Abstract
Prostate adenocarcinoma is the second most frequent cancer worldwide and is one of the leading causes of male cancer-related deaths. However, it varies greatly in its behaviour, from indolent non-progressive disease to metastatic cancers with high associated mortality. The aim of this study was to identify predictive biomarkers for patients with localised prostate tumours most likely to progress to aggressive disease, to facilitate future tailored clinical treatment and identify novel therapeutic targets. The expression of 602 genes was profiled using oligoarrays, across three prostate cancer cell lines: CA-HPV-10, LNCaP and PC3, qualitatively identifying several potential prognostic biomarkers. Of particular interest was six transmembrane epithelial antigen of the prostate (STEAP) 1 and STEAP 2 which was subsequently analysed further in prostate cancer tissue samples following optimisation of an RNA extraction method from laser captured cells isolated from formalin-fixed paraffin-embedded biopsy samples. Quantitative analysis of STEAP1 and 2 gene expression were statistically significantly associated with the metastatic cell lines DU145 and PC3 as compared to the normal prostate epithelial cell line, PNT2. This expression pattern was also mirrored at the protein level in the cells. Furthermore, STEAP2 up-regulation was observed within a small patient cohort and was associated with those that had locally advanced disease. Subsequent mechanistic studies in the PNT2 cell line demonstrated that an over-expression of STEAP2 resulted in these normal prostate cells gaining an ability to migrate and invade, suggesting that STEAP2 expression may be a crucial molecule in driving the invasive ability of prostate cancer cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) is a disease that affects men ranging from 45 to 80+ years of age, but is most prevalent in men over the age of 75 years [1]. It is generally a slow growing cancer that is usually asymptomatic in its early stages and in many men follows an indolent course without the development of metastatic disease. Nonetheless, the PCa incidence rate has increased world-wide in recent years, although this may be due in part to improved screening and detection methods [2]. PSA testing to detect PCa at an early stage has resulted in increasing numbers of men being diagnosed with small sized tumour volumes, enhancing the curative treatment of these tumours [3]. PSA levels are used alongside clinical stage and prostate biopsy Gleason score to determine the risk of recurrence and/or progression after initial diagnosis of non-metastatic disease [4]. Consequently, this risk stratification can allow clinicians to recommend treatment options accordingly. However, the D’Amico risk categories lack sufficient accuracy to predict prognosis for many patients and various nomograms or scoring systems such as artificial neural networks to predict outcome are not sufficiently robust to tailor treatment. Whilst the Gleason scoring system can provide some prognostic value, it does not accurately discriminate between patients with tumours that are likely to progress or remain indolent [5]. Additionally, following primary treatment, PSA levels may indicate biochemical recurrence and/or progression. However, there is still an urgent need for predictive biomarkers able to identify those patients most at risk of developing progressive PCa to ensure their treatment is tailored appropriately.
Metastasis accounts for the main cause of cancer mortality and is a process whereby the primary site tumour cells gain the ability to spread to a secondary site [6]. The main stages involved in metastasis include degradation of the basement membrane, invasion through the extracellular matrix (ECM), intravasation, evasion of anoikis, extravasation, and finally establishment of secondary site carcinoma through proliferation, angiogenesis, and micro-environment activation. The ECM is centrally involved in this process as despite being non-cellular, it provides a physical scaffold and facilitates necessary signalling for tissue morphogenesis, differentiation and homeostasis [7]. Mounting evidence has suggested that alterations in the tumour microenvironment are principally controlled by matrix metalloproteinases (MMPs). MMPs can degrade a variety of cell adhesion molecules resulting in the modulation of cell–cell and cell-ECM interactions [8]. The role and expression of MMPs and its inhibitor tissue inhibitor metalloproteinases (TIMP) varies within different cancers and on the neighbouring cells that are induced in a paracrine manner by cancer cells [8]. Fibroblasts are stimulated to release MMPs by cancer cells that release interleukins and growth factors [9]. MMPs can therefore promote or inhibit development or progression of cancer. In PCa, MMPs -2, -7, -9, -13 and -14 have been shown to be involved in PCa and their over-expression promotes PCa growth and metastasis [10–13]. Furthermore, recent evidence has demonstrated that the protein six transmembrane epithelial antigen of the prostate (STEAP2) increases ERK signalling, a molecule whose downstream effectors are MMPs [14]. Its functions are thought to involve the endocytotic and secretory pathways [15], and it also acts as a ferrireductase and cupric reductase which stimulates the uptake of iron and copper into the cell [16]. STEAP2 has been shown to affect PCa cell growth, as its down regulation results in the up-regulation of some cell cycle inhibitors including CDKIs and p21 both in vitro and in vivo [14]. STEAP2 also affects cell proliferation, and when down-regulated, apoptosis levels are increased. Reports focusing on the STEAP2 status in human tumours are very limited and no previous studies have assessed its potential role in the promotion of tumour invasiveness. However, one investigation has demonstrated its up-regulation within prostate tumours compared to normal glands in 46 tissue sections taken from 13 prostatectomy tissue samples, suggesting a possible role in carcinogenesis [15]. Thus, further functional studies are warranted.
Developing a detailed understanding of the metastatic process is critical as this knowledge may lead to the identification of novel molecular biomarkers with a central role in tumour invasion. This will not only provide tools to enable the early identification of patients that have developed molecular changes that support metastatic spread, but could also lead to new therapeutic drug targets. Such advances would therefore have a dramatic impact as they will result in the tailored clinical management of patients. Consequently, the aim of this study was to identify potential prognostic biomarkers for aggressive PCa that may be capable of distinguishing between localised and metastatic tumours that in turn may be applied to the clinical setting.
Materials and methods
Cell culture
To evaluate potential biomarkers for PCa progression in the present study, a model based on five prostate cell lines with increasing metastatic potential was utilised. PNT2 (normal prostate epithelial cells) was the control model for gene expression analysis, to which variations in expression within CA-HPV-10 (localised PCa), LNCaP (lymph node metastasis), DU145 (brain metastasis) and PC3 (bone metastasis) cells were directly compared. All cell lines were obtained from the American Type Tissue Culture (ATTC, UK). PNT2, LNCaP, DU145 and PC3 were cultured in RPMI 1640 (Gibco, UK) supplemented with 10 % foetal bovine calf serum (FBS) and 0.5 % penicillin/streptomycin. CA-HPV-10 cells were grown in Keratinocyte media supplemented with human recombinant EGF and Bovine Pituitary Extract supplements, the quantity of both as specified by the manufacturer (Life Technologies, UK). Cells were grown to 80 % confluency prior to sub-culture or analysis.
Patient tissue samples
Ethical approval for this collection was obtained from the Swansea Local Research Ethics Committee (Ref NO: 07/WMW02/59, approved in August 2007). Formalin fixed paraffin embedded (FFPE), PCa samples from patients (n = 18) were categorised into four groups as follows: low risk localised (LRL), high risk localised (HRL), locally advanced, and metastatic disease. The groupings were defined according to the patients’ Gleason score, initial pre-operative PSA and stage (Table 1). The tissue obtained from these patients consisted of transrectal ultrasound (TRUS) guided biopsies with an equivalent haematoxylin and eosin (H&E) slide that was analysed by a consultant pathologist to ensure that sufficient material was present for the study (minimum 20 % tumour involvement). Normal prostate TRUS tissue samples from patients (n = 4) who displayed no prostate disease were used as controls.
Group 1 (Table 1) included four patients, aged 48–66 years (median age 58 years), diagnosed with Gleason score 6 tumours, a presenting PSA of 4.7–6.1 ng/mL (median PSA 5.63 ng/mL) and with clinically ≤T2 tumours. Group 2 comprised of four patients with clinical T2 stage, aged between 64 and 70 years (median age 67 years), a presenting PSA of 22–33 ng/mL (median initial PSA 27 mg/mL) and Gleason scores ranged from 7 to 10 (median Gleason score 8). Group 3 included four patients with an age range of 57–72 years (median age 64 years), PSA of 2.8–10 ng/mL (median PSA 6.3 ng/mL), Gleason score of 8 or 9, and all had evidence of extra capsular spread. Group 4 patients had evidence of metastasis and/or death as a result of PCa, aged 55–83 years (median age 70.4 years) and had an initial PSA value of 27.0–600 ng/mL (median PSA 220.4 ng/mL). Group 5 contained four patients with no evidence of cancer on prostatectomy sections.
Laser capture microdissection (LCM)
FFPE patient tissue sample blocks were sectioned at 5 µm thickness and placed onto a specialised LCM PEN membrane slide (Carl-Zeiss). Slides were de-paraffinised using xylene, followed by a series of ethanol washes and subsequently H&E stained. Once dry, areas of malignant cells within each of the biopsies present on the slide were identified and laser captured into individual eppendorf tubes with the PALM LCM system (Carl-Zeiss).
RNA extraction
Total RNA was extracted from all cell lines using the Illustra RNA spin Mini Kit (GE Healthcare, UK), with exception of CA-HPV-10 cells, which were subjected to the Trizol method for RNA extraction according to manufacturer’s instructions (Life Technologies, UK). Three biological replicates for each of the cell lines were obtained.
For the laser captured patient tissue samples, total RNA was extracted using the MasterPure Kit (Illumina, WI) according to manufacturer’s recommendations, and pooled with patients of the same group (Table 1) due to limited RNA yields following laser capture.
Gene expression profiling
Initial Oligoarray screening was conducted using four cell lines: PNT2, CA-HPV-10, LNCaP and PC3. Using the Truelabelling Amp 2.0 kit for RNA amplification (SABiosciences, UK), 1 µg total RNA was converted into cRNA according to manufacturer’s instructions, and applied to the following oligoarrays (SABiosciences, UK): ECM and adhesion molecules, PCa biomarkers, WNT Signalling, and TGF-β/BMP signalling pathway; screening a total of 602 genes (see Supplementary Fig. 1 for full gene list). The protocol was carried out according to manufacturer’s instructions, and each array contained eight housekeeping genes as an internal control to ensure appropriate hybridisation during the assay.
cDNA synthesis
For all cell lines (PNT2, CA-HPV-10, LNCaP, DU145 and PC3) 1 µg of total RNA was used for generation of cDNA using the Retroscript Kit (Ambion, UK) according to manufacturer’s instructions. For patient tissue samples, following amplification, 1 µg of cRNA was required for cDNA generation using the QuantiTect Reverse Transcription Kit (Qiagen, UK). A gene specific cDNA generation was performed for all tissue samples using the following primer sequences: STEAP1 forward- CATTATTCCCAGATTCTTTGATTGTC, Reverse- CGCTTGAATATTGTTGCTGCATA; STEAP2 forward- GCAATGCTTTAAACTGGAGAGAAT, reverse- AAAGTTTGGTGGTGTATAAAATCTGT; HPRT forward- GACTGTAGATTTTATCAGACTGA, reverse-TGGATTATACTGCCTGACCAA. STEAP2 primer sets were designed and provided by PrimerDesign Ltd (Cat# SY-hu-600, UK) whilst HPRT primers were designed and optimised in-house, and purchased from Sigma-Aldrich (UK).
Quantitative real time PCR (qPCR) analysis
qPCR was performed using iQ SYBR Green Supermix (Bio-Rad, UK) on a MYiQ qPCR machine (Bio-Rad, UK). Primers were used with a starting concentration of 15 pm and the reference gene was HPRT. Cycles for STEAP1 and HPRT were as follows: 95 °C for 3 min (1 cycle), 94 °C for 30 s followed by 60 °C for 10 s and 72 °C for 15 s (40 cycles), and 95 °C for 30 s (1 cycle). Melt curve analysis was performed for each reaction to ensure amplification specificity. Custom designed STEAP2 primers were purchased from Primer Design Ltd (UK, Cat# SY-hu-600), with the qPCR protocol as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 60 s. Fold gene expression was assessed using the 2−ΔΔCT method.
Western blot analysis
For each of the five cell lines, a concentration of 50 µg of protein was loaded onto an 8 % gel. Antibody dilutions for each protein were as follows: STEAP1 1:100, STEAP2 1:200, and β-actin loading control 1:2,500 (all antibodies supplied by Santa Cruz, CA). Protein bands were visualised on the membranes following transfer using a chemiluminesence approach with the Immun-star™ WesternC™ kit (Bio-Rad, UK) according to manufacturer’s instructions, while membrane imaging and densitometry was performed on the ChemiDoc (Bio-Rad, UK). Each protein band was normalised against the housekeeping protein β-actin.
STEAP2 transfection
Entry clones and destination vectors were purchased (Invitrogen, UK) in which the entry vector contained the gene sequence for STEAP2 (CloneID IOH37819, Invitrogen, UK). The ORF is in the pENTR221 Gateway vector and the Destination vector utilised was Gateway® pcDNA™ DEST40 Vector (Invitrogen, UK). For transfection efficiency positive control, pENTR™—gus was used, while the negative control contained only the destination vector without the entry clone. An LR recombination reaction (a recombination reaction between attL and attR sites) between the entry clone and destination vector was performed using the Gateway® LR Clonase® II Enzyme mix (Invitrogen, UK) according to manufacturer’s instructions, resulting in the final plasmid construct for transfection.
Plasmids containing the STEAP2 gene were transfected into 5α select electroporation competent E. coli (BioLine, UK) using a GenePulser XCell (Bio-Rad, UK). Transfected bacteria were grown on ampicillin agar plates and plasmid DNA isolated using the QIAquick PCR Purification Kit (Qiagen, UK). One day prior to transfection, PNT2 cells were seeded in 6 well plates at 0.5 × 106 cells in 2 mL of growth media ensuring that the cells were 50–80 % confluent in time for transfection. In order to produce transient expression cell lines, 650 ng of isolated plasmid DNA was diluted in 500 µl serum free RPMI 1640 media. Using the Lipofectamine™ LTX & Plus Reagent (Invitrogen, UK) according to manufacturer’s instructions, 3 µL Lipofectamine LTX was added to the diluted plasmid DNA and incubated for 2 h at room temperature and subsequently applied to the cultured cells. After 24 h of incubation in the Lipofectamine LTX reagent, cells were washed with PBS and 1 µg Neomycin antibiotic was applied for 24 h at 37 °C for cell selection. STEAP2 up-regulation in the purified population of transfected cells was subsequently confirmed by qPCR.
Migration assay
Selected transfected cells, and non-transfected PNT2 cells, were grown to 80 % confluency in the presence of 0.5 µg/mL neomycin. A uniform and equal scratch was made along the length of all wells containing the STEAP2 transfected cells, negative scrambled vector control, and the non-transfected PNT2 negative control cells. Images of the scratches were taken at time points 0, 3 and 6 h and this experiment was repeated in triplicate.
Invasion assay
Transwells with 8 µm pore size filters (Millipore, UK) covered with Matrigel diluted 1:3 with RPMI 1640 media containing no supplements were placed into 6-well plates. To the lower chamber, RPMI 1640 containing 10 % FBS was added and to the upper chamber 2.5 × 104 cells were seeded in RMPI 1640 without supplements. Plates were incubated at 37 °C in a 5 % CO2 atmosphere for 24 h. Cells on the upper surface of the Matrigel layer were removed, while remaining cells that had invaded through the Matrigel layer were stained with haematoxylin and scored under a light microscope. Non-transfected normal PNT2 epithelial cells were used as negative controls, PC3 cells were used as positive controls due to their highly invasive ability, and all experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS software version 16.0 for Windows. For qPCR data as the relative expression levels for the three replicates for each gene within each cell line was not normally distributed, these fold values were log transformed and tested for distribution. The log transformed relative expression levels were normally distributed and therefore used for comparisons with a one way ANOVA with Tukey’s and Dunnet’s post hoc test. This test was also used to determine the significance of the percentage of wound closure within the scratch assay, and the percentage of cell invasion.
Results
By utilising five prostate cell lines and screening >600 genes for variations in transcriptional expression patterns, the aim of this study was to identify potential new biomarkers that may distinguish between localised, locally advanced and metastatic PCa.
Screening of 602 genes identified variation in STEAP1 and STEAP2 expression in metastatic PCa cells
RNA extracted from PNT2, CA-HPV-10, LNCaP, and PC3 cell lines was successively applied to four oligoarrays focused on genes for ECM and adhesion molecules, PCa biomarkers, WNT signalling, and TGF-β/BMP signalling pathways, screening a total of 602 genes (supplementary Fig. 1). Variations in gene expression within these arrays (normalised to multiple housekeeping genes on each array as controls for hybridisation), were qualitatively analysed by visually comparing gene spot intensities to the control PNT2 cells (supplementary Fig. 2). This analysis subsequently identified several genes of potential interest; in total, 24 genes were down-regulated and 42 genes up-regulated within the cancer cell lines (Fig. 1). Of these, 28 genes had highly notable visual differences in gene expression that were associated within the metastatic cells only (and were not substantially altered in the localised CA-HPV-10 tumour cells). However, two genes were selected for further validation due to vast differences in gene spot intensities compared to PNT2. These genes included: STEAP1 and STEAP2 due to their marked increase in expression within the metastatic cell lines only and not within the localised cancer cell line or PNT2.
Heat map of gene expression changes in CA-HPV-10, LNCaP and PC3 cells in relation to PNT2 gene expression. From the 603 genes screened using the four different pathway specific arrays, as indicated by the gene spots, a total of 24 genes were noted to be down-regulated and 42 genes were up-regulated in the prostate cancer cells within the locally advanced CA-HPV-10 cell line, lymph node metastasis cell line: LNCaP, and the bone metastasis cell line PC3, as compared to PNT2 cells. The oligoarray gene spot intensities were used to form a heat map where red indicates a general up-regulation of the gene, green a down-regulation, and black is indifferent expression compared to the PNT2 gene spot intensity. The original gene spot array image can be seen in supplementary Fig. 2
For qPCR analysis a third metastatic cell line, DU145, was included to assess the expression of STEAP1 and 2 across three metastatic cell lines. Quantification using qPCR demonstrated STEAP1 expression was statistically significantly up-regulated in all four cancer cell lines compared to PNT2; thus, as the increased expression was also present in the localised tumour cell line (CA-HPV-10), this alteration was not specific to invasive PCa alone. In contrast, STEAP2 expression was elevated significantly in only the metastatic cell lines (Table 2) and not in the localised cells, suggesting a potential role for STEAP2 in the promotion of localised PCa to an aggressive state.
Protein expression levels of STEAP1 and STEAP2 in prostate cancer cells
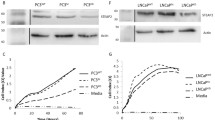
As expression patterns at the transcriptional level are not always mirrored following translation, the protein levels of STEAP1 and STEAP2 were analysed in all five cells lines. STEAP1 protein expression was slightly elevated in the more aggressive metastatic cell lines DU145 and PC3, compared to PNT2 cells (Fig. 2) which is in agreement with the RNA levels quantified. However, interestingly, the protein expression of STEAP2 was significantly increased in the metastatic cell lines DU145 and PC3 (28-fold), in comparison to PNT2 protein levels. The data obtained for STEAP2 protein levels also correlated with the findings observed at the transcript level further demonstrating a potential role for STEAP2 in particular, in the promotion of localised prostate cancer to an aggressive state.
Protein Expression levels of STEAP1 and STEAP2 in PNT2, DU145 and PC3 cells. STEAP2 was highly expressed in the more aggressive metastatic cell lines DU145 and PC3 with a 27 fold increase in both compared to PNT2 when analysing with densitometry. Slight up-regulation of STEAP1 was also observed in DU145 and PC3 (2.52 and 3.38 increase respectively) compared to PNT2 (n = 3)
Expression of STEAP1 and STEAP2 in TRUS biopsy patient tissue samples
TRUS biopsies are often the first tissue sample obtained from patients with suspected PCa and thus it is important to identify biomarkers that are detectable within this sampling tissue. Due to the heterogeneous nature of this disease, TRUS biopsies contain both tumour and non-tumour tissue, therefore the use of LCM allows the isolation of specific cancerous cell populations for analysis. Consequently, LCM is critical for gene expression profiling as it prevents the tumour cell expression profiles from being masked or diluted by the natural transcriptional activity of the surrounding normal tissue. However, a caveat of this technique is that very limited quantities of RNA are extracted from the small tumour cell numbers isolated by LCM and this problem is exacerbated by the substantially limited size of the FFPE TRUS biopsy tissue samples, which are typically 5 µm thick, by 1 cm long and 1.2 mm wide. Thus, in the present study, we have described an enhanced method for RNA extraction from laser captured FFPE tissue samples and the subsequent processing steps to facilitate gene expression analysis.
As the data generated in the cell lines predominantly highlighted STEAP1 and STEAP2 in prostate carcinogenesis, their gene expression was analysed in 18 PCa FFPE TRUS biopsy patient tissue samples and four control prostate RNA extracts that originated from un-diseased FFPE prostate tissue. The expression profile of these two genes of interest was assessed in pooled patient samples placed into one of four groups: low risk localised (LRL), high risk localised (HRL), locally advanced, and metastatic tumours and the test gene expression profiles were compared to that seen within the control normal tissue samples.
Interestingly, STEAP1 expression was increased in LRL patients (4.45-fold), HRL (8.15-fold) and metastatic patients (7.45-fold) compared to the normal prostate tissue samples, but its expression was not substantially elevated in the locally advanced patients (2.03-fold) (Fig. 3) (p = 0.007, 0.011, 0.011 and 0.185 respectively). With regard to STEAP2 gene expression, patients in the HRL (1.98-fold) and metastatic (1.71-fold) groups demonstrated normal expression levels of STEAP2 compared to the normal, non-cancer patients, whilst in the low risk localised and locally advanced PCa group of patients there was a 5.77 (p = 0.022) and 14.10-fold (p = 0.003) increase respectively in expression compared to benign prostate tissue.
Relative STEAP1 and STEAP2 gene expression levels in prostate tumour TRUS biopsy tissue samples as compared to normal prostate tissue. a STEAP1 expression was significantly up-regulated in the low risk localised tumours (p = 0.007), high risk localised tumours (p = 0.011) and metastatic patients (p = 0.011) but not within the locally advanced patients (p = 0.185) (n = 3). b STEAP2 expression was significantly elevated in patients within the low risk localised (p = 0.022) and a large up-regulation noted within the locally advanced tumours (p = 0.003) (n = 3). Four to five patients included within each group were pooled together for this analysis
Over expression of STEAP2 results in increased migration and invasion
The data generated from the PCa cell line-based analyses and the gene expression studies in FFPE TRUS biopsy specimens identified STEAP2 as a potential biomarker for invasive PCa. However, in order to determine the mechanistic role that STEAP2 plays in driving prostate carcinogenesis, further analysis was conducted to determine if an up-regulation of the gene in PNT2 normal prostate epithelial cells would enable the gain of migratory or invasion characteristics. A transient transfection approach was utilised to promote over-expression of STEAP2 in PNT2 cells, which was confirmed by qPCR. A 2.2 fold increase in STEAP2 expression was recorded in the transfected cells compared to their parental non-transfected PNT2 control cells. Nonetheless, this nominal up-regulation of STEAP2 in PNT2 cells caused an increase in their capacity for migration as compared to that of non-transfected PNT2 cells (Fig. 4a). In STEAP2 transfected cells the mean percentage of wound closure over 6 h, across three replicates was 63.7 %, which was a statistically significant increase in migratory capacity compared to 40.2 % closure observed with the non-transfected PNT2 cells (p = 0.014) (Fig. 4b). PNT2 cells transfected with the scrambled vector demonstrated a wound closure of 37.1 %, which was very similar to that of the non-transfected cells. Indeed, the difference between wound closure in cells containing the scrambled vector was not statistically different to the non-transfected cells (p = 0.877), but was statistically significantly lower than the STEAP2 transfected cells (p = 0.024). This clearly demonstrated that STEAP2 over-expression resulted in the normal PNT2 cells gaining an increased propensity for migration.
Wound closure rates of PNT2 cells expressing varying levels of STEAP2. a Representative light microscopy images taken at 0, 3 and 6 h time-points following the creation of a scratch wound in PNT2 cell monolayers (images taken with ×10 magnification). b In comparison to both the non-transfected PNT2 cells and the scrambled vector cells, it can be clearly seen that STEAP2 transfected PNT2 cells demonstrated an increased migratory ability as the scratch closed at a faster rate (p = 0.014). c Representative light microscope images taken at 24 h of cell invasion across the basement membrane. Non-Transfected (a) and scrambled vector (b) had very few invasive cells after 24 h, whereas and up-regulation of STEAP2 (c) resulted in an increase of cells with invasive capacity, although not statistically significant. PC3 cells were used as a positive control for invasion (d) (n = 3 for all cell types)
It was also pertinent to consider whether STEAP2 over-expression increased the invasive ability of the normal prostate PNT2 cells. To investigate invasion, PC3 cells were selected as a positive control given their inherent invasive capacity (Fig. 4c). The mean number of PC3 cells, across the three triplicates, invading through the Matrigel layer was 6 % over 24 h (Fig. 4d). Non-transfected PNT2 cells demonstrated a substantially lower invasive ability with only 0.7 % of cells migrating across the basement membrane over the same time period (significantly lower than that of PC3 cells (p = 0.037)). Interestingly however, PNT2 cells over-expressing STEAP2 demonstrated an increase in the number of cells gaining the ability to invade the basement membrane, with 2.3 % of cells traversing the barrier. Although this increase in cell invasion was not statistically significant (p = 0.851), it was 3.2-fold higher than that observed in the control cells, suggesting an increase in migratory capacity following over-expression of STEAP2.
Of interest is whether there is any correlation between STEAP2 expression and clinicopathological status of these patient groups (Table 3). High expression of STEAP2 was noted at the gene expression level within the LRL and locally metastatic patients and not within the HRL or metastatic group. The two patient groupings where high STEAP2 expression was seen, did not have PSA values that were statistically different (p = 0.721), with both having similar PSA values observed at time of diagnosis when the TRUS biopsies were obtained. When taken together, the PSA values of both LRL and locally advanced patients do have statistically different PSA values compared to the HRL (p = 0.002) and the metastatic group (p = 0.001).
Discussion
Incidence rates of PCa are increasing, possibly due in part to the implementation of PSA screening detecting more patients at earlier stage of the disease. However, even with PSA monitoring, clinical stage and Gleason score, when taken together do not adequately identify those tumours that will potentially progress to metastatic disease- which is the main cause of death from malignancy. Metastasis is a crucial step in the progression of cancer. Not only does it make patient treatment more problematic, but it is also associated with poor prognosis. It is important therefore to identify molecules that play a focal role in driving prostate carcinogenesis, particularly those that promote its spread to secondary sites. They could serve as biomarkers for predicting disease progression or alternatively could be targets for potential future therapies. In this study, over 600 genes were screened using oligoarrays that served to first identify those that displayed the most prominently altered expression levels compared to the non-cancerous prostate epithelial cell line, PNT2. This allowed the targeting and subsequent analysis of STEAP1 and STEAP2 in metastatic PCa cell lines, DU145 and PC3, as well as their analysis in patient TRUS biopsy tissue samples (although within a small subset of patient numbers).
Of particular interest was the high gene expression of STEAP2 in metastatic cancer cell lines as compared to PNT2. Furthermore, elevated STEAP2 protein expression was observed in the more aggressive and metastatic cell lines DU145 and PC3 compared to the PNT2 cell line. Thus, it was of particular interest to assess the possibility that STEAP2 may promote a phenotype that supports metastasis. By adopting transient transfection techniques, STEAP2 was over-expressed in the normal prostate epithelial PNT2 cells, and these modified cells were subsequently examined using invasion and migration assays. The migration assay demonstrated that PNT2 cells with elevated STEAP2 gene expression were able to migrate at a quicker rate than the wild-type cells. This was an exciting finding as this study is the first to demonstrate the effect that STEAP2 has on inducing migration in non-cancerous cells that are forced to over-express the gene, thus indicating STEAP2 may have an important role in carcinogenesis. This supports a previous study in normal monkey kidney fibroblast cells (COS-7) transfected with STEAP2, which resulted in higher cell proliferation rates [14]. Wang and colleagues also knocked down STEAP2 in LNCaP cells and demonstrated that this resulted in increased apoptosis [14].
The ability of PNT2-STEAP2 transfected cells to invade through a basement membrane was also considered. Strikingly, a trans-well migration/invasion assay demonstrated that up-regulation of STEAP2 in PNT2 cells caused an increase in the number of cells invading through the matrix compared to wild-type PNT2 cells. The transient transfection technique employed in this investigation resulted in a 2.2-fold increase in STEAP2 expression in PNT2, which is of particular importance given the invasion response observed at this nominal level of over-expression. In the clinical samples assessed, higher levels of STEAP2 over-expression were observed that may therefore be associated with the promotion of greater invasive and migratory abilities. PC3 cells demonstrated a higher invasive capacity than PNT2-STEAP2 transfected cells, but this is to be expected given their higher nascent levels of STEAP2 expression coupled to the fact that many signalling pathways would be destabilised in PC3 due to the genetically unstable nature of these cancerous cells. In contrast, the invasion responses observed in the transfected PNT2 cells would be solely due to the STEAP2 up-regulation, and thus it may be expected that the level of invasion would not be as pronounced as observed for PC3. Consequently, this cumulative migration and invasion data demonstrates that STEAP2 must play a role in promoting cancer cells to invade the local microenvironment, leading to neoplastic progression.
Meta-analysis on publicly available data on the International Cancer Genome Consortium (ICGC) resulted in a total of 67 donors affected by STEAP1 variation across a total of 23 projects accounting for 14 different cancers. There were also 75 mutations identified across all of these projects. The top three cancers with the highest rate of STEAP1 variance include malignant lymphoma where 9 % of patients had varied STEAP1 expression, serous ovarian cancer (5.4 %) and liver cancer patients (3.85 %).
A recent study by Yamamoto et al. demonstrated that STEAP1 was an important molecule for tumour progression. Disruption of the intra-cellular communication between the tumour cells and adjacent tumour associated stromal cells can be achieved when STEAP1 is knockdown within tumour cells [17]. They also observed that STEAP1 was important in the communication between DU145 cells and its surrounding tumour associated stromal cells. This cross-talk leads to the growth of the tumour in vivo, making STEAP1 an interesting therapeutic target [17]. Furthermore, association between STEAP1 expression with features of worse prognosis such as biochemical relapse, high grade tumour and seminal vesicle invasion has been reported [2]. Thus indicating STEAP1 could be an independent prognostic marker of biochemical relapse, particularly as over-expression of STEAP1 in prostate tumours resulted in a decreased time to metastasis and PCa specific death. In other cancers, there is a significant association between high STEAP1 expression and reduced overall survival, such as in colorectal cancer, diffuse large B cell lymphoma, and acute myeloid leukaemia. Furthermore, over-expression of STEAP1 has been observed in malignant plasma cells compared to normal bone marrow plasma cells [18]. The role of STEAP1 in cancer proliferation and invasion may be via its regulation of Na+, k+, and Ca+ concentrations [19–22]. An invasive PCa phenotype has been associated with higher levels of voltage gated Na+ channels, leading to androgen independent stages [22–24]. Consequently, progression of PCa to an androgen insensitive stage results where Ca2+ and K+ levels have been modulated [25, 26].
Although STEAP2 is not highlighted in the 336 cases presented in The Cancer Genome Atlas, evaluation of publicly available data on other databases including the ICGC identified a number of studies where STEAP 2 expression was varied within patients. The three cancers with the highest variance in STEAP2 expression were in oesophageal cancer (18 % of patients), malignant lymphoma (18.2 %), and serous ovarian cancer (16.1 %). A small PCa data set that involved 11 patients resulted in 9.1 % having altered STEAP2 expression. Where data was available, the clinical outcome of patients with altered STEAP2 expression across all cancers resulted in 41 patients having complete remission, 22 relapsed, 21 progressed, 7 remained stable, and 3 had partial remission. A total of 184 mutations in the STEAP2 gene were also noted across all cancer projects.
Previous reports indicate STEAP2 is directly involved in influencing PCa cell proliferation directed partially by the regulation of cell cycle-related gene expression [14]. It has also been shown that STEAP2 is a survival factor in PCa cells as its knockdown resulted in increased apoptosis within LNCaP cells. The apoptotic pathway in which STEAP2 exerts its effect is not currently understood although the extrinsic apoptotic pathway may be a possibility due to STEAP2 localisation at the cell membrane [14, 27]. Under normal conditions signals that are extracellular activate the receptors on the cell surface, initiating a signalling cascade that results in the formation of death-inducing signalling complex (DISC) promoting apoptosis [14]. STEAP2 may therefore be inhibiting the extrinsic apoptotic pathway and possibly decreasing DISC stability.
Altered signalling profiles of the MAPK/ERK pathway has been reported in metastatic cells compared to those that are non-metastatic [28–30], and the effect of STEAP2 on this pathway in PCa cell lines was assessed by Wang and colleagues. They ectopically expressed STEAP2 in DU145 cells and observed ERK activation in response to EGF, which was significantly increased in response to STEAP2. Therefore it was hypothesised that the role of STEAP2 in proliferation and anti-apoptotic function could possibly be ERK mediated [14]. The phosphorylation of ERK results in its translocation into the nucleus where it activates a variety of transcription factors, including activating protein-1 (AP-1). The MMPs −1, −3, −7, −9, −11, −13, and uPA all have consensus sequences for AP-1 [31]. MMPs have the ability to degrade the ECM which is an essential and important step in the progression of cancer. ERK expression has been implicated in PCa [29], as ERK expression increases in a linear manner in epithelial cells from normal prostate, to BPH to PCa [32]. Additionally, elevated ERK levels were observed in high grade and advanced stage prostate tumours which were associated with poor prognostic features [33, 34]. The activation of the ERK pathway by STEAP2 results in the partial arrest of the cell cycle at G0–G1 phase in cancer cells which increases proliferation and tumour development [14], and correlates with our findings following the increased migratory capacity observed in STEAP2 transfected prostate cells.
It is of importance to note that tumours are heterogeneous, the nature of which has been recently reviewed in detail [35, 36]. Consequently, individual TRUS biopsies harboured differing clones of tumour cells, which would have likely demonstrated varying molecular profiles and Gleason grades. In this study LCM was applied to isolate all tumour specific cells within the tissue sections and these samples were subsequently pooled for qPCR analysis. As a result, it is likely that different clones that expressed STEAP2 to varying degrees may have been present within the same tumour. Thus, the gene expression outputs for STEAP2 may have been diluted in cases where there were mixed clonal populations, some of which highly expressed STEAP2, while others may have had more nominal transcriptional levels. It would therefore be pertinent in future studies to assess whether or not STEAP2 is expressed more highly at the invasive front, and whether the differing tumour sub-clones do indeed present with varying or similar transcriptional levels of STEAP2.
In summary, the present study has demonstrated for the first time that over-expression of STEAP2 results in increased migration and invasion of normal prostate epithelial cells, highlighting its potential importance in prostate carcinogenesis. It is hypothesised that one pathway in which STEAP2 could be involved, is through the promotion of an invasive phenotype via the up-regulation of ERK that in turn stimulates the transcription factor AP-1. AP-1 may then activate the MMPs, which have previously been shown to promote PCa progression (MMPs −2, −7, −9, −13 and −14) resulting in the degradation of the basement membrane (11–14). Further studies interrogating this hypothesis are required to assess the correlations between MMPs implicated in PCa and the expression levels of STEAP2. Additionally, future research should focus on examining the relationship between STEAP2 expression and clinical outcome in a large patient cohort to validate its use as a biomarker of aggressive disease.
Abbreviations
- BMP:
-
Bone morphogenic protein
- DISC:
-
Death inducing signalling complex
- ECM:
-
Extracellular matrix
- ERK:
-
Extracellular signal-regulated kinases
- FFPE:
-
Formalin fixed paraffin embedded
- H&E:
-
Haematoxylin and eosin
- HRL:
-
High risk localised
- LCM:
-
Laser capture microdissection
- LRL:
-
Low risk localised
- BPH:
-
Matrix metalloproteinase
- MMP:
-
Benign prostate hyperplasia
- MAPK:
-
Mitogen activated protein kinase
- PCa:
-
Prostate cancer
- PSA:
-
Prostate specific antigen
- TIMP:
-
Tissue inhibitor metalloproteinase
- TGF:
-
Transforming growth factor
- TRUS:
-
Transrectal ultrasound
- STEAP:
-
Six transmembrane epithelial antigen of the prostate
References
Cancer Statistics http://www.cancerresearchuk.org/cancerinfo/cancerstats/types/prostate/incidence/#age. Accessed 15 Mar 2013
Ihlaseh‐Catalano SM, Drigo SA, Jesus C, Domingues MAC, Trindade Filho JCS, Camargo JLV et al (2013) STEAP1 protein overexpression is an independent marker for biochemical recurrence in prostate carcinoma. Histopathology 63(5):678–685
Wright JL, Lange PH (2007) Newer potential biomarkers in prostate cancer. Rev Urol 9(4):207–213
D’Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA et al (1998) Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 280(11):969–974
Partin AW, Mangold LA, Lamm DM, Walsh PC, Epstein JI, Pearson JD (2001) Contemporary update of prostate cancer staging nomograms (Partin Tables) for the new millennium. Urology. 58(6):843–848
Duffy M, McGowan P, Gallagher W (2008) Cancer invasion and metastasis: changing views. J Pathol 214(3):283–293
Frantz C, Stewart KM, Weaver VM (2010) The extracellular matrix at a glance. J Cell Sci 123(24):4195–4200
Gialeli C, Theocharis AD, Karamanos NK (2011) Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 278(1):16–27
Murphy G (2008) The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer 8(12):932–941
Brehmer B, Biesterfeld S, Jakse G (2003) Expression of matrix metalloproteinases (MMP-2 and-9) and their inhibitors (TIMP-1 and-2) in prostate cancer tissue. Prostate Cancer Prostatic Dis 6(3):217–222
Pang ST, Flores-Morales A, Skoog L, Chuan YC, Nordstedt G, Pousette A (2004) Regulation of matrix metalloproteinase 13 expression by androgen in prostate cancer. Oncol Rep 11(6):1187–1192
Cao J, Chiarelli C, Richman O, Zarrabi K, Kozarekar P, Zucker S (2008) Membrane type 1 matrix metalloproteinase induces epithelial-to-mesenchymal transition in prostate cancer. J Biol Chem 283(10):6232–6240
Millimaggi D, Mari M, D’Ascenzo S, Carosa E, Jannini EA, Zucker S et al (2007) Tumor vesicle-associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia (New York, NY) 9(4):349
Wang L, Jin Y, Arnoldussen YJ, Jonson I, Qu S, Mælandsmo GM et al (2010) STAMP1 is both a proliferative and an antiapoptotic factor in prostate cancer. Cancer Res 70(14):5818
Korkmaz KS, Elbi C, Korkmaz CG, Loda M, Hager GL, Saatcioglu F (2002) Molecular cloning and characterization of STAMP1, a highly prostate-specific six transmembrane protein that is overexpressed in prostate cancer. J Biol Chem 277(39):36689–36696
Ohgami RS, Campagna DR, McDonald A, Fleming MD (2006) The Steap proteins are metalloreductases. Blood 108(4):1388
Yamamoto T, Tamura Y, Kobayashi J-i, Kamiguchi K, Hirohashi Y, Miyazaki A et al (2013) Six-transmembrane epithelial antigen of the prostate-1 plays a role for in vivo tumor growth via intercellular communication. Exp Cell Res 319(17):2617–2626
Moreaux J, Kassambara A, Hose D, Klein B (2012) STEAP1 is overexpressed in cancers: a promising therapeutic target. Biochem Biophys Res Commun 429(3):148–155
Gomes IM, Maia CJ, Santos CR (2012) STEAP proteins: from structure to applications in cancer therapy. Mol Cancer Res 10(5):573–587
Challita-Eid PM, Morrison K, Etessami S, An Z, Morrison KJ, Perez-Villar JJ et al (2007) Monoclonal antibodies to six-transmembrane epithelial antigen of the prostate-1 inhibit intercellular communication in vitro and growth of human tumor xenografts in vivo. Cancer Res 67(12):5798
Lalani EN, Laniado ME, Abel PD (1997) Molecular and cellular biology of prostate cancer. Cancer Metastasis Rev 16(1–2):29–66
Smith P, Rhodes NP, Shortland AP, Fraser SP, Djamgoz M, Ke Y et al (1998) Sodium channel protein expression enhances the invasiveness of rat and human prostate cancer cells. FEBS Lett 423(1):19–24
Bennett ES, Smith BA, Harper JM (2004) Voltage-gated Na + channels confer invasive properties on human prostate cancer cells. Pflügers Archiv 447(6):908–914
Laniado ME, Lalani EN, Fraser SP, Grimes JA, Bhangal G, Djamgoz M et al (1997) Expression and functional analysis of voltage-activated Na+ channels in human prostate cancer cell lines and their contribution to invasion in vitro. Am J Pathol 150(4):1213
Laniado ME, Fraser SP, Djamgoz M (2001) Voltage-gated K+ channel activity in human prostate cancer cell lines of markedly different metastatic potential: distinguishing characteristics of PC-3 and LNCaP cells. Prostate 46(4):262–274
Prevarskaya N, Skryma R, Bidaux G, Flourakis M, Shuba Y (2007) Ion channels in death and differentiation of prostate cancer cells. Cell Death Differ 14(7):1295–1304
De Mattia F, Gubser C, van Dommelen MMT, Visch HJ, Distelmaier F, Postigo A et al (2009) Human Golgi antiapoptotic protein modulates intracellular calcium fluxes. Mol Biol Cell 20(16):3638–3645
Kaarbo M, Kokk TI, Saatcioglu F (2007) Androgen signaling and its interactions with other signaling pathways in prostate cancer. BioEssays 29(12):1227–1238
Arnoldussen YJ, Saatcioglu F (2009) Dual specificity phosphatases in prostate cancer. Mol Cell Endocrinol 309(1):1–7
Krueger JS, Keshamouni VG, Atanaskova N, Reddy KB (2001) Temporal and quantitative regulation of mitogen-activated protein kinase (MAPK) modulates cell motility and invasion. Oncogene 20(31):4209
Reddy KB, Nabha SM, Atanaskova N (2003) Role of MAP kinase in tumor progression and invasion. Cancer Metastasis Rev 22(4):395–403
Royuela M, Arenas MI, Bethencourt FR, Fraile B, Paniagua R (2002) Regulation of proliferation/apoptosis equilibrium by mitogen-activated protein kinases in normal, hyperplastic, and carcinomatous human prostate. Hum Pathol 33(3):299–306
Gioeli D, Mandell JW, Petroni GR, Frierson HF, Weber MJ (1999) Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res 59(2):279
Uzgare AR, Kaplan PJ, Greenberg NM (2003) Differential expression and/or activation of P38MAPK, erk1/2, and jnk during the initiation and progression of prostate cancer. Prostate 55(2):128–139
Merlo LM, Pepper JW, Reid BJ, Maley CC (2006) Cancer as an evolutionary and ecological process. Nat Rev Cancer 6(12):924–935
Grunewald T, Herbst SM, Heinze J, Burdach S (2011) Understanding tumor heterogeneity as functional compartments-superorganisms revisited. J Transl Med 9(1):79
Acknowledgments
Funding for this work was kindly provided by grants to SHD from the National Institute for Social Care and Health Research (NISCHR) and the Swansea Prostate Cancer Research Fund.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10585_2014_9679_MOESM1_ESM.doc
Supplemental Fig. 1: List of all genes screened in the oligoarray analysis. A comprehensive list of genes analysed in the oligoarray analysis displayed based on the signalling pathway array type they were positioned upon. Supplementary material 1 (DOC 91 kb)
10585_2014_9679_MOESM2_ESM.doc
Supplemental Fig. 2: Oligoarray gene spot intensities for all four cell lines. Using oligoarrays tailored to four various pathways: WNT signalling pathway, TGFβ/BMP signalling pathway, ECM and adhesion molecules, and PCa biomarkers; over 600 genes were analysed for qualitative identification of potential biomarkers of aggressive PCa. Supplementary material 2 (DOC 72 kb)
Rights and permissions
About this article
Cite this article
Whiteland, H., Spencer-Harty, S., Morgan, C. et al. A role for STEAP2 in prostate cancer progression. Clin Exp Metastasis 31, 909–920 (2014). https://doi.org/10.1007/s10585-014-9679-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-014-9679-9