Abstract
Ovarian cancer is a severe malignant tumour of the female genital organs. Six-transmembrane epithelial antigen of the prostate 1 (STEAP1) expression is correlated with the occurrence and progression of multiple cancers. Here, we assessed STEAP1 expression in ovarian cancer and explored the relationship between STEAP1 and ovarian cancer progression. We used immunohistochemistry and public databases to test STEAP1 expression in normal human ovarian tissues, benign ovarian tumours, and ovarian cancer. The expression of STEAP1 and epithelial-to-mesenchymal transition (EMT)-related genes was analysed using immunocytochemistry, quantitative reverse transcription polymerase chain reaction, and western blotting in ovarian cancer cell lines. Lentivirus was used to knockdown and overexpress STEAP1. Invasion, migration, growth, clonogenicity, and apoptosis were assessed using transwell assay, growth curve, plate clone formation assay, and flow cytometry. We used a tumour xenograft to verify the relationship between STEAP1 and in vivo ovarian cancer cell growth. Matrix metalloproteinase-2 (MMP2) and matrix metalloproteinase-9 (MMP9) activities were examined using Matrix metalloproteinase zymography assay. STEAP1 was highly expressed in the human ovarian cancer tissues and a highly invasive ovarian cancer cell line. Overexpression of STEAP1 was related to poor prognosis in ovarian cancer patients. Down-regulation of STEAP1 suppressed the invasion, migration, proliferation, clonogenicity, EMT progression in human ovarian cancer cells and xenograft tumour growth in vivo, but it enhanced apoptosis. In human ovarian cancer, the STEAP1 gene is highly expressed, and its function is correlated with human ovarian cancer cell metastasis and growth. STEAP1 may be a possible target for suppressing ovarian cancer metastasis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant tumours are the second cause of death worldwide (2017). It is reported that ovarian cancer ranks fourth in incidence and third in mortality among gynaecological tumours, and the survival rate of ovarian cancer is only 38–40% in 5 years (Bray et al. 2018; Allemani et al. 2015). Numerous patients are diagnosed with ovarian cancer at a late stage or experience recurrence due to nonspecific symptoms at an early stage, and the current chemotherapy regimens are prone to resistance development (Oberaigner et al. 2012; Vaughan et al. 2011). Higher malignancy of ovarian tumour with recurrence and metastasis predicts poor prognosis. Hence, it is important to explore invasion and metastasis mechanisms in ovarian cancer to seek possible molecular targets, improve cancer prognosis, and attenuate cancer recurrence and metastasis.
Hubert discovered a protein in prostate cancer tissue, named six-transmembrane epithelial antigen of the prostate 1 (STEAP1) (Hubert et al. 1999). And the protein, localized to the cell membrane particularly at cell junctions, is a six-transmembrane protein with both the N- and C-termini on the cytoplasmic side (Barroca-Ferreira et al. 2018). STEAP1 protein is highly expressed in multiple cancer tissues like prostate cancer (Gomes et al. 2018), lung cancer (Hayashi et al. 2011), Ewing sarcoma (Grunewald et al. 2012a), colorectal cancer (Nakamura et al. 2018), breast cancer(Maia et al. 2008), and gastric cancer (Wu et al. 2018). STEAP1, as a transmembrane protein, is thought to be involved in intercellular communication (Yamamoto et al. 2013). Moreover, it participates in tumour cell growth regulation, tumour aggressiveness, apoptosis, small molecule transport, and iron metabolism (Barroca-Ferreira et al. 2018). STEAP1 also contributes to cancer treatment. Some strategies and measures, such as DNA vaccines for STEAP1, anti-STEAP1 mAbs, and STEAP1 expression inhibition, have been used in cancer therapy research, including clinical trials and in vivo studies (Carrasquillo et al. 2019; Chen et al. 2019; O Donoghue et al. 2019), especially in prostate cancer.
The relationship between invasion and metastasis of ovarian cancer and STEAP1 expression has not been studied until now. This study is the first to illustrate the possible roles and mechanisms of STEAP1 in vivo and in vitro ovarian cancer invasion and metastasis, and assessed STEAP1’s potential as a target for improving ovarian cancer prognosis.
Materials and methods
Cell culture
We obtained human ovarian cancer cell lines ES-2, CAOV-3, and OVCAR-3 from Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd. The human ovarian cancer epithelial cell line SKOV-3 was purchased from Shanghai Institute for Biological Sciences. The ES-2 cell line was cultured in McCoy’s 5A Medium (BOSTER) with 10% foetal bovine serum (FBS; Clarkbio) and 1% penicillin–streptomycin solution (Merck). Caov-3 was grown in Dulbecco’s Modified Eagle’s Medium (Sigma) with 10% FBS (Clarkbio) and 1% penicillin–streptomycin solution (Merck). The OVCAR-3 cell line was grown in RPMI-1640 (Sigma) with 20% FBS (Clarkbio), 0.01 mg/ml bovine insulin (Solarbio), and 1% penicillin–streptomycin solution (Merck). The SKOV-3 cell line was grown in RPMI-1640 (Sigma) with 10% FBS (Clarkbio) and 1% penicillin–streptomycin solution (Merck). All cell lines were cultured at 37 °C with 5% CO2.
Tissue sample collection, immunohistochemistry and survival analysis
594 ovarian tissue samples were collected at Shandong provincial hospital from 2012 to 2018, including 65 human normal ovarian tissues, 145 benign tumour tissues, and 384 malignant tumour tissues. All 384 malignant tumour tissues were classified into three histological types (serous cystadenocarcinoma, mucinous cystadenocarcinoma, and endometrioid carcinoma), according to the WHO classification of tumours and pathological features (Kurman et al. 2014). All sampled ovarian cancer patients were diagnosed according to the tumour, node, metastasis staging system, and they had not undergone any treatment before tissue sample collection.
Tissue samples were fixed with 10% formalin for 72 h to denature proteins in tissues, embedded in paraffin, and processed into 4-μm-thick paraffin sections. Then sections were de-paraffinized in xylene for 5 min and we rehydrated the sections with different concentrations of alcohol (100%, 95%, 85%,70% for 5 min). Antigen retrieval was performed in citrate–citrate solution at 125 °C for 2 min. According to the mouse SP kit instruction, sections were incubated with 3% hydrogen peroxide (H2O2) for 20 min at 37 °C, blocked with serum, incubated with anti-STEAP1 antibody (1:100, ab207914, Abcam; diluted with PBS) for 24 h at 4 °C, washed, incubated with secondary antibody for 30 min at 37 °C, stained with the enzyme–substrate 3,3′-diaminobenzidine tetrahydrochloride (DAB). Negative control was performed with PBS, replacing anti-STEAP1 primary antibody, and incubated for 24 h at 4 °C. According to the instruction of primary antibody (specificity of the primary antibody: https://www.abcam.cn/steap1-antibody-j2d2-n-terminal-ab207914.html), human prostate hyperplasia tissues were treated as a positive control. STEAP1 antigens in the tissues were brown after combining with DAB. Finally, the light microscope (BX63F, OLIMPUS, Japan) and OLIMPUS imaging software cellSens 1.16 were used to acquire images of immunohistochemistry. The brightness and contrast ratio of all images was automatically adjusted by the software.
Three pathologists graded each section independently from 0 to 7 and translated the scores into the sum indexes (−), (+), (++), or (+++). According to the percentage of stained cells, a sample was scored 0, 1, 2, 3 or 4. In addition, a sample was scored 0, 1, 2 or 3 according to staining intensity. The sum of stained cells score and staining intensity score was the score for each section.
384 malignant ovarian tumour patients were divided into two groups (low expression, high expression) according to STEAP1 expression in immunohistochemistry. Kaplan–Meier analysis was performed for survival analysis of the two groups. All patients survival conditions were derived from follow-up records.
Analysis of public databases
The Kaplan–Meier Plotter database and Oncomine database were utilized in this study. The relationship between STEAP1 expression and ovarian cancer patient prognosis was investigated in the Kaplan–Meier Plotter database (http://kmplot.com/analysis/index.php?p=service&cancer=ovar). The Oncomine database (https://www.oncomine.org/resource/main.html) used to analyse STEAP1 expression in ovarian cancer tissues and normal ovarian tissues.
Immunocytochemistry
Cells were seeded into 24-well plates with cell slides and cultured in the medium for 24 h at 37 °C. The cells on the slide were washed with phosphate buffer solution (PBS) three times, fixed with 4% paraformaldehyde for 15 min, and washed with PBS for 5 min. Then cells on the slide were incubated with 3% H2O2 for 20 min at 37 °C, washed, and blocked with goat serum for 30 min. Cells on the slide were also incubated with anti-STEAP1 primary antibody (1:200, ab207914, Abcam; diluted with PBS) for 24 h at 4 °C to mark the antigen STEAP1, washed, and incubated with secondary antibody for 30 min at 37 °C, stained with DAB. Negative control was performed with PBS, replacing anti-STEAP1 primary antibody, incubated for 24 h at 4 °C. According to the instruction of primary antibody (https://www.abcam.cn/steap1-antibody-j2d2-n-terminal-ab207914.html), LNCap cells were treated as a positive control. STEAP1 antigens in the cells were brown. The more cells were browned, the darker the brown, the higher we thought STEAP1 expression was. Hematoxylin is used to stain the nucleus. Finally, acquisition of immunocytochemistry images was also performed by OLIMPUS BX63F microscope (Japan) and OLIMPUS imaging software cellSens 1.16. The brightness and contrast ratio of all images was automatically adjusted by the software.
RT-qPCR
Total RNA was extracted from cell lines using TRIZol reagent (TaKaRa Biotechnology, China), and RNA was resuspended at a concentration of 500 ng/μl. Total RNA (2 μl) was reverse transcribed to cDNA in a 20-μl reaction system (PrimeScript RT reagent Kit with gDNA Eraser; TaKaRa; China). RT-qPCR was performed in a 96-well plate with three technical replicate wells. A 20-μl reaction system in each well included 10 μl SYBR Premix EX Taq, 0.4 μl PCR Forward Primer, 0.4 μl PCR Reverse Primer, 2 μl cDNA, and 7.2 μl dH2O. RT-qPCR and analysis (relative quantitation gene expression) were performed using the Light Cycler 480 System (Applied Biosystems Inc; Roche, Inc). Specific primers were designed using Light Cycler Probe Design software (Roche Diagnostics, Basel, Switzerland) and synthesized by TaKaRa Biotechnology Co, Ltd. The primer sequences are listed in Table S1.
Invasion and migration assays
A Boyden chamber was used to perform transwell assays to assess the in vitro invasion and migration of cell lines. Boyden chamber, in which 50 μl mixture mixed with Matrigel (BD Biosciences) and culture medium without FBS (1:7 dilution) was spread out, was placed in the 24-well plate for 2 h at 37 °C to assess invasion. Cells (1.5 × 105) suspended in medium without FBS were seeded into the Boyden chamber mixture. To keep the exterior and interior level of the transwell consistent, we added 900 µl of medium with 20% FBS into the 24-well plate as a chemotactic factor. Cells were incubated for 24 h at 37 °C. Cells on the upper surface of the transwell were wiped, cells on the lower surface were soaked in 4% paraformaldehyde, stained with crystal violet, and washed gently. Finally, the membrane was cut from the transwell and fixed on the slide. Cells were counted using the inverted microscope (BX63F; Olympus, Japan) in five random fields. The migration assays were performed like the invasion assay but without Matrigel.
Western blotting
Radioimmunoprecipitation assay buffer containing 1 mM phenylmethylsulfonyl fluoride on ice was used to extract cell protein. Protein concentration was detected using the BCA method. 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate 40 µg of protein sample in each lane. The separated protein samples were transferred to a PVDF membrane and blocked with 5% bovine serum albumin. To bind the antigen (target protein) to the antibody, the membrane with target protein was incubated with Antibody Dilution Buffer P0256, Beyotime, China) containing anti-target-protein primary antibody (1:1000; diluted with Antibody Dilution Buffer) for 14 h at 4 °C. And then, after incubating with secondary antibody (BOSTER) for 1 h at indoor temperature, the blots were visualized using enhanced chemiluminescence (Pierce ECL Western blot analysis substrate; Millipore, Inc) and Chemiluminescence model of Amersham Imager 600 (General Electric Co, USA). The grey-scale value was detected using ImageJ 1.46r (National Institutes of Health, USA) and was normalized according to the β-actin grey-value. Anti-STEAP1 primary antibody was purchased from Abcam (ab207914), anti-EMT-related proteins primary antibody (ZEB1, E-cadherin, N-cadherin, Vimentin, Snail, Slug) from Cell Signaling Technology (#9782: https://www.cellsignal.com/products/primary-antibodies/epithelial-mesenchymal-transition-emt-antibody-sampler-kit/9782?site-search-type=Products), anti-Twist primary antibody from Santa Cruz Biotechnology (sc-81417: https://www.scbt.com/p/twist-antibody-twist2c1a?requestFrom=search).
Knockdown and overexpression of STEAP1 gene
Lentivirus carrying a recombining vector with the STEAP1 gene (25886-1) or two different small hairpin RNAs (shRNAs:60361-1, 60362-1) (all purchased from Gene Chem Inc, Shanghai, China) were used to perform overexpression or knockdown, respectively. According to the protocol, the suitable multiplicity of infection (MOI) was ascertained using a preliminary assay. In the formal assay, cells were seeded into 24-well plate, and lentivirus was added into the medium at a suitable MOI (overexpression: 120; knockdown: 100) the next day. After 12 h, the medium with lentivirus was substituted for fresh medium with FBS. Cells with green fluorescence were observed under the Image Xpress Micro Confocal (Molecular Devices) after culturing for 72 h. The transfection effect was evaluated using RT-qPCR, western blotting and immunocytochemistry.
Clone formation assay
Clone formation assay was performed to evaluate cell clonogenicity. Eight hundred single cells from each well were added into a six-well plate with three replicate wells and cultured for 10 days in 2 ml complete medium at 37 °C. Cells were fixed with 4% paraformaldehyde, stained with crystal violet-solution, and washed with running water after 10 days. Colony clusters were counted under the inverted microscope (Olympus), and a cluster of 50 cells or more was identified as a colony. The average value for three wells was recorded.
Generation of growth curves
A growth curve assay was performed to estimate cell growth. 1 × 104 cells were seeded into a 24-well plate with three replicate wells before culturing for 7 days 37 °C. The number of cell in the wells was counted on each day. The average of three wells was used to plot the growth curve.
Cell apoptosis analysis using flow cytometry
Cells were added into a six-well plate. After growing to 70% confluence, all cells (including dead cells) were digested with trypsin and centrifuged for 5 min at 1000 rpm/min. After discarding the supernatant, the cells were cleaned twice with PBS before being suspended with 300 μl 1 × buffer. Cell suspension solution (100 μl) was added to a glass tube with 5 μl 7-AAD and 5 μl PE fluorochrome. The glass tubes with cells and fluorochrome were placed in a dark for 15 min. Finally, 400 μl 1 × buffer was added into each glass tube, the cells were blended, and the fluorescence value of 10,000 cells was measured using flow cytometry. The entire process was performed gently and repeated three times.
Matrix metalloproteinase zymography assay
The activities of MMP2 and MMP9 were monitored using the matrix metalloproteinase zymography assay. Cells in logarithmic growth were seeded into six-well plates. After growing to 90% confluence, complete medium was replaced with 1 ml free-FBS medium and the cells were cultured for 24 h. According to the MMP zymography assay kit instructions (Applygen Technologies, China), the mixture of free-FBS medium and 2 × SDS-PAGE non-reducing buffer (1:1) was electrophoresed using 8% SDS-PAGE with collagenase substrate at 4 °C with 20 mA constant current and approximately 30 μl mixture per lane. Do not stop electrophoresis until the 2 × SDS-PAGE non-reducing buffer reached the bottom of the gel. The gelatum with MMP was washed twice with 1 × buffer A for 30 min, incubated with 1 × buffer B for 48 h at 37 °C, dyed with 0.25% Coomassie brilliant blue for 2 h, and destained in destaining buffer (10% acetic acid and 30% methanol) until bands became apparent. Gels were visualized in the Gel Imaging Model of Amersham Imager 600 (General Electric Co, USA), and the white blot is the MMP enzyme reaction area. The grey-scale value of white blot was detected using ImageJ 1.46r (National Institutes of Health, USA).
Tumour xenografts in nude mice
20 BALB/C-nu/nu nude mice (Beijing Vital River Laboratory Animal Technology Co, Ltd) were randomly divided into two groups (5 mice in one group), including ES-2, ES-2 shRNA, OVCAR-3 and OVCAR-3 EX for tumorigenicity assays. Approximately 1.0 × 107 cells were inoculated into each mouse. Mice were fed in a sterile animal facility (provided by Shandong University Laboratory Animal Center) for 7 weeks, and tumour size was measured weekly using a vernier caliper. The tumours were dissected after 7 weeks, and tumour size was calculated using the formula V = length × width2 × 0.5.
Statistic analysis
Statistical analysis was performed using SPSS 20. Measurement data are expressed as “mean ± SE” and analysed using two independent samples two-tailed t test (two groups), ANOVA (three and more groups, between groups with SNK or Dunnett-t test). However, enumeration data were analysed by χ2 test or Fisher exact test. Kaplan–Meier analysis was performed for survival analysis. *P < 0.05 (two-sided) was considered statistically significant.
Results
STEAP1 expression in human ovarian cancer tissues and normal ovarian tissues
STEAP1 expression was divided into “STEAP1 low (−/+)” and “STEAP1 high (++/+++)” based on staining. STEAP1 expression was low in normal ovarian tissues and benign tumours, and expressed highly in ovarian cancer (Fig. 1a–e). Moreover, STEAP1 mostly localized to the cancer cell membrane and cytoplasm. High STEAP1 expression was positively correlated with the poor differentiation of tissues, higher clinical stage, and lymph node metastasis (P < 0.01; Table 1), but it was not significantly correlated with histological types (P = 0.15; Table 1).
Six-transmembrane epithelial antigen of the prostate 1(STEAP1) expression in human ovarian cancer tissues and normal ovary tissues, and analysis of public databases. a Normal human ovarian tissue. b Normal human ovarian tissue (epithelium). c Benign ovarian tumour. d High differentiation of ovarian carcinoma. e Low differentiation of ovarian carcinoma. f STEAP1 mRNA expression in ovarian cancer tissues and normal ovarian tissues. In the Lu Ovarian Statistics of Oncomine database, STEAP1 expression in ovarian cancer tissues was higher than in normal ovarian tissues. g The high expression of STEAP1 was correlated with poor prognosis (PFS) of ovarian cancer patients in the Kaplan–Meier Plotter database. h Kaplan–Meier survival analysis of ovarian cancer patients. The progression-free survival rate of low expression group (n = 111) was significantly higher than that of the high expression group (n = 273), P < 0.01
Using the Oncomine database, we revealed a fact that STEAP1 was significantly highly expressed in serous adenocarcinoma, mucinous adenocarcinoma, and endometrial adenocarcinoma (Fig. 1f; Table S2). Using the Kaplan–Meier Plotter database, we showed that the progression-free survival (PFS) rate of ovarian cancer patients with STEAP1 up-regulation was significantly lower than that of patients with STEAP1 down-regulation [HR = 1.2 (1.06–0.37), log-rank P = 0.0056; Fig. 1g]. What’s more, results of survival analysis showed that the progression-free survival rate of low expression group (n = 111) was significantly higher (P<0.01; Fig. 1h) than that of high expression group (n = 273). This result suggested that STEAP1 was associated with ovarian cancer patient prognosis, and it might be a potential target to improve patient prognosis.
Different expression of STEAP1 in four human ovarian cancer cell lines with different migration and invasive abilities
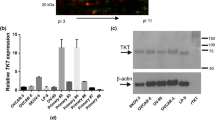
STEAP1 had low mRNA expression in the OVCAR-3 cell, moderate mRNA expression in the Caov-3 and SKOV-3 cell lines, and high mRNA expression in the ES-2 cell line (Fig. 2a). Western blot and immunocytochemistry showed that STEAP1 had the lowest protein expression in the OVCAR-3 cell line, followed by that in the Caov-3 and SKOV-3 cell lines. The highest protein expression was in the ES-2 cell line (Fig. 2b–d). Migration and invasion assays indicated that the ES-2 cell line had high migration and invasion abilities. The Caov-3 and SKOV-3 cell lines were moderately invasive, and the OVCAR-3 cell line had low invasive abilities (Fig. 2e–g). The highly invasive ES-2 cell line had high STEAP1 expression, the moderately invasive Caov-3 and SKOV-3 cell lines had the moderate expression of STEAP1, and the less invasive OVCAR-3 cell line had low STEAP1 expression. These results suggested that STEAP1 expression may be related to the ovarian cancer cells invasion and migration.
STEAP1 expression and migration and invasion abilities of four human ovarian cancer cell lines. a The mRNA expression of STEAP1 in OVCAR-3, Caov-3, SKOV-3, and ES-2 human ovarian cancer cell lines was detected using RT-qPCR, *P < 0.05 compared to OVCAR-3. b Expression of STEAP1 protein in OVCAR-3, Caov-3, SKOV-3, and ES-2 human ovarian cancer cell lines was detected using western blotting. c Bar graph of STEAP1 protein expression of four ovarian cancer cell lines in western blotting, *P < 0.05. d STEAP1 protein expression in four human ovarian cancer cell lines was determined using immunocytochemistry. e Images of migration abilities of four ovarian cancer cell lines measured using Boyden chambers without Matrigel. f Images of invasive abilities of four ovarian cancer cell lines measured by Boyden chambers with Matrigel. g Histograms of cell number in the migration and invasion assays from our images; *P < 0.05 compared to OVCAR-3
Verification of STEAP1 overexpression and knockdown
Based on the above experimental results, we knocked down STEAP1 using two different shRNAs in the ES-2 cell line, which had strong invasion abilities, and we exogenously overexpressed STEAP1 using lentivirus in OVCAR-3, which had weak invasion abilities. We verified the transfection effect using RT-qPCR and western blot. We found that STEAP1 expression decreased by 75% in ES-2, and we obtained stable ES-2 negative control (ES-2 NC) and ES-2 knockdown (ES-2 shRNA1, ES-2 shRNA2) cell lines (Fig. 3a–d), and transfection effects of two different shRNAs were the same. STEAP1 overexpression in OVCAR-3 was increased by 5 times, and stable OVCAR-3 negative control (OVCAR-3 NC) and OVCAR-3 up-regulation (OVCAR-3 EX) cell lines were obtained (Fig. 3e, f). The same transfection effect was also verified using immunocytochemistry (Fig. 3g).
Verification of knockdown and overexpression of STEAP1 in ovarian cancer cell lines. ES-2 NC ES-2 cell line transfected with negative control lentivirus sequences, ES-2 shRNA1 ES-2 cell line transfected with lentivirus construct LV-STEAP1-RNAi(60361-1), ES-2 shRNA2 ES-2 cell line transfected with lentivirus construct LV-STEAP1-RNAi(60362-1), OVCAR-3 NC OVCAR-3 cell line transfected with negative control lentivirus sequences, OVCAR-3 EX OVCAR-3 cell line transfected with lentivirus construct LV-STEAP1(25886-1). a Knockdown effect of ES-2 shRNA1 at the RNA level was verified using RT-qPCR, *P < 0.05. b Knockdown effect and bar graph of ES-2 shRNA1 at the protein level was verified using western blotting, *P < 0.05. c Knockdown effect of ES-2 shRNA2 at the RNA level was verified using RT-qPCR, *P < 0.05. d Knockdown effect and bar graph of ES-2 shRNA1 at the protein level was verified using western blotting, *P < 0.05. e Overexpression effect at the RNA level was verified using RT-qPCR, *P < 0.05. f Overexpression effect and bar chart of graph of OVCAR-3 EX at the protein level was verified using western blotting, *P < 0.05. g Transfection effects were determined using immunocytochemistry. STEAP1 expression in ES-2 shRNA1 and ES-2 shRNA2 cells were all significantly weaker than in ES-2 cells. STEAP1 expression in OVCAR-3 EX cells was significantly stronger than in OVCAR-3 cells
STEAP1 effects on migration and invasion abilities of human ovarian cancer cell lines in vitro
We tested the invasion and migration in the transfected cell lines after successful transfection. We found that the invasion and migration in the ES-2 shRNA1 and ES-2 shRNA2 were much lower than that of the ES-2, and the invasion and migration in the OVCAR-3 EX cell line was much stronger than that of the OVCAR-3 cell line (Fig. 4a, b). The invasion and migration of ES-2 shRNA1 were the same as that of ES-2 shRNA2. These results indicated that STEAP1 degradation suppressed the invasion and migration ability of ovarian cancer cells in vitro, but STEAP1 overexpression enhanced the invasion and migration ability of ovarian cancer cells in vitro.
Effects of STEAP1 on migration and invasion abilities. a Images and bar chart of migration abilities of transfected cells and non-transfected cells measured using Boyden chambers without Matrigel. *P < 0.05. b Images and bar chart of invasive abilities of transfected cells and non-transfected cells measured using Boyden chambers with Matrigel. *P < 0.05
Effects of STEAP1 on EMT progression
Differences in EMT-related genes expression between non-transfected and transfected cell lines were tested using RT-qPCR and western blotting. The results showed that Vimentin (encoded by VIM), N-cadherin (encoded by CDH2), ZEB1, Twist, Snail, and Slug (EMT-related genes) expression were inhibited in the ES-2 shRNA1. On the other hand, E-cadherin (encoded by CDH1) was up-regulated in the ES-2 shRNA1. In contrast, Vimentin, N-cadherin, ZEB1, Twist, Snail, and Slug were up-regulated in the OVCAR-3 EX, but the E-cadherin gene was down-regulated (Fig. 5a, b, d). There were no significant differences in the expression of EMT-related genes between non-transfected cell lines and negative control cell lines. These results indicated that STEAP1 repression inhibited the EMT process, but STEAP1 overexpression promoted the EMT process.
Effects of STEAP1 on epithelial-to-mesenchymal transition progress, activities of MMP2 and MMP9, growth, and clonogenicity of human ovarian cancer cell lines. a The mRNA expression of EMT-related genes and MMP2 and MMP9 in transfected cells and non-transfected cells using RT-qPCR, *P < 0.05. b Protein expressions of EMT-related genes in transfected cells and non-transfected cells using western blotting. c Activity of MMP2 and MMP9 in transfected cells and non-transfected cells. d Bar charts of EMT-related genes expression in transfected cells and non-transfected cells using western blotting, *P < 0.05. e Bar charts of MMP2 and MMP9 activity in transfected cells and non-transfected cells using Matrix metalloproteinase zymography assay, *P < 0.05. f The plate clone formation assay was performed to estimate STEAP1’s effect on cell clonogenicity. STEAP1 down-regulation decreased the clonogenicity of the ES-2 cell line, and STEAP1 up-regulation strengthened the clonogenicity of the OVCAR-3 cell line. g The growth curves of ES-2 and ES-2 shRNA1 cells. STEAP1 knockdown inhibited the growth of ES-2 cell lines, *P < 0.05. h The growth curves of OVCAR-3 and OVCAR-3 EX cells. STEAP1 overexpression promoted the growth of OVCAR-3 cell lines, *P < 0.05
Effects of STEAP1 on MMP2 and MMP9 activity
The transcription and activity of MMP2 and MMP9 were determined using RT-qPCR and matrix metalloproteinase zymography assay, respectively. The mRNA expression of MMP2 and MMP9 was significantly suppressed in ES-2 shRNA1 cells, but an expression of MMP2 and MMP9 in OVCAR-3 EX cells increased significantly (Fig. 5a). As shown in Fig. 5c, e, the activity of MMP2 and MMP9 was weaker in the ES-2 shRNA1 compared to that in the ES-2. The activity of MMP2 and MMP9 in the OVCAR-3 EX was stronger than that in the OVCAR-3. The MMP2 and MMP9 activity in negative control cell lines did not differ significantly from that in the non-transfected cell lines. This suggests that reduced STEAP1 expression weakened the activity of MMP2 and MMP9, but the overexpression of STEAP1 enhanced MMP2 and MMP9 activity.
Effects of STEAP1 on clonogenicity, growth, and cell apoptosis of human ovarian cancer cell lines
The clone formation assay also showed that the number and size of colonies were significantly less compared to that in the ES-2 cell line after STEAP1 knockdown. On the contrary, the number and size of colonies were significantly more and larger than that in the OVCAR-3 cell line after STEAP1 overexpression (Fig. 5f). From the growth curves, we revealed that cell growth decreased significantly after STEAP1 knockdown and increased significantly after STEAP1 overexpression (Fig. 5g, h). The cell apoptosis ratio in the ES-2 shRNA1 was higher than that in the ES-2, and the apoptosis ratio in the OVCAR-3 EX was lower than in the OVCAR-3 (Fig. 6a, b). In summary, decreasing STEAP1 expression inhibited ovarian cancer cell growth and clonogenicity, and it also reinforced apoptosis in the ovarian cancer cell line. Increasing STEAP1 expression boosted ovarian cancer cell growth and clonogenicity, and also blocked apoptosis in the ovarian cancer cell line.
STEAP1 effects on cell apoptosis in vitro, and tumour growth in vivo in nude mice. a Cell apoptosis was detected using flow cytometry, Q1 cell debris, Q2 late apoptotic cells, Q3 normal growing cells, Q4 early apoptotic cells. b Bar chart of apoptosis rate in ES-2, ES-2 shRNA1, OVCAR-3, and OVCAR-3 EX groups, *P < 0.05. c Tumour images and growth curve of ES-2 and ES-2 shRNA1 groups in nude mice (five nude mice every group). STEAP1 knockdown hampered the growth of highly invasive tumours in nude mice, *P < 0.05. d Tumour images and growth curve of OVCAR-3 and OVCAR-3 EX groups in nude mice (five nude mice every group). STEAP1-overexpression promoted the growth of weakly invasive tumours in nude mice, *P < 0.05
STEAP1 effects on tumour growth in nude mice
The ovarian cancer cell lines ES-2, ES-2 shRNA1, OVCAR-3, and OVCAR-3 EX all developed tumours in vivo in nude mice. The tumour growth speed and the average volume of tumours in the ES-2 shRNA1 group were significantly reduced compared to those in the ES-2 group, and the results of tumour growth in OVCAR-3 were opposite after STEAP1-overexpression. The above results indicated that STEAP1 expression was correlated with tumour growth in nude mice (Fig. 6c, d; Table S3).
Discussion
Here, we systematically explored the relationship between STEAP1 expression and the processes of invasion and metastasis in ovarian cancer. We confirmed the high expression of STEAP1 in ovarian cancer. STEAP1 was involved in the growth, clonogenicity, and apoptosis of ovarian cancer cells, and it may regulate the migration and invasion ability in ovarian cancer cells by regulating the EMT process.
Many reports have confirmed that STEAP1 is highly expressed in many cancer tissues (prostate cancer, lung cancer, bladder cancer, and breast cancer). Here, we found that STEAP1 is lowly expressed in normal ovarian tissues and benign tumours, and highly expressed in ovarian cancer. These results agree with previous studies of ovarian cancer cell lines (Hubert et al. 1999). We also found that STEAP1 expression was strongly correlated with tumour cell differentiation and lymph node metastasis. We analysed the prognosis of patients with ovarian cancer, and we showed that high STEAP1 expression was a risk factor for prognosis using the Kaplan–Meier Plotter public database and survival analysis of our data. It has been reported that the level of STEAP1 expression in prostate cancer is strongly correlated with tumour stage, pathological grade, and lymph node metastasis (Challita-Eid et al. 2007). High expression is also associated with tumour invasion, short-term recurrence, and poor prognosis in previous prostate cancer studies (Ihlaseh-Catalano et al. 2013).
However, immunohistochemistry in primary colorectal cancer did not reveal that STEAP1 expression level was associated with cancer stage and grade, and survival analysis showed that low expression is a risk factor for patient prognosis in colorectal cancer patients (Lee et al. 2016). In another colon cancer study using a public database, it was found that the up-regulation of STEAP1 transcription was associated with poor overall survival (Nakamura et al. 2018). The opposite was also found in the survival analysis of two Ewing’s sarcoma patients. Cheung (Cheung et al. 2007) found that high expression of STEAP1 is a risk factor for favourable prognosis of Ewing’s sarcoma. In contrast, Grunewald (Grunewald et al. 2012b) used multivariate analysis to find that low expression of STEAP1 is a risk factor for prognosis of Ewing’s sarcoma. Similarly, our previous two studies on breast cancer and endometrial cancer were contrary to the results of this ovarian cancer study (Sun et al. 2019; Xie et al. 2019). STEAP1 showed low expression in breast and endometrium cancer, and its low expression was positively correlated with tumour poor differentiation, high stage, and lymph node metastasis. Survival analysis showed that low expression of STEAP1 was a risk factor for breast and endometrial cancer patients. Based on the above reports and our findings, the inconsistent expression of STEAP1 in different cancer tissues and the discrepant patient prognoses may be related to different tissue types and tumour microenvironments. The difference in microenvironments leads to STEAP1 expression and cancer prognosis differences. Also besides, understanding STEAP1’s effect on patient prognosis requires verification by more rigorous studies with larger sample sizes.
Here, we revealed a fact that STEAP1 is highly expressed in the highly invasive ovarian cancer cell line ES-2, it is lowly expressed in the less invasive ovarian cancer cell line OVCAR-3, and it is moderately expressed in the moderately invasive ovarian cancer cell lines SKOV-3 and Caov-3. After transfection with lentivirus, we found that decreased STEAP1 expression inhibited the invasion and migration ability, growth, and colony formation ability of ovarian cancer cells in vitro. It also inhibited the growth of nude mouse xenograft tumours and promoted the occurrence of cell apoptosis. STEAP1 overexpression enhanced the invasion and migration ability, growth, and colony formation ability of ovarian cancer cells in vitro. It also inhibited apoptosis in ovarian cancer cells. These results indicate that STEAP1 may be involved in the regulation of invasion and migration, growth, cell clonogenicity, apoptosis, and xenograft tumour growth ability of ovarian cancer cells, and they also suggest that STEAP1 may be a potential target for treating ovarian cancer.
Similar results were demonstrated in prostate cancer, Ewing’s sarcoma, gastric cancer, colorectal cancer, and lung cancer studies. Gomes (Gomes et al. 2018) found that reduction of STEAP1 expression inhibited cell growth and induced apoptosis in LNCaP prostate cancer cells. Challita-Eid et al. (2007) used monoclonal antibodies against STEAP1 to treat xenograft tumours of human prostate cancer and bladder cancer, finding that the antibody significantly inhibited the in vivo growth of xenograft tumours. Yamamoto et al. (2013) promoted STEAP1 expression by transfecting the human prostate cancer cell line DU145 and human oral squamous cell carcinoma cell line OSC30 with STEAP1. They indicated that the growth of tumours in mice was not related to in vitro cancer cell growth. In Ewing’s sarcoma, Grunewald et al. (2012a) found that STEAP1 silencing inhibited growth, invasion, in vitro sarcoma cell colony formation, and tumour growth and metastasis abilities in vivo in mice. However, there was no noticeable effect on apoptosis, necrosis, or cell cycle arrest. In a study of gastric cancer, Wu et al. (2018) found that STEAP1 knockdown inhibited the in vitro migration, invasion, proliferation, and inhibited the growth of tumours in vivo in a mouse xenograft tumour model, suggesting that STEAP1 is related to the proliferation, migration, invasion, and tumorigenicity of gastric cancer cells. In a study of lung cancer, Zhuang et al. (2015) used small interfering RNA (siRNA) knockdown of STEAP1 expression in lung cancer vascular endothelial cells, which significantly decreased the in vitro cell migration and vascular formation abilities. Hayashi et al. (2011) successfully induced the T-cell immune response of lung cancer cells in vitro using the synthetic peptide STEAP281–296, indicating that STEAP1 can be an effective target for CD4+ T-cell for immunotherapy.
Invasion and metastasis of cancer is a tumour characteristic that is an important indicator of poor prognosis of cancer patients (Hanahan and Weinberg 2011). EMT is a crucial process of tumour invasion and metastasis. In the process of EMT, epithelial cells lose their apical-basal polarity and the ability to adhere to other cells, turning into fusiform mesenchymal cells (Loret et al. 2019). This process also promotes the ability of tumour cells to spread across the basal membrane/stromal cells, along with bloody circulation to other sites, and to survive and grow to form new metastatic lesions (Chaffer and Weinberg 2011). Vimentin, N-cadherin, and E-cadherin are core markers of EMT, and EMT-related transcription factors such as Twist, Slug, Snail, ZEB1 and others also regulate EMT progression (De Craene and Berx 2013; Goossens et al. 2017; Lamouille et al. 2014; Puisieux et al. 2014). What’s more, MMP2 and MMP9 are related to tumour cell EMT (Dongre and Weinberg 2019). E-cadherin expression was down-regulated during EMT or invasion and migration enhancement in tumour cells, but the Vimentin, N-cadherin, Twist, Slug, Snail, ZEB1, MMP2, and MMP9 were up-regulated.
Recently, Loret summarized EMT’s contribution to the occurrence, development, metastasis, and drug resistance of ovarian cancer (Loret et al. 2019). Meanwhile, our trial also showed that the invasion and migration ability of tumour cells in vitro changed after STEAP1 knockdown or overexpression, so we investigated changes in EMT-related genes. Here, E-cadherin expression was up-regulated, and Vimentin, N-cadherin, Twist, Slug, Snail, ZEB1, MMP2, and MMP9 were down-regulated after STEAP1 expression was decreased. The EMT process, invasion ability, and migration ability of the ovarian cancer cells were weakened. After exogenously increasing STEAP1 in ovarian cancer cells, E-cadherin was down-regulated and Vimentin, N-cadherin, Twist, Slug, Snail, ZEB1, MMP2, and MMP9 was up-regulated. The EMT process and the invasion and migration abilities were enhanced. Based on the above results, we speculate that STEAP1 may drive the invasion and migration ability of ovarian cancer cells by regulating the EMT process. Wu et al. (2018) found that STEAP1 may be the key factor driving EMT in gastric cancer metastasis. EMT occurred in metastatic gastric cancer and STEAP1 was up-regulated compared to mRNA levels of normal gastric tissues. Furthermore, improvement of the EMT process of gastric cancer cells induced STEAP1 up-regulation. Although results of this study are not consistent with that of studies about breast cancer and endometrial carcinoma (Sun et al. 2019; Xie et al. 2019), the relationship between STEAP1 and invasion and metastasis of cancer is verified. Invasion and metastasis of cancer is a very complex process, involving multiple genes expression and regulation (Chaffer and Weinberg 2011; Thiery and Sleeman 2006). And because of the differences in tumour histology, cell types and microenvironment, one gene may play different roles in the development of tumours. What’s more, we do not know mechanisms that STEAP1 regulates EMT, growth, proliferation, apoptosis of ovarian cancer, the details of how STEAP1 regulates the EMT process must be further studied. Our study is the first to link STEAP1’s potential function with EMT, revealing a possible mechanism by which STEAP1 could regulate the invasion and migration ability of ovarian cancer cells.
In conclusion, our study showed that high STEAP1 expression in ovarian cancer has an association with severe clinical pathological characteristics and poor prognosis of ovarian cancer patients. High STEAP1 expression promotes the growth of ovarian cancer cells, and it may enhance the invasion and migration ability of cells by promoting EMT. STEAP1 is a potential target for ovarian cancer treatment, and more research is needed to confirm our results.
References
Allemani C et al (2015) Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 385:977–1010. https://doi.org/10.1016/S0140-6736(14)62038-9
Barroca-Ferreira J et al (2018) Targeting STEAP1 protein in human cancer: current trends and future challenges. Curr Cancer Drug Targets 18:222–230. https://doi.org/10.2174/1568009617666170427103732
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Carrasquillo JA et al (2019) Imaging metastatic castration-resistant prostate cancer patients with89 Zr-DFO-MSTP2109A anti-STEAP1 antibody. J Nucl Med. https://doi.org/10.2967/jnumed.118.222844
Chaffer CL, Weinberg RA (2011) A perspective on cancer cell metastasis. Science 331:1559–1564. https://doi.org/10.1126/science.1203543
Challita-Eid PM et al (2007) Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumour xenografts in vivo. Cancer Res 67:5798–5805. https://doi.org/10.1158/0008-5472.can-06-3849
Chen X, Wang R, Chen A, Wang Y, Wang Y, Zhou J, Cao R (2019) Inhibition of mouse RM-1 prostate cancer and B16F10 melanoma by the fusion protein of HSP65 and STEAP1 186-193. Biomed Pharmacother 111:1124–1131. https://doi.org/10.1016/j.biopha.2019.01.012
Cheung IY, Feng Y, Danis K, Shukla N, Meyers P, Ladanyi M, Cheung NKV (2007) Novel markers of subclinical disease for ewing family tumours from gene expression profiling. Clin Cancer Res 13:6978–6983. https://doi.org/10.1158/1078-0432.ccr-07-1417
De Craene B, Berx G (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13:97–110. https://doi.org/10.1038/nrc3447
Dongre A, Weinberg RA (2019) New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 20:69–84. https://doi.org/10.1038/s41580-018-0080-4
O Donoghue JA et al (2019) Pharmacokinetics and biodistribution of a [89 Zr]Zr-DFO-MSTP2109A anti-STEAP1 antibody in metastatic castration-resistant prostate cancer patients. Mol Pharm 16:3083–3090. https://doi.org/10.1021/acs.molpharmaceut.9b00326
Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016 (2017). Lancet 390:1151–1210. https://doi.org/10.1016/S0140-6736(17)32152-9
Gomes IM, Rocha SM, Gaspar C, Alvelos MI, Santos CR, Socorro S, Maia CJ (2018) Knockdown of STEAP1 inhibits cell growth and induces apoptosis in LNCaP prostate cancer cells counteracting the effect of androgens. Med Oncol 35:40. https://doi.org/10.1007/s12032-018-1100-0
Goossens S, Vandamme N, Van Vlierberghe P, Berx G (2017) EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer 1868:584–591. https://doi.org/10.1016/j.bbcan.2017.06.006
Grunewald TGP et al (2012a) STEAP1 is associated with the invasive and oxidative stress phenotype of ewing tumors. Mol Cancer Res 10:52–65. https://doi.org/10.1158/1541-7786.mcr-11-0524
Grunewald TGP et al (2012b) High STEAP1 expression is associated with improved outcome of Ewing’s sarcoma patients. Ann Oncol 23:2185–2190. https://doi.org/10.1093/annonc/mdr605
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. https://doi.org/10.1016/j.cell.2011.02.013
Hayashi S et al (2011) Six-transmembrane epithelial antigen of the prostate and enhancer of zeste homolog 2 as immunotherapeutic targets for lung cancer. J Transl Med 9:191. https://doi.org/10.1186/1479-5876-9-191
Hubert RS et al (1999) STEAP: a prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA 96:14523–14528. https://doi.org/10.1073/pnas.96.25.14523
Ihlaseh-Catalano SM, Drigo SA, de Jesus CM, Domingues MA, Trindade Filho JC, de Camargo JL, Rogatto SR (2013) STEAP1 protein overexpression is an independent marker for biochemical recurrence in prostate carcinoma. Histopathology 63:678–685. https://doi.org/10.1111/his.12226
Kurman RJ, Carcangiu LM, Herrington CS, Young RH (eds) (2014) WHO classification of tumours of female reproductive organs. IARC Press, Lyon
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. https://doi.org/10.1038/nrm3758
Lee CH et al (2016) The prognostic role of STEAP1 expression determined via immunohistochemistry staining in predicting prognosis of primary colorectal cancer: a survival analysis. Int J Mol Sci 17:e592. https://doi.org/10.3390/ijms17040592
Loret N, Denys H, Tummers P, Berx G (2019) The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers 11:838. https://doi.org/10.3390/cancers11060838
Maia CJB, Socorro S, Schmitt F, Santos CRA (2008) STEAP1 is over-expressed in breast cancer and down-regulated by 17β-estradiol in MCF-7 cells and in the rat mammary gland. Endocrine 34:108–116. https://doi.org/10.1007/s12020-008-9113-7
Nakamura H et al (2018) Six-transmembrane epithelial antigen of the prostate 1 protects against increased oxidative stress via a nuclear erythroid 2-related factor pathway in colorectal cancer. Cancer Gene Ther 26:313–322. https://doi.org/10.1038/s41417-018-0056-8
Oberaigner W, Minicozzi P, Bielska-Lasota M, Allemani C, de Angelis R, Mangone L, Sant M (2012) Survival for ovarian cancer in Europe: the across-country variation did not shrink in the past decade. Acta Oncol 51:441–453. https://doi.org/10.3109/0284186x.2011.653437
Puisieux A, Brabletz T, Caramel J (2014) Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 16:488–494. https://doi.org/10.1038/ncb2976
Sun J, Ji G, Xie J, Jiao Z, Zhang H, Chen J (2019) Six-transmembrane epithelial antigen of the prostate 1 is associated with tumour invasion and migration in endometrial carcinomas. J Cell Biochem 120:11172–11189. https://doi.org/10.1002/jcb.28393
Thiery JP, Sleeman JP (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7:131–142. https://doi.org/10.1038/nrm1835
Vaughan S et al (2011) Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer 11:719–725. https://doi.org/10.1038/nrc3144
Wu YY, Jiang JN, Fang XD, Ji FJ (2018) STEAP1 regulates tumorigenesis and chemoresistance during peritoneal metastasis of gastric cancer. Front Physiol 9:1132. https://doi.org/10.3389/fphys.2018.01132
Xie J, Yang Y, Sun J, Jiao Z, Zhang H, Chen J (2019) STEAP1 inhibits breast cancer metastasis and is associated with epithelial–mesenchymal transition procession. Clinical Breast Cancer 19(1):e195–e207. https://doi.org/10.1016/j.clbc.2018.08.010
Yamamoto T et al (2013) Six-transmembrane epithelial antigen of the prostate-1 plays a role for in vivo tumour growth via intercellular communication. Exp Cell Res 319:2617–2626. https://doi.org/10.1016/j.yexcr.2013.07.025
Zhuang X et al (2015) Identification of novel vascular targets in lung cancer. Br J Cancer 112:485–494. https://doi.org/10.1038/bjc.2014.626
Acknowledgements
Our study was funded by Nature Science Foundation of Shandong Province (ZR2017MH060) and Key R&D Plan of Shandong Province (2018GSF118049). These funds provided only financial support and were not involved in the study or manuscript writing.
Funding
This study was funded by the Nature Science Foundation of Shandong Province (ZR2017MH060) and Key R&D Plan of Shandong Province (2018GSF118049).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors in this study declare that they have no conflict of interests.
Ethical approval
This study was approved by the Medical Ethics Committee of Human and Animal Institution of Shandong University. All methods were performed in accordance with the relevant guidelines and regulations.
Informed consent
We obtained the written informed consents of the patients involved in the study. All mouse experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of the People’s Republic of China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiao, Z., Huang, L., Sun, J. et al. Six-transmembrane epithelial antigen of the prostate 1 expression promotes ovarian cancer metastasis by aiding progression of epithelial-to-mesenchymal transition. Histochem Cell Biol 154, 215–230 (2020). https://doi.org/10.1007/s00418-020-01877-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-020-01877-7