1994) Conservation of genome structure between rice and wheat. Bio/technology 12:276–278
Leckie F, Mattei B, Capodicasa C, Hemmings A, Nuss L, Aracri B, De Lorenzo G, Cervone F (1999) The specificity of polygalacturonase-inhibiting protein (PGIP): a single amino acid substitution in the solvent-exposed β- strand/β-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J 18:2352–2363
Li R, Rimmer R, Yu M, Sharpe AG, Seguin-Swartz G, Lydiate D, Hegedus DD (2003) Two Brassica napus polygalacturonase inhibitory protein genes are expressed at different levels in response to biotic and abiotic stresses. Planta 217:299–308
Lin W, Li Z (2002) The partial structure of wheat polygalacturonase-inhibiting protein. Zhongguo Shengwu Huaxue Yu Fenzi Shengwu Xuebao 18:197–201
Manfredini C, Sicilia F, Ferrari S, Pontiggia D, Salvi G, Caprari C, Lorito M, DeLorenzo G (2005). Polygalacturonase-inhibiting protein 2 of Phaseolus vulgaris inhibits BcPG1, a polygalacturonase of Botrytis cinerea important for pathogenicity, and protects transgenic plants from infection. Physiol Mol Plant Pathol 67:108–115
Mattei B, Bernalda MS, Federici L, Roepstorff P, Cervone F, Boffi A (2001) Secondary structure and post-translation modifications of the leucine-rich repeat protein PGIP (polygalacturonase-inhibiting protein) from Phaseolus vulgaris. Biochemistry 40:569–576
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25
Milner Y, Avigad G (1967) A copper reagent for the determination of hexuronic acids and certain ketohexoses. Carbohydr Res 4:359–361
Oeser B, Heidrichm PM, Muller U, Tudzynski P, Tenberge KB (2002) Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genet Biol 36:176–186
Parker JE, Coleman MJ, Szabo V, Frost LN, Schmidt R, van der Biezen EA, Moores T, Dean C, Daniels MJ, Jones JDG (1997) The Arabidopsis Downy mildew resistance gene Rpp5 shares similarity to the Toll and INterleukin-1 receptors with N and L6. Plant Cell 9:879–894
Powell AL, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant–Microbe Interact 13:942–950
Saitou N, Nei M (1987) The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sears E R (1966) Nullisomic-tetrasomic combination in hexaploid wheat. In: Riley R, Lewis KR (eds) Chromosome manipulation and plant genetics. Oliver and Boyd, Edinburg, pp 29–45
Sella L, Castiglioni C, Roberti S, D’Ovidio R, Favaron F (2004) An endo-polygalacturonase (PG) of Fusarium moniliforme escaping inhibition by plant polygalacturonase-inhibiting proteins (PGIPs) provides new insights into the PG–PGIP interaction. FEMS Microbiol Lett 240:117–124
Sneath P, Sokal R (1973) Numerical Taxonomy. Freeman, San Francisco
Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R et al (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827
Tai T, Tanksley S, (1991) A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue. Plant Mol Biol Rep 8:297–303
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–80
Acknowledgments
Research supported by MIUR (Ministero dell’Istruzione, dell’Università e della Ricerca; grants PRIN [Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale] 2005 and FIRB [Fondo per gli Investimenti della Ricerca di Base] 2002–2005) and by the European Community (grant QLK1-2000-00811).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Waugh.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Janni, M., Di Giovanni, M., Roberti, S. et al. Characterization of expressed Pgip genes in rice and wheat reveals similar extent of sequence variation to dicot PGIPs and identifies an active PGIP lacking an entire LRR repeat. Theor Appl Genet 113, 1233–1245 (2006). https://doi.org/10.1007/s00122-006-0378-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0378-z