Abstract
Multiple sclerosis (MS) is a demyelinating neurological disorder with a highly heterogeneous clinical presentation and course of progression. Disease-modifying therapies are the only available treatment, as there is no known cure for the disease. Careful selection of suitable therapies is necessary, as they can be accompanied by serious risks and adverse effects such as infection. Magnetic resonance imaging (MRI) plays a central role in the diagnosis and management of MS, though MRI lesions have displayed only moderate associations with MS clinical outcomes, known as the clinico-radiological paradox. With the advent of machine learning (ML) in healthcare, the predictive power of MRI can be improved by leveraging both traditional and advanced ML algorithms capable of analyzing increasingly complex patterns within neuroimaging data. The purpose of this review was to examine the application of MRI-based ML for prediction of MS disease progression. Studies were divided into five main categories: predicting the conversion of clinically isolated syndrome to MS, cognitive outcome, EDSS-related disability, motor disability and disease activity. The performance of ML models is discussed along with highlighting the influential MRI-derived biomarkers. Overall, MRI-based ML presents a promising avenue for MS prognosis. However, integration of imaging biomarkers with other multimodal patient data shows great potential for advancing personalized healthcare approaches in MS.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) is the primary cause of neurological disability afflicting young adults, with a worldwide prevalence of more than 2.5 million people [1, 2]. Although there is uncertainty surrounding the exact cause of MS, its multifactorial nature has been established, arising from a combination of various factors including genetics, vitamin D deficiency, Epstein–Barr infection, smoking and obesity [3, 4]. MS affects the central nervous system (CNS), characterized by demyelinating lesions throughout the brain, optic nerves and spinal cord, and followed by neurodegeneration [5, 6].
Two main disease courses are observed in MS. The first involves cycles of relapse and remission, known as relapse-remitting MS (RRMS), which often commences in younger individuals, and is followed by slow, gradual progression at later stages known as secondary progressive MS (SPMS). The second course, which only represents around 10% of cases [7], is more common in older adults, bypassing the cyclic nature and following a continuous course of disease progression, termed primary progressive MS (PPMS) [8].
The earliest manifestation of MS is known as clinically isolated syndrome (CIS), which resembles relapses occurring in MS but manifests as an isolated incident and does not necessarily progress to overt MS [9]. Recent evidence also points to the possibility of a prodromal phase in disease evolution, in which non-specific neurological symptoms occur but do not qualify as CIS or definite MS [10]. While not officially part of the MS spectrum, radiologically isolated syndrome (RIS) involves the discovery of white matter (WM) lesions indicative of demyelinating disease without accompanying clinical symptoms [11, 12]. More than fifty percent of individuals with RIS develop clinical symptoms within a decade [13].
Despite the central focus on lesion formation and dissemination in MS [14], disability progression often occurs irrespective of new findings of lesions [15]. Two distinct processes contribute to disability accumulation in MS, namely relapse-associated worsening and progression independent of relapse activity (PIRA) [16, 17]. Due to the significant contribution of PIRA and the limited success of immunomodulatory therapy in MS, the current autoimmune dogma has been challenged, suggesting the disease is essentially caused by a diffuse smoldering process with focal inflammation as a secondary manifestation [15].
MS profoundly affects quality of life through disability accumulation [18] and a wide range of symptoms. While there is no cure, a broad spectrum of disease-modifying therapies (DMTs) are available for slowing down disease progression and disability accumulation [19, 20].
Hence, predicting a patient’s course of disease and risk of progression is pertinent to properly evaluate the benefits of early intervention while minimizing the adverse effects associated with DMT administration, particularly highly effective treatments [21, 22]. However, the heterogeneous nature of MS disease course precludes an accurate prediction with existing prognostic indices and established biomarkers [23], which take a ‘one size fits all’ approach without accounting for individual differences. Artificial intelligence (AI) applications are now extensively being researched for the purpose of personalized healthcare [24], including their use in MS [25].
Magnetic resonance imaging (MRI) has proven to be a pivotal imaging biomarker in MS, playing an essential role in diagnosis, monitoring, prognosis, and outcome assessment [26, 27]. However, expert visual inspection of images is laborious and time-consuming, bringing about the need for automated tools to extract beneficial biomarkers [28]. Furthermore, despite the leaps in MRI biomarker extraction, their association with clinical disability in MS remains moderate at best [29]. To overcome this issue, current research is now combining MRI predictive power with AI, particularly machine learning (ML) [30].
Significant milestones in the MS disease course are the conversion of CIS to clinically definite MS and subsequent signs of increased disease activity reflecting the exacerbation of MS-related disabilities. The most prominent of these disabilities with the greatest impact on quality of life are motor and cognitive impairments [31], with the Expanded Disability Status Scale (EDSS) representing the current standard for assessing MS-related disability [32]. However, the EDSS heavily emphasizes ambulation as a disability measure, with limited contribution of cognition and upper limb function, particularly toward the higher end of the scale [33], necessitating additional assessments for a comprehensive picture of disability in MS patients. Furthermore, these aspects of disability should be independently assessed as they do not necessarily manifest together [34].
The current review provides a comprehensive overview of ML models integrating MRI biomarkers for the task of predicting disease progression in MS. Various reviews on the applications of ML in MS exist in the literature [28, 35,36,37]. However, this work provides a more focused perspective integrating disease progression, MRI biomarkers and ML methods. As such, this paper is meant to serve as a reference for both clinicians and researchers for this application.
Methods, scope and structure
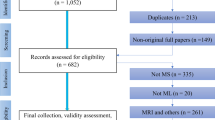
To identify relevant studies for this review, Web of Science, Scopus and PubMed were searched for original research papers published between January 2015 and July 2024 with the following keywords: “machine learning” OR “artificial intelligence” OR “neural networks” OR “deep learning” AND “multiple sclerosis” OR “MS”) AND (“MRI” OR “magnetic resonance imaging”). Additional records were identified through hand search of relevant studies and review papers.
Research papers were included if they adhered to the following criteria: (1) Applied ML techniques in predictive tasks for MS disease progression, (2) incorporated MRI biomarkers as predictors and (3) involved predictive modeling with longitudinal data, using baseline data to forecast future outcomes. The results of the search are outlined in Fig. 1.
Through the identified body of research, studies investigating the value of ML algorithms in predicting MS disease course included the five following areas: predicting the conversion of CIS to MS, cognitive outcome, EDSS-related disability, motor disability and disease activity.
The current review is structured in accordance with the existing literature into the aforementioned five tasks. Sections “Magnetic resonance imaging biomarkers in MS” and “Overview of machine learning” aim to provide an overview of MRI-derived biomarkers (see Fig. 2) and ML methods for unfamiliar readers, and subsequent sections discuss each of the MS predictive tasks including the utilized ML models, MRI-derived predictors and model performance.
MRI-based biomarkers for MS. Traditional biomarkers include lesion features, such as lesion count, volume and location, in addition to the volume of various brain structures. More recent biomarkers include the central vein sign (CVS), paramagnetic rim lesions (PRL), diffusely abnormal white matter (DAWM) and normal appearing white matter (NAWM). NAWM is not visible on conventional MRI, but can be measured indirectly through structural integrity features such as those summarized above. Additionally, biomarkers derived from network neuroscience, including structural and functional connectivity measures, are useful in characterizing MS
Magnetic resonance imaging biomarkers in MS
MRI has proven to be a pivotal imaging biomarker in MS, playing an essential role in diagnosis, monitoring, prognosis, and outcome assessment [26, 27, 38, 39]. Conventional MRI modalities utilized in MS include T1 (T1w) and T2-weighted imaging (T2w), and fluid-attenuated inversion recovery (FLAIR). WM lesions have a characteristic hyperintense appearance on T2w and FLAIR. Some MS lesions are also visible on T1w as hypointensities and are commonly termed as black holes [40,41,42]. When employing gadolinium-based enhancement with T1w images (T1w-Gd), active lesions can be captured through the leakage of Gd through the compromised blood brain barrier, creating a hyperintense appearance [43].
Classic MRI biomarkers usually involve quantification through the number and volume of these lesions. However, these conventional lesion biomarkers, such as the number of lesions, are only moderately associated with MS clinical disability and disease progression at best [29], commonly referred to as the clinico-radiological paradox (CLRDP), primarily due to their failure to capture the diffuse effects of the disease on the WM beyond the focal lesions [40, 44]. Resolving this paradox is at the forefront of contemporary MS biomarker research for both cognitive and physical outcomes of MS [29, 45,46,47,48].
MS prolonged inflammatory activity can be determined through the presence of an iron-laden paramagnetic rim (PRL) surrounding chronic lesions, visualized with susceptibility-based MRI. Another viable biomarker of chronic inflammation is the slowly expanding lesions (SELs) [49, 50], that exhibit a linear expansion on consecutive T1w and T2w scans due to chronic demyelination. SELs contribute to MS disease trajectory and are of particular interest due to their association with smoldering disease activity [15]. The central vein sign is a relatively new imaging biomarker, appearing due to the perivenular nature of MS lesions, and can be captured in ultra-high field MRI studies and T2* sequences [51]. It appears as a central line hypointensity, manifesting partially or completely within the lesion. Its primary function is in diagnosis, as it can successfully discriminate MS from disorders with a similar presentation of WM hyperintensities, such as neuromyelitis optica spectrum disorder and cerebral small vessel disease [52, 53]. However, some recent studies have raised the possibility that CVS may be a prognostic biomarker of RIS converting to MS [54]. Originally thought to be restricted to the WM, MS lesions also impact the gray matter (GM), including both cortical and deep subcortical regions [5, 6, 55, 56]. Within this context, multiple measures have been explored, such as cortical volume/thickness (whole vs. regional), whole brain parenchymal volume, subcortical volume (i.e., thalamus), as elaborated in the subsequent chapters focusing on predicting the course of MS disease. Furthermore, GM pathology has been observed earlier in the disease than WM damage and may also contribute to the CLRDP seen when only accounting for focal WM lesions [57], and is better correlated with physical and cognitive disabilities in patients [58, 59]. GM damage cannot be visualized clearly using T2w and FLAIR sequences due to the low myelin content and poor contrast generated by demyelination [55, 60, 61]. Thus, there have been increasing efforts to develop alternatives aiding the quantification of GM damage. Double inversion recovery offers superior sensitivity for the detection of cortical lesions by contrast enhancement through suppression of WM and cerebrospinal fluid (CSF) signals, though its acquisition is time-consuming and suffers from artifacts [62, 63]. Alternatively, phase-sensitive inversion recovery can be used with comparable performance while being acquired in a clinically feasible timeframe [63].
While MRI investigations concerning MS have centered on the brain, it's crucial to note that the spinal cord also plays a significant role in MS-related disability. However, conducting MRI scans of the spinal cord presents more challenges due to its smaller anatomical size and heightened susceptibility to artifacts compared to brain MRI, explaining the lack of inclusion of this biomarker in the prediction of MS disease outcomes [64].
Further, some of the more advanced MRI biomarkers, that quantify damage beyond focal lesions, are the diffusely abnormal WM (DAWM) and the normal appearing WM (NAWM). NAWM appears healthy on conventional MRI, but damage within can be visualized using advanced MRI (e.g., Diffusion weighted (DWI)/ Diffusion tensor imaging (DTI)-based fractional anisotropy (FA) and mean diffusivity (MD)) [65] and histopathology [66]. On the other hand, DAWM is widespread, appearing isointense to GM on conventional MRI, and is commonly situated near the occipital horns of the lateral ventricles (LV) [66,67,68,69]. Another property of integrity that can be used as an indirect measure of myelin content in brain tissue is the magnetization transfer ratio (MTR) [70], which has been shown to relate to motor disability in MS [71].
Finally, an alternative approach to representing lesion-inflicted damage in MS is through structural connectivity, which can be measured by tractography with DWI and DTI. Using this method, the brain is represented as a mathematical graph with GM structures as nodes and WM tracts as connecting edges. As such, WM lesions lead to disruptions in neuronal connectivity, and these structural disconnections are associated with cognitive and motor impairment in MS patients [72, 73]. In addition to structural MRI, useful biomarkers in MS can be obtained from functional MRI (fMRI) through measures of functional connectivity (FC) [74, 75]. Recent research on FC emphasizes the notable effects on deep GM regions, particularly the thalamus, where both increased FC (with hippocampus, motor, occipital, temporal cortical region) and decreased FC (with frontal cortical regions) was observed [75].
Overview of machine learning
Big data is at the center of modern healthcare, revolutionizing the way we understand and approach medical challenges. The advent of AI has empowered the processing of vast datasets in an unprecedented manner, enabling healthcare professionals to effectively analyze patient data and make informed decisions [76,77,78]. ML is one of the most utilized branches of AI, particularly in healthcare applications [79]. ML avoids explicit programming in favor of a task- or data-driven approach in which a specific algorithm is fit to the supplied data with the purpose of learning features that may be generalized to make predictions on new, unseen data [80]. The two main branches of ML are supervised and unsupervised learning [81].
The major factor distinguishing supervised from unsupervised ML is the availability of labeled data. Supervised learning involves mapping input data to output labels using a predefined mathematical function, including either classification or regression. Classification tasks aim to predict discrete labels, while regression is used to predict continuous output [82]. Classification can be used to predict a binary outcome, known as binary classification, or for three or more outcomes, known as multi-class classification [83]. Contrarily, unsupervised ML is employed to learn underlying patterns from unlabeled data, comprising primarily of dimensionality reduction and clustering tasks [80].
The primary problem in ML is overfitting, whereby the model memorizes patterns—including noise—from the data it is trained on, leading to subpar performance on unseen data. The main causes of overfitting, which impairs the models' capacity for generalization, are insufficient training data and unduly complicated models. Reducing learnable parameters (model complexity) and obtaining additional training data via augmentation techniques are two ways to mitigate overfitting [84, 85].
Examples of supervised learning algorithms include linear (LR) and logistic regression (LoR), support vector machine (SVM), K-nearest neighbors (KNN) and naïve Bayes (NB). LR is the most basic form of regression, mapping predictors linearly to a continuous outcome variable [86]. Generalized linear models (GLM) are a class of models that extend LR by allowing for different types of distributions for the dependent variable, using a link function to model the relationship between the dependent and independent variables. LoR is a type of GLM that assumes a linear relationship between the predictor variables and the logit of the outcome, and produces output bounded between 0 and 1. This is useful for classification tasks, as a threshold can be applied to the output to produce binary classes of 0 and 1. When more than 2 classes are used, it is known as multinomial LoR [87].
SVMs can be used for both classification and regression tasks, utilizing a subsample of the training data known as the support vectors to find an optimal hyperplane capable of maximizing the separation between the classes of interest. Depending on the linear separability of the dataset, various kernel functions can be used such as linear, radial and polynomial kernels [88, 89]. Another algorithm for classification and regression is KNN. This model does not explicitly map variables to their labels, but instead relies on the proximity of data points in the feature space. When a new data point is introduced to the model, its label is predicted based on the labels or values of a predetermined number, or k, of its nearest neighbors [90]. On the other hand, NB, which is based on Bayes theorem, is primarily used for classification [91].
Additional, tree-based algorithms are decision trees (DT) and random forest (RF). DTs work by continuously dividing the data into smaller subsets according to various attribute values until a predefined stopping condition is reached. This iterative partitioning creates a tree-like diagram where each node signifies a decision point or a split based on a particular attribute [92,93,94]. RFs are an extension of DT, which combine a number of DTs as base learners to form a more robust model by averaging the predictions of individual trees [95, 96]. This technique is known as ensemble learning, which can also aid in alleviating overfitting [97]. Another form of ensembles is boosting, which iteratively adds weaker base learners, or DTs in this case, with each new tree added to correct the errors made by the previous ones, thereby improving the model's overall accuracy and robustness. Examples of this are Adaptive Boosting (AdB), Gradient Boosting (GB) and eXtreme Gradient Boosting (XGBoost) [95, 96].
Unsupervised ML, as previously referred to, identifies underlying patterns or structures within the data without requiring labeled outcomes. Clustering algorithms, such as k-means clustering, partition datapoints into groups based on similarities or distances from each other, aiming to minimize within group variance and maximize intergroup variance [98, 99]. For example, Gaussian Mixture Models (GMMs) extend the concept of Gaussian Probability (GP) using multiple Gaussian distributions to model complex data structures. While GP describes a single Gaussian, GMMs combine several components, each with their mean and covariance [98].
Alternatively, dimensionality reduction techniques transform datasets into a lower-dimensional space while retaining as much of the original variability or essential information as possible [98, 99]. Examples of this are principal component analysis (PCA) and the uniform manifold approximation and projection (UMAP). Where PCA finds linear combinations of the variables, UMAP captures nonlinear relationships by preserving the local and global structure of the data in the reduced space [100].
The aforementioned algorithms are part of traditional ML, requiring feature engineering to curate raw data into useful features that can greatly influence model performance [101]. Additionally, the emergence of high dimensional datasets brought about the well-known “curse of dimensionality”, when features are highly sparse and exceed the number of training points. High dimensionality can also contribute to overfitting, thus requiring feature selection methods to select a smaller subset of the variables, or projection into a smaller feature space through dimensionality reduction techniques discussed above [102]. Feature selection can be carried out in three different manners according to the level of integration with the model. Filter methods are done independently prior to model training, and rely on statistical techniques such as correlation analysis or information gain. Wrapper methods, on the other hand, are employed as part of the training process. Feature selection relies on the performance of the model, such as recursive feature elimination (RFE), where features with the least importance are systematically removed, and the model is re-evaluated until the optimal subset of features is identified. Finally, embedded methods are integrated into the training algorithm of the model, such as the addition of regularization, which penalizes added complexity by shrinking or eliminating the contribution of some features, thereby automatically performing feature selection. Examples include LASSO and ridge regularization, often combined with LR and LoR modeling [103,104,105].
Deep learning (DL) is a branch of ML that differs from traditional techniques by removing the necessity for feature engineering [101]. In essence, DL can be seen as a revival of the artificial neural network, where artificial neurons are arranged in stacked layers [106]. Modern DL architectures include multilayer perceptrons (MLP), convolutional neural networks (CNN), recurrent neural networks (RNN), generative adversarial networks (GAN), long short-term memory networks (LSTM), gated recurrent units (GRU), and encoder-decoder, among others [107].
CNNs are the most popular architecture for computer vision applications, consisting of a series of layers that obtain latent features from an image in a process known as feature extraction, which is done through convolutional operations that apply filters to the input image [108]. Residual Networks (ResNet) is a deep CNN architecture that uses skip connections, or residual learning, to allow for deeply stacked layers without suffering performance degradation [109]. Another form of deep CNN consisting of either 16 or 19 layers is the VGG-Net architecture, which works by capturing fine-grained patterns through small convolutional filters [110]. Specifically designed for image segmentation, the U-Net architecture is another variation of the CNN, in which an encoder–decoder structure is used to first compress the input image into a lower-dimensional representation and then reconstruct it with high spatial resolution, enabling precise localization of features within the image [111].
A specialized form of DL models, known as the graph neural network (GNN), is useful for handling data structured as graphs, composed of a series of vertices and edges, with edges representing the relationships between vertices. Such models are especially useful in neurological disorders, where different brain regions and their connectivity patterns can be modeled as graphs. Examples of GNN architectures include graph convolutional networks (GCN) and graph attention networks (GAT) [112,113,114,115].
Despite rapid developments in ML and DL algorithms, numerous challenges persist in healthcare predictive tasks. Medical datasets often suffer from class imbalance due to the relatively low prevalence of certain conditions. Most classification algorithms do not perform well on imbalanced datasets, often showing bias toward the majority class [116]. To address this, popular methods include up-sampling of the minority class, such as the synthetic minority oversampling technique (SMOTE) [117].
Another major challenge in disease risk prediction is the black box nature of many predictive models. Commonly used models, such as RF, SVM, and DL models, are effective but often lack interpretability. This lack of transparency can hinder their utility in healthcare, where clear and understandable explanations are crucial for supporting medical professionals in decision-making. To circumvent this, various interpretability methods have been developed, including Shapley Additive exPlanations (SHAP), which borrows from game theory to assess the importance of predictive features after model training [118, 119].
Medical image analysis has undergone a transformation with the wake of DL, producing impressive outcomes in tasks like registration, segmentation, and classification [108, 120]. Neuroradiology is at the forefront of ML and DL applications, and MS ranks among the ten most actively explored neurological diseases in this area of research [37]. Applications have included MS diagnosis, disease course, disability progression, in addition to treatment responses [35, 36, 121]. Figure 3 displays the most popular ML algorithms utilized for prognostic tasks in MS with the inclusion of MRI-based biomarkers.
The application of MRI-based biomarkers and ML methods for prognostic tasks in MS. The most popular ML algorithms include logistic regression (LR), support vector machine (SVM), random forest (RF), multilayer perceptrons (MLP) and convolutional neural networks (CNN). Prognostic applications include predicting the conversion of clinically isolated syndrome (CIS) to MS, cognitive outcome, EDSS-related disability, motor outcome and disease activity
Machine learning studies for the prediction of multiple sclerosis progression with MRI biomarkers
This section offers a discussion of the most recent studies integrating MRI biomarkers into ML models for the prognosis of MS progression. Disease prognosis is divided into five main categories: conversion of CIS to overt MS, cognitive outcome, EDSS-related disability, motor disability and disease activity. The model architectures and MRI-derived biomarkers are discussed, and details for included studies are displayed in Tables 1, 2, 3, 4, 5, 6. Additionally, Fig. 4 shows the frequency of use for different ML model types across the five prognostic applications, and Fig. 5 illustrates biomarkers used throughout the discussed studies, specifically those related to MS lesion imaging.
Proposed ML models and their frequency according to application in MS disease prognosis. The most popular algorithms include random forest (RF), support vector machine (SVM) and convolutional neural networks (CNN). Additional models include AdaBoost decision trees (AdB-DT), deep belief networks (DBN), decision trees (DT), graph neural networks (GNN), ensemble modeling, K-nearest neighbors (KNN), light gradient boosting (L-GBT), logistic regression (LR), multiple kernel learning (MKL), multilayer perceptrons (MLP), residual networks (ResNet), ridge regression (RR), uniform manifold approximation and projection (UMAP), U-Nets and extreme gradient boosting (XGBoost)
Conversion of clinically isolated syndrome to overt multiple sclerosis
CIS describes the condition preceding MS, in which patients present with a single episode suggestive of CNS demyelination, most often impacting the optic nerve, brain stem or spinal cord [11, 122]. For a diagnosis of CIS, episodes must persist for at least 24 h without qualifying for a diagnosis of MS, and devoid of signs and symptoms indicative of an alternative condition [123]. While 85% of MS patients originally present with CIS, around 20% of those with CIS never progress to overt MS [124], implying a heterogeneous and unpredictable course of disease.
Timely administration of DMTs can delay CNS damage and prevent future disabilities [124, 125], but comes with several risks, such as serious infections and DMT-specific infections [126,127,128]. Therefore, caution is required for selecting patients most likely to benefit from early intervention, which is currently problematic due to the low specificity of the 2017 McDonald criteria [14] alone for identification of early MS [124]. Therefore, advancing methods to predict CIS to MS conversion is vital for personalized patient care, including the utilization of ML with MRI biomarkers for this purpose.
In [129,130,131,132], traditional ML algorithms were employed for predicting CIS conversion to MS. In Zhang et al. [129] and Bendfeldt et al. [131], MRI-derived lesion geometric features were highlighted as features of importance. The first study [129] utilized a RF model incorporating lesion shape features, with minimum sphericity, surface–volume ratio as well as mean lesion volume, achieving a superior specificity compared to the 2010 McDonald criteria (50% versus 22%). The second study [131] employed linear and kernel-based SVMs, integrating Minkowski functionals of lesion geometry from T1w-Gd lesion masks (volume, surface area, mean breadth and Euler-Poincare characteristic). Combining these features with clinical variables achieved an accuracy of 70.4% in patients under interferon beta-1b treatment, particularly when summarizing lesion characteristics for the entire brain as opposed to specific regions of interest.
For Wottschel et al. [130] and Rasouli et al. [132], lesion location was accounted for as a MRI predictor. In [130], the average distance of lesions from the brain center and the shortest distance from the vertical axis horizontally were selected as influential features through forward RFE and subsequent SVM modeling. In [132], an XGBoost model was employed using clinical, demographic, and brain/spinal cord MRI lesion location to predict conversion to MS within 10 years. The model achieved an area under the receiver operating characteristic curve (AUROC) of 0.858 on test data, with periventricular and infratentorial lesions emerging as the strongest predictors as determined by SHAP.
Tayyab et al. [133] introduced two aspects of novelty to their predictive model. First, the volumes of deep GM nuclei were included as predictor variables along with T2-w lesion load, brain parenchymal fraction (BPF) and anatomical location of CIS onset. Second, instead of excluding participants with uncertain outcome labels, they were included in a probabilistic RF model, which models outcome as a probability rather than a deterministic label. This approach outperformed models trained by either excluding subjects with uncertain labels or assigning post hoc labels. In terms of feature importance, thalamic and lesion volumes were the highest-ranking predictors. This model achieved a particularly high recall at 0.923, however, the AUROC was modest at 0.76.
The challenges of MRI data heterogeneity in multicenter datasets were highlighted in [134]. Despite the inclusion of biomarkers that are more robust to MRI variations, including GM probability and cortical thickness, predictive modeling with linear SVM and RFE produced a large variation in accuracies between single-center experiments, extending from 0.73 to 0.93 when using leave-one-out cross-validation scheme. Furthermore, models utilizing combined datasets from multicenter data achieved the lowest accuracy when patients from all centers were included in the analysis with a value of 0.70. However, on RFE, a consistent subset of features was selected including deep GM, frontal, temporal and limbic lobe-derived features.
Recently, studies have gravitated toward utilizing latent imaging features obtained from segmented lesion masks via DL models, rather than traditionally extracted MRI lesion features that are heavily reliant on domain expertise and are prone to bias. An example of this application is evident in the prediction of CIS disease progression as demonstrated in [135,136,137,138].
Two related studies [136, 137] explored the use of CNNs for predicting the two-year outcome of CIS based on lesion masks and other features. The first study [137] focused on using preprocessed lesion masks, incorporating downsampling and Euclidean distance transform as inputs to the CNN model, achieving an accuracy of 0.729 on sevenfold cross-validation. The subsequent study [136] extended this approach by adding demographic, clinical, and additional MRI parameters as detailed in Table 1. This enhanced model, which also utilized latent features from lesion masks preprocessed in the same manner as [137], achieved a higher accuracy of 0.75 and particularly improved specificity, outperforming the previous model and other ML techniques like multivariate LoR and RF. Although the gain in accuracy was not large, the comparative results emphasize the importance of incorporating a broader range of features alongside lesion masks for more accurate and balanced predictions in CIS outcomes.
On the other hand, Afzal et al. [135] directly fed T2w images into the CNN network, mitigating overfitting with data augmentation and a smaller network architecture. Due to limited data availability, model evaluation was carried out with the baseline MRIs of only 6 subjects to predict conversion to CDMS after one year. Prediction accuracy ranged from 0.833 to 1.0 when varying the data subset utilized for training. However, the extremely small sample size raises concerns about the study's generalizability and the potential for overfitting, even with the applied mitigation strategies. The reported high accuracy, especially given the limited data, suggests that the model might have learned specific characteristics of the training set rather than generalizable patterns. Additionally, the variation in accuracy depending on the training subset used indicates a lack of consistency, further questioning the robustness of the model.
Furthermore, a study by Pareto et al. [138] attempted to predict a second clinical event in CIS patients based solely on features derived from T1w MRIs. However, the performance did not outperform random guessing, suggesting the need for other MRI modalities and clinical variables to enhance the separation of patient subgroups.
While the discussed predictive models for the conversion from CIS to overt MS present promising avenues for early intervention, comparing their efficacy poses significant challenges. The reliance on cross-validation in these studies raises concerns about potential overfitting and the true generalizability of the models to unseen data. Moreover, the studies use various prediction intervals and definitions for conversion to MS that further complicates comparisons. Different criteria were used across the studies to classify patients as having CIS according to differing versions of the McDonald criteria. The 2005 McDonald criteria [139] required more stringent clinical and MRI evidence for diagnosing MS, often resulting in more patients being classified with CIS until a second clinical event occurred. The 2010 [140] and 2017 [14] updates relaxed these requirements, allowing for earlier diagnosis based on MRI findings alone. Therefore, many CIS cases diagnosed according to earlier criteria would have qualified for a diagnosis of MS.
Cognitive outcome
Cognitive impairment (CI) is a common disability affecting individuals with MS, with as many as 70% of patients impacted [141], and may be the earliest neurological symptom in patients with RIS [142, 143]. The aspects of cognition most often altered in MS are working memory and information processing speed (IPS), in addition to executive functions, complex attention, and visuospatial ability [144, 145]. Despite the debilitating effects of cognitive decline in this group of patients, the bulk of research efforts is focused on physical disability, and the cognitive domain remains largely underrepresented [146]. Depending on the course of the disease, CI may develop insidiously over time, or present as an abrupt decline during disease relapses [147].
Treatment of existing cognitive decline in MS has largely shown to be ineffective, and compensatory solutions for its manifestations are scarce, rendering prevention of its occurrence the best course of action [148, 149]. Furthermore, clinical trials with cognition as a primary endpoint are rare [150, 151]. Therefore, early prediction of susceptible individuals for inclusion can aid in enhancing the predictive power of these trials to produce reliable assessments of experimental therapies [152].
Damage to the GM and WM of the brain are likely the cause of CI in MS patients. However, the etiology is complex, and conventional MRI biomarkers of structural integrity do not necessarily correlate with symptoms of CI in this subpopulation [141]. Studies in functional brain networks have revealed a possible compensatory effect to structural damage offered by dynamic changes in brain connectivity, with functional reorganization potentially masking the clinical expression of CI [153]. While GM damage, WM lesions, and changes in structural and functional connectivity all offer insights to the heterogenous presentation of CI in MS, cortical atrophy has been shown to be the strongest MRI predictor of future cognitive decline [147].
A series of studies [154,155,156] used statistical models to identify MRI biomarkers predicting CI in MS patients. Key findings included the role of various brain volumes as predictors of CI, with cortical GM volume (GMV) emerging as a strong predictor of CI across different MS stages. Furthermore, various cross-sectional studies [157,158,159] explored the use of ML and MRI biomarkers for the prediction of the Symbol Digit Modalities Test (SDMT) scores [160]. SDMT, which can be carried out within 5 min, is considered the gold standard for quick cognitive assessment in MS. However, it can only assess a single aspect of CI, namely IPS [142]. A variety of ML models were employed including XGBoost [157], RF [158] and SVM [159], all using a combination of clinical and MRI predictors for analysis. These studies reiterated the significance of cortical and deep GMVs for the prediction of CI, in addition to the increased connectivity of the cingulo-opercular task control network in resting-state fMRI [158].
Three studies [152, 161, 162] involved predictive modeling across different timeframes for various aspects of cognitive decline, revealing the limited contribution of lesion volume alone [152] and emphasizing the added benefit of integrating multimodal MRI features. Labiano-Fontcuberta et al. [152] constructed a longitudinal ML model designed specifically for routine clinical practice, thus utilizing features that are feasibly attainable in a clinical setting. A single aspect of cognitive functioning was measured through the Processing Speed Test (PST), serving as a substitute for the SDMT. The only MRI biomarker employed was T2 lesion load, along with multiple clinical and demographic predictors as detailed in Table 2. Various linear, nonlinear and tree-based models were utilized along with SMOTE for class-balancing, with the tree-based models XGBoost and RF performing best on both training and testing data, achieving an AUROC of 0.94 and 0.90 respectively. Given the emphasis on clinical application in this study, variable importance scores were computed for each of the predictors, placing T2 lesion load on the lower end of variable importance.
In the remaining two studies [161, 162], multiple aspects of cognition were assessed employing a larger number of MRI biomarkers. While [162] utilized lesion volume and 76 cortical/subcortical GMVs, Al-iedani et al. [161] offered the most advanced MRI biomarkers including DTI-derived measures, MRS-derived neurometabolite concentrations from the hippocampus, prefrontal cortex and posterior cingulate cortex, in addition to lesion and structural volumes. Given the large number of predictor variables employed, both studies leveraged multivariable GLMs with regularization to select the best subset of features for subsequent modeling. The variables selected for [162] were best for the prediction of IPS (Acc = 73%) and verbal memory (Acc = 79%), whereas those utilized in [161] did not perform well for IPS, yielding an R2 of only 0.31 as opposed to a value of 0.54 for the remaining cognitive tests. However, the results are not necessarily comparable given the heterogeneous cohort in [162] involving various MS disease stages, as opposed to [161] which only constituted RRMS. Furthermore, a train-test split was not employed in [161], thus lacking information on model generalizability. Nonetheless, the most promising biomarkers included the FA of NAWM and Glutathione levels in the prefrontal cortex for [161], and cingulate and medial temporal lobe volumes for the models in [162].
A scarcity is evident for applications of ML to predictive modeling of cognitive decline, and this is further accentuated for longitudinal predictive tasks. The feasibility of implementing various cognitive tests in a clinical setting is challenged by time constraints, emphasizing the need for standardized testing protocols to ensure comparable and reliable results. The influence of the disease stage on predictive accuracy warrants further exploration, as does the need for consensus on defining CI, including the choice of thresholds and the number of cognitive domains exhibiting decline. Additionally, the integration of motor components, especially in patients with upper limb disabilities, remains a critical consideration for accurate predictions given the reliance of these testing procedures on upper limb mobility [163]. Addressing these nuances will enhance the practicality and effectiveness of ML models in predicting cognitive outcomes in MS patients.
EDSS-related disability
The Expanded Disability Status Scale (EDSS) [164] is the most commonly employed instrument for assessing disease severity and tracking disease progression in MS [32]. It consists of a scale ranging from 0 to 10, with 0 indicating no neurological abnormalities, and 10 representing death due to MS. Seven functional systems are assessed, including the pyramidal, cerebellar, brainstem, sensory, bowel/bladder, mental and visual functions, in addition to an 8th category signifying the presence of other symptoms outside of the aforementioned categories [165]. The final EDSS step is established based on the interplay between the eight functional systems and ambulatory function [33]. Despite its widespread use, the reliability of EDSS has been questioned due to several limitations: emphasis on ambulation at the higher end of the scale [33], nonlinear nature of the scale [166], variable duration at each stage [165], and underrepresentation of vision, cognition and upper limb function at values of 4 and above [167]. Nonetheless, since its inception in 1983, the EDSS has been valuable for objective long-term monitoring of MS patients [166].
Numerous studies have investigated the relationship between MRI biomarkers and EDSS in MS [168,169,170], revealing moderate relationships with T2 lesion volume (T2LV) and its change [168], cerebral atrophy rate [170], and various brain volumes [168]. This relationship is more pronounced, however, in the earlier stages of the disease [171]. Unconventional biomarkers were also associated with EDSS progression, such as signal intensity variations of lesions through inversion recovery ultrashort echo time (IR-UTE) [172], volume of MS lesions present at baseline but atrophied on follow-up [173], and structural cortical networks [174]. Therefore, various studies have incorporated MRI biomarkers in ML models for prediction of disease progression as determined through EDSS [175,176,177,178,179,180], which are summarized in Table 3.
Combining MRI volumetric, connectivity and radiomics features with clinical variables, Pontillo et al. [181] utilized a range of ML regression models for the prediction of EDSS. With over 125,000 extracted features, a combination of collinearity, variance, LASSO regression and correlation analyses were employed to reduce the predictor variables to only 9 features, which included a subset of radiomics features obtained from the cerebellum, prefrontal cortex, and deep GM structures. The best-performing models were SVM and ridge regression, achieving a MAE of 1.112 and 1.155 on external validation, respectively [181]. Tommasin et al. [182] included FA maps of NAWM in 1000 RF classifier models that were trained to assess EDSS worsening in addition to cerebellar and thalamic volumes, among others detailed in Table 3. Classifiers trained with radiological features alone outperformed those trained on clinical data or combined clinical/radiological variables, achieving an AUROC of 0.92. The only variable identified as influential in all model iterations was T2 lesion load, while the remaining MRI features were only prominent in less than 40% of trained models.
By modeling MS disease trajectories as changes in EDSS, Campanioni et al. [183] employed regressors and classification models to forecast disability accrual using baseline MRI features, patient age and sex. Regression models aimed to predict the change in EDSS, normalized by time interval, while the classification models were for the purpose of predicting EDSS values at 5 different timepoints, where categories represented the individual scores on the scale. XGBoost models performed best for both tasks, and SHAP identified the most influential feature across most models as the number of brain lesions on MRI.
The value of ensemble modeling for prediction of future decline, as measured through EDSS, was assessed by authors in [175,176,177], with all models utilizing BPF and T2LV as MRI-derived predictors. Zhao et al. [175] focused on optimizing SVM models with strategies like undersampling, bagging, and penalizing false negatives, finding that BPF was a predictor for non-progressive MS, while T2LV predicted EDSS progression. Interestingly, longitudinal changes in EDSS, ambulation index and FS scores were also incorporated into the model. When limiting predictors to clinical and demographic data, an overall accuracy of 0.55 and a sensitivity of 0.78 were achieved, while including MRI biomarkers improved these scores to 0.67 and 0.81, respectively. This work was extended in [177] using two independent datasets (EPIC and CLIMB) with both homogeneous and heterogeneous ensembles. Similarly, longitudinal changes in clinical variables were employed, revealing that, while GMV and ventricular CSF were strong predictors in homogeneous models, these clinical features remained the most influential across all models.
In [176], the authors aimed to develop a prognostic tool for predicting EDSS-based disease progression in SPMS patients, also leveraging ensemble learning. Various ensemble methods were considered, including LoR, linear SVM, and tree-based models, such as RF and AdaBoost decision trees (AdB-DT). Tree-based models showed superior performance, particularly in specificity, compared to parametric models. Among parametric models, BPF was more effective than T2LV, whereas tree-based models performed better with T2LV. Nonetheless, clinical features like baseline EDSS and 9-Hole Peg Test scores were more predictive than MRI-based features.
Roca et al. [178] extended ensemble modeling by combination with DL, constructing a model consisting of a RF regressor, UMAP and CNN for latent feature extraction from FLAIR images and lesion masks. MRI biomarkers fed into the RF and UMAP included whole brain and WM tract-based lesion load, in addition to LV volume. By combining the aforementioned features with age in the ensemble regressor, MSE values of 3 were achieved on unseen test data. The performance of the various models varied across EDSS scores, with most models performing best in the intermediate range of the EDSS and more prone to error at the higher and lower ends of the scale. Overall, RF models utilizing engineered features performed better in predicting EDSS than latent features extracted by CNN. Notably, the standout performers among these features were the lesion load in the posterior corona radiata L tract and the volume of the LV [178].
In [179], multicenter clinical trial data was also utilized to build a classifier to identify progressors in terms of EDSS over one year. This approach relied entirely on DL, namely CNN blocks consisting of parallel pathways with various resolutions that are concatenated before being passed to the next layer. Five MRI modalities were included as input (T2w, T1p, T1c, PDw and FLAIR), in addition to T2w and T1w-Gd lesion masks. When including the 5 modalities alone, the AUROC achieved was 0.66, while including the lesion masks boosted the performance to 0.701. Dular et al. [184] also utilized multimodal MRI in a DL architecture, employing transfer learning for enhanced latent feature extraction. A VGG network was trained for the task of brain age estimation, and the resulting pretrained model was then leveraged for the task of disease progression classification according to changes in EDSS. This study, however, performed poorly in terms of precision and recall, which can be seen in Table 3.
Utilizing the DL architecture ResNet50, Taloni et al. [180] constructed a classifier discriminating MS progressors from non-progressors over the course of 4 years. Rather than implementing a 3D-based model on patient MRIs, 2D slices were extracted in the coronal, sagittal and axial planes, then further divided into 4 slabs of slices per plane according to anatomical location and structural similarity. The model training unfolded in a dual-phase approach: initially, a pretrained ResNet50 underwent three rounds of fine-tuning—once for each plane—to discern and classify slices based on their slab location. The outcome of this process yielded 3 Brain Scan Optimized Models (BSOM). The 3 models were then expanded to a total of 12 by passing the slices of each slab through the finetuned ResNet50, this time with the prediction of EDSS-progressors as the target. Overall, models exploiting individual slices for prediction performed better than those incorporating 3D slabs of MRI data for prediction, particularly axially directed slices incorporating information on the lateral and third ventricles.
Leveraging a multimodal deep neural network, Zhang et al. [185] integrated various encoder architectures into a single DL model for the classification of patients based on three-year EDSS scores. ResNet, 1D CNNs, and graph attentional convolutional encoders were used to generate embeddings from MRIs, structured electronic health records and unstructured clinical notes, respectively. Given the integration of longitudinal data, a bidirectional GRU was integrated into the pipeline as the decoder structure. Including MRI embeddings improved AUROC from 0.8078 to 0.8380 for classification of patients reaching a milestone of EDSS greater than 4 [185].
It is crucial to note that there were varied definitions of sustained disability increase across the discussed ML models (Table 3), highlighting the need for standardized criteria in future research. Furthermore, prediction accuracies may benefit from the inclusion of spinal cord parameters. Spinal cord imaging is not recommended for routine monitoring of MS patients [186], hence the lack of their incorporation in the aforementioned studies. Given the motor-centric focus of the EDSS, it is unsurprising that significant correlations have been found between spinal cord imaging features and this disability scale [187, 188].
Motor disability
The most recognized symptoms of MS are the occurrence of motor impairments, which impart a heavy impact on patient quality of life. Frequently manifesting deficits include ambulation, upper limb (UL) movements, balance [189, 190] and gait impairment, which is the clinical hallmark of MS [191]. However, MS patients are not characterized by a typical gait due to the wide dissemination of lesions throughout the central nervous system, which can impact gait in various manners [192]. Balance impairment is a pivotal concern, as it increases the risk of falls in patients with MS and hinders independence in daily life [193].
Walking impairment was reported to have the greatest impact on quality of life by both MS patients and physicians [191], with as many as 75% of patients reporting reduced mobility as a consequence [192]. In a cross-sectional study by Buckova et al. [194], fMRI-derived FC measures combined with a SVM model were predictive of walking ability as measured by the Multiple Sclerosis Walking Scale (MSWS).
Despite the wide focus on lower limb function, UL impairments are significantly experienced in MS, ranging from fine to gross movements [195, 196]. The gold standard for assessing distal UL function, or manual dexterity, is the nine-hole peg test (9HPT). 9HPT measures manual dexterity by recording the amount of time taken to place and remove nine pegs from nine holes within a peg board, assessing both hands separately [197, 198]. In Cordani et al. [199], the MD of premotor cortex transcallosal fibers, GM, WM and whole brain volumes were predictive of 9HPT scores in a cross-sectional cohort using RF modeling.
The only recent study, to our knowledge, assessing longitudinal motor impairment in MS patients is Mostafa et al. [200], which assessed changes in UL function as determined by 9HPT scores over 144 weeks for a cohort of PPMS patients in an ocrelizumab clinical trial. A variety of ML models were used along with demographic, clinical and MRI biomarkers. Specifically, brain volume, T2LV and Gd + lesion count were employed as the MRI-derived features. All models performed similarly in spite of algorithmic complexity, however, there was a notable drop in performance on testing (AUROC = 0.63–0.67) in comparison to training (AUROC = 0.74–0.89) when experiments with 6 different splits of the data were performed, most likely reflecting a problem of overfitting. However, this study provides a critical contribution to the field by the inclusion of a relatively large cohort of PPMS patients. The focus on PPMS patients addresses a crucial gap in research focused on prediction of progression in this less frequently studied but significant subgroup of MS patients [201].
Due to combined cognitive and motor deficits, MS patients are at increased risk of experiencing falls, which adversely affects daily life on various physical, psychological, and social fronts. Fall history is the greatest predictor of future events. However, this impairs preventative strategies and does not offer an understanding of the mechanism behind fall occurrences [202, 203]. Furthermore, the multifactorial nature of falls encompassing motor, sensory and cognitive deficits hinders the use of a single clinical measure for accurate risk assessment.
In [71], classification models distinguishing fallers from non-fallers in a cohort of RRMS patients were constructed using multiple LR with forward stepwise selection of variables based on Akaike Information Criterion (AIC). To enhance fall prediction accuracy, the study proposed a comprehensive approach integrating clinical measures and advanced imaging techniques, specifically corticospinal tract (CSI) FA and magnetization transfer ratio (MTR). In addition, MRI measures, clinical measures of walking assessment and vibration sensation were incorporated into the modeling. Inclusion of CST measures in the model raised the accuracy from 50 to 73.8%, where CST MTR was selected in the final model. The major drawback of this study was the retrospective prediction of falls, using data collected after the fall occurrence rather than baseline data prior to the event.
Notably, a significant gap exists in our ability to predict future motor progression through baseline MRI and clinical data. While cross-sectional studies and retrospective analyses contribute valuable insights into the current state of motor impairment, they fall short in providing robust models for forecasting the course of MS over time. The lack of predictive models based on longitudinal data hampers our capacity to identify individuals at risk for escalating motor deficits early on, hindering the timely implementation of targeted interventions. This underscores the urgent need for prospective, longitudinal studies that integrate comprehensive baseline assessments.
Disease activity
Considering the complexity of the MS disease course, evaluating disease activity requires a holistic approach incorporating various clinical, imaging and serum biomarkers [204]. Commonly used to assess disease activity is the No Evidence of Disease Activity (NEDA), termed NEDA-3 due to its combined assessment of 3 aspects of the disease. To be classified as NEDA-3, a patient must exhibit no evidence of relapses, MRI activity or EDSS progression [205, 206]. NEDA-4 has incorporated brain volume loss as an additional criterion [205]. Accurate prediction of future disease activity can aid in determining patients most likely to benefit from new therapeutics [207].
In [208], LV intensity was recognized as an important predictive feature of future disease activity as assessed by NEDA-3. A combination of clinical variables, MRI-derived features (T2w lesion count, WMV, GMV, CSF volume and total brain volume) and flattened, cropped FLAIR images centralized around the LV were utilized for modeling. MKL was employed for the prediction task, with each of the three feature subsets represented as a radial basis function kernel. The model combining all three kernels revealed an AUROC of 0.70 and balanced accuracy of 0.61, improving these metrics from 0.63 and 0.55 achieved by the model excluding LV-cropped images [208]. Also considering NEDA as an endpoint, Andorra et al. [209] utilized RF modeling integrating clinical, MRI, genetic and omics data for two-year prediction of disease activity. MRI measures included whole brain volume, GMV, WMV, T2LV and number of T1w-Gd lesions. The dataset was high dimensional, including 100 features, requiring dimensionality reduction techniques including correlation analysis, PCA and selection using feature importance. A balanced accuracy and AUROC of 0.6 and 0.79 were obtained. However, MRI features were not ranked among the top 10 predictors [209].
In a small study conducted by Du et al. [210], multimodal MRI and PET radiomics features were extracted from WM lesions and employed in subsequent ML modeling. Various combinations of ML models and feature elimination methods were tested, with the best-performing combination being SVM with RFE with an AUROC of 0.96. The individual best-performing modality was T2w, outperforming both PET and T1w radiomics features. However, the combination of all modalities provided the best performance by a wide margin.
Similarly, Basu et al. [211] aimed to predict disease activity using XGBoost modeling in a cohort of patients enrolled in cladribine clinical trials. The MRI features involved total number and volume of T1w, T2w and T1w-Gd lesions, in addition to combined unique lesion count of the various modalities and new T1w lesions. Disease activity was also defined by a composite score involving EDSS, relapses and MRI activity. By combining the MRI-derived features with various neurological, hematological, and biochemical testing results, the XGBoost model achieved an AUROC of 0.8 and balanced accuracies of 0.75–0.8. Using SHAP, the most influential MRI predictors were the multimodal unique lesion count and new T1w lesions.
Two studies defined disease activity by combining EDSS with cognitive outcome [212]. Storelli et al. [213] developed a DL algorithm utilizing baseline T2- and T1-weighted MRI scans to predict disease worsening in MS patients over 2 years. The algorithm was based on a CNN designed to predict clinical worsening, cognitive deterioration, or both, using EDSS and SDMT as outcome measures. The model demonstrated high predictive accuracy, reaching up to 85.7%, and outperformed two expert physicians (70%). In Nateboom et al. [212], motor disability progression was also evaluated through the 9HPT and Timed 25-Foot Walk Test (T25FW), in addition to EDSS and SDMT. This study was conducted on two separate cohorts, as described in Table 5, utilizing combinations of clinical and MRI-derived global and regional volumes. However, evaluation on the test set yielded poor results for both cohorts, most likely due to the large class imbalance present in the datasets.
Monitoring disease activity through lesion activity on MRI is particularly advantageous, as it can allow for earlier determination of treatment efficacy [214]. The relationship between individual MS lesion patterns and clinical disability is a research topic of rising interest, though routine clinical MRI modalities have limitations in the inflammatory characterization of lesions. T2w images are sufficiently sensitive for the detection of WM lesions, however, more advanced modalities are required for the differentiation of lesion status, such as T1w-Gd imaging and quantitative T1 relaxation time mapping (T1-RT) [215,216,217,218,219].
The evolution of WM and GM lesions in MS entails cycles of inflammation and demyelination, remyelination, and axonal loss, progressing through four main stages [220]. Early lesions are characterized by an active, inflammatory demyelination with infiltration of macrophages, lymphocytes and microglia surrounding a central vein [221] [222]. Once the inflammatory cells retreat to the border of the lesion, rendering the lesion center demyelinated and hypocellular, the lesion is considered chronic active. This is followed by a period of inactivity, and finally remyelination or repair of the lesion, known as lesion resolution [220].
Interest in chronic active lesions has gained momentum due to their high prevalence [220], association with clinical disability and specificity to MS in comparison to other neurological conditions [223]. These lesions are also known as smoldering lesions or SELs, due to the active demyelination occurring at the lesion borders [216, 220], which can be detected by the presence of a paramagnetic rim, primarily characterized on 7 T MRI [224]. SELs are of particular interest due to their association with smoldering disease activity, which is theorized to be the true pathological basis of MS [15].
Longitudinal analysis of serial MRIs in MS poses a significant challenge due to the diversity introduced by varying MRI machines and acquisition protocols, in addition to the nuances of registration in the case of pathological brains [225]. Various methods have been developed to overcome this challenge, which substantially improve downstream prediction tasks reliant on accurate segmentation and detection of new and enlarging lesions (NEL). However, advances in segmentation architectures are not discussed in the current work, and interested readers are encouraged to refer to comprehensive review papers for an in-depth discussion of this particular field of research [225,226,227].
Numerous studies have investigated ML for the prediction of MS lesion emergence and expansion, with various textural and geometric descriptors of lesions leveraged as model features. In [228], GMMs were employed to categorize subtypes of lesion features from textural information, including RIFT, local binary pattern and intensity features. For each patient, a “bag of lesions” representation was constructed based on the lesion type probabilities obtained from the GMM, which was then fed into a RF model to predict whether the subject will develop new or enlarging T2 lesions within the next 24 months. In [229], however, the emphasis of the ML model was on the prediction of individual unenhanced lesion evolution in terms of expansion/shrinkage using FLAIR radiomics-based features. Given the large number of available attributes, various combinations of feature selection and ML algorithms were evaluated, demonstrating non-linear SVM with ReliefF as the best-performing model at an accuracy of 0.827.
Similar to [228], Sepahvand et al. [230] predicted MRI activity status within 2 years using baseline images (T1w, T2w, FLAIR), in addition to T2w lesions masks. The method relied on latent features extracted through a CNN for prediction, achieving a maximum accuracy, sensitivity and specificity of around 80%. Providing the segmented masks to the network proved crucial in these experiments, as utilizing only MRI modalities as input diminished accuracy to only 26.31%, primarily due to the reduced sensitivity for detection of active cases (8.45%) [230]. The authors later developed a follow-up model, named NE Subnet, for the segmentation of new and enlarging T2w lesions using a U-Net with subtraction imaging, included as an attention mechanism, which improved the detection of small-scale changes and differentiation from noise. Classification of future disease activity was then carried out on the basis of this model, achieving a higher sensitivity of around 0.977, with a trade-off in specificity (0.693) [231].
Alternatively, Acar et al. [232] predicted the emergence of new lesions utilizing 3D discrete wavelet transforms of whole FLAIR images, where images were treated as 2D signals. Specifically, the second-level LLL (low pass filtering) sub-bands were utilized as the feature vector for subsequent ML modeling. Out of the 10 different wavelet families analyzed, the best-performing features were extracted from the Symlet8 family when combined with a SVM model, achieving a measure of 0.95 in precision, recall and F1 score.
More recently, treatment-modified course of disease activity was predicted using a combination of baseline mixed MRI modalities and clinical data. The goal was to develop a clinical tool for evaluating the efficacy of various treatment options for MS, weighing the associated risk with the predicted benefit in terms of reduction in NEL. MRI-specific features were extracted using a ResNet encoder, concatenated with clinical features, and finally passed through a multihead MLP, with the various output heads representing the treatment-specific outcome in terms of future disease activity. Implementation of this architecture for regression provided superior results to classification, where regression predicted the difference in lesion count from baseline, while classification simply predicted future disease activity status based on the minimal evidence of disease activity on future T2 sequences (MEDA-T2), which applies a threshold of 3 NEL for recommendation of MEDA-T2 status. Based on this model, a clinical tool was implemented in which a patient’s predicted NEL according to treatment arm was employed for improved treatment recommendation [233].
In [234], the authors harnessed the power of GNNs to forecast inflammatory disease activity within a period of 1–2 years, defined by the emergence of new or enlarged inflammatory lesions on MRI within the specified timeframe. Self-supervised features were extracted from all segmented MRI lesions, and various GNNs were employed to generate a classification of inflammatory status, with individual lesions representing graph nodes, and spatial locations of lesions characterized through edge information. To enhance the interpretability of the model, a self-pruning module was introduced. This module played a pivotal role in identifying and highlighting the specific lesions that significantly contributed to the final prediction. Using this method, the best-performing GNN was the GCN, and outperformed other traditional ML and DL methods. The results from [228,229,230,231, 233, 234] are summarized in Table 6.
Predicting disease activity in MS is an extensive topic of interest, highlighted by the large number of studies discussed in this section. While NEDA and MRI activity are of growing interest as endpoints for clinical trials, EDSS remains the primary endpoint most commonly used for clinician assessment [235, 236]. Furthermore, NEDA has also been suggested for monitoring the effectiveness of DMTs to avoid the accumulation of irreversible disability [237]. However, the focus on the inflammatory aspect in this measure has raised doubts regarding its suitability in PPMS patients with limited MRI inflammatory activity. Finally, the definition of disease activity in terms of timeframes, and whether to include only new lesions or both new and enlarging lesions differs between studies, further complicating standardization and hindering comparisons between studies [236].
Conclusions
The integration of ML and MRI biomarkers for predictive tasks in MS progression presents a promising avenue for advancing clinical insights. Though the primary emphasis of this review centers around brain imaging, it is evident that relying on a singular measure is insufficient for obtaining predictions that are accurate enough to be deployed in clinical practice. The integration of multimodal data is likely the best solution for a personalized medicine approach in MS prognosis [238].
The integration of network neuroscience with ML may prove advantageous in overcoming the CRDP given the recognition of MS as a disconnection syndrome. This can be done by integrating both structural and functional connectivity-derived measures into ML models [29, 239]. Additionally, combining graph representations of neurological disorders such as MS with DL in graph neural network models leverages the advantages of spatially aware geometric deep learning [112]. Finally, a multiresolution, multimodal representation of brain connectivity could potentially improve prognostic accuracy [240].
Furthermore, the use of ML and DL comes with a number of limitations that need to be addressed. First, the quality of the data is paramount to the success of any AI model, requiring extensive datasets sufficiently representing the heterogeneity of population characteristics, disease course and scanning parameters [241]. Moreover, there remains a considerable gap, particularly for DL, between AI and medical applications due to a lack of consideration for explainability, which is a necessary ethical and legislative consideration for any clinical decision support system [119]. In addition, the lack of consensus regarding clinical endpoints for measuring disease progression in MS studies hinders direct comparison of model architectures and the predictive power of utilized features. Finally, only a few of the discussed models were externally validated. Prognostic models cannot be deployed in clinical practice before undergoing external validation on different datasets from the ones on which they were trained. Given that performance is usually inferior outside of the training population, this is essential to avoid adverse effects by making clinical decisions based on incorrect predictions [242].
Abbreviations
- 9-HPT:
-
Nine-hole peg test
- AI:
-
Artificial intelligence
- AUPRC:
-
Area under precision–recall curve
- AUROC:
-
Area under the receiver operating characteristic curve
- BPF:
-
Brain parenchymal fraction
- CI:
-
Cognitive impairment
- CIS:
-
Clinically isolated syndrome
- CNN:
-
Convolutional neural network
- CNS:
-
Central nervous system
- CLRDP:
-
Clinico-radiological paradox
- CSF:
-
Cerebrospinal fluid
- DL:
-
Deep learning
- DMT:
-
Disease-modifying therapies
- DT:
-
Decision trees
- DTI:
-
Diffusion tensor imaging
- DWI:
-
Diffusion weighted imaging
- EDSS:
-
Expanded disability status scale
- fMRI:
-
Functional MRI
- FA:
-
Fractional anisotropy
- FC:
-
Functional connectivity
- FLAIR:
-
Fluid-attenuated inversion recovery
- GM:
-
Gray matter
- GMM:
-
Gaussian mixture model
- GMV:
-
Gray matter volume
- GRU:
-
Gated recurrent unit
- KNN:
-
K-nearest neighbor
- LR:
-
Linear regression
- LoR:
-
Logistic regression
- LSTM:
-
Long short-term memory
- LV:
-
Lateral ventricles
- MAE:
-
Mean absolute error
- MD:
-
Mean diffusivity
- ML:
-
Machine learning
- MLP:
-
Multi-layer perceptron
- MRI:
-
Magnetic resonance imaging
- MS:
-
Multiple sclerosis
- MSWS:
-
Multiple sclerosis walking scale
- MT:
-
Magnetization transfer imaging
- MTR:
-
Magnetization transfer ratio
- PASAT:
-
Paced auditory serial addition test
- PCA:
-
Principal component analysis
- PDw:
-
Proton density weighted imaging
- PIRA:
-
Progression independent of relapse activity
- PPMS:
-
Primary progressive MS
- ResNet:
-
Residual networks
- RF:
-
Random forest
- RNN:
-
Recurrent neural network
- RRMS:
-
Relapse-remitting MS
- SDMT:
-
Symbol digit modalities test
- SHAP:
-
Shapley additive explanations
- sNfL:
-
Serum neurofilament light chain
- SPMS:
-
Secondary progressive MS
- SVM:
-
Support vector machine
- T1w:
-
T1-weighted imaging
- T2LV:
-
T2w lesion volume
- T2w:
-
T2-weighted imaging
- T25w:
-
Timed 25 walk
- UL:
-
Upper limb
- WM:
-
White matter
- XGBoost:
-
Extreme gradient boosting
References
Attfield KE, Jensen LT, Kaufmann M et al (2022) The immunology of multiple sclerosis. Nat Rev Immunol 22:734–750. https://doi.org/10.1038/s41577-022-00718-z
Hauser SL, Cree BAC (2020) Treatment of multiple sclerosis: a review. Am J Med 133:1380-1390.e2. https://doi.org/10.1016/j.amjmed.2020.05.049
Yang JH, Rempe T, Whitmire N et al (2022) Therapeutic advances in multiple sclerosis. Front Neurol. https://doi.org/10.3389/fneur.2022.824926
Dobson R, Giovannoni G (2019) Multiple sclerosis—a review. Eur J Neurol 26:27–40. https://doi.org/10.1111/ene.13819
Lassmann H (2018) Multiple sclerosis pathology. Cold Spring Harb Perspect Med 8 (3):a028936. https://doi.org/10.1101/cshperspect.a028936
Filippi M, Bar-Or A, Piehl F et al (2018) Multiple sclerosis. Nat Rev Dis Primers. https://doi.org/10.1038/s41572-018-0041-4
Kuhlmann T, Moccia M, Coetzee T et al (2023) Multiple sclerosis progression: time for a new mechanism-driven framework. Lancet Neurol 22:78–88. https://doi.org/10.1016/S1474-4422(22)00289-7
Lassmann H (2019) Pathogenic mechanisms associated with different clinical courses of multiple sclerosis. Front Immunol 22:78–88. https://doi.org/10.3389/fimmu.2018.03116
Brownlee WJ, Miller DH (2014) Clinically isolated syndromes and the relationship to multiple sclerosis. J Clin Neurosci 21:2065–2071. https://doi.org/10.1016/j.jocn.2014.02.026
Marrie RA, Allegretta M, Barcellos LF et al (2022) From the prodromal stage of multiple sclerosis to disease prevention. Nat Rev Neurol 18:559–572. https://doi.org/10.1038/s41582-022-00686-x
Hosseiny M, Newsome SD, Yousem DM (2020) Radiologically isolated syndrome: a review for neuroradiologists. Am J Neuroradiol 41:1542–1549. https://doi.org/10.3174/ajnr.A6649
De Stefano N, Giorgio A, Tintoré M et al (2018) Radiologically isolated syndrome or subclinical multiple sclerosis: MAGNIMS consensus recommendations. Mult Scler 24:214–221. https://doi.org/10.1177/1352458517717808
Benito-León J, del Pino AB, Aladro Y et al (2023) Abnormal functional connectivity in radiologically isolated syndrome: a resting-state fMRI study. Mult Scler J 29:1393–1405. https://doi.org/10.1177/13524585231195851
Thompson AJ, Banwell BL, Barkhof F et al (2018) Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 17:162–173. https://doi.org/10.1016/S1474-4422(17)30470-2
Giovannoni G, Popescu V, Wuerfel J et al (2022) Smouldering multiple sclerosis: the ‘real MS.’ Ther Adv Neurol Disord. https://doi.org/10.1177/17562864211066751
Lublin FD, Häring DA, Ganjgahi H et al (2022) How patients with multiple sclerosis acquire disability. Brain 145:3147–3161. https://doi.org/10.1093/brain/awac016
Tur C, Carbonell-Mirabent P, Cobo-Calvo Á et al (2023) Association of early progression independent of relapse activity with long-term disability after a first demyelinating event in multiple sclerosis. JAMA Neurol 80:151–160. https://doi.org/10.1001/jamaneurol.2022.4655
Gil-González I, Martín-Rodríguez A, Conrad R, Pérez-San-Gregorio MÁ (2020) Quality of life in adults with multiple sclerosis: a systematic review. BMJ Open. https://doi.org/10.1136/bmjopen-2020-041249
Fox RJ, Mehta R, Pham T et al (2022) Real-world disease-modifying therapy pathways from administrative claims data in patients with multiple sclerosis. BMC Neurol. https://doi.org/10.1186/s12883-022-02738-7
Liu Z, Liao Q, Wen H, Zhang Y (2021) Disease modifying therapies in relapsing-remitting multiple sclerosis: a systematic review and network meta-analysis. Autoimmun Rev 20:102826. https://doi.org/10.1016/j.autrev.2021.102826
Langer-Gould AM, Smith JB, Gonzales EG et al (2023) Multiple sclerosis, disease-modifying therapies, and infections. Neurol Neuroimmunol Neuroinflamm. https://doi.org/10.1212/NXI.0000000000200164
Zingaropoli MA, Pasculli P, Iannetta M et al (2022) Infectious risk in multiple sclerosis patients treated with disease-modifying therapies: a three-year observational cohort study. Mult Scler J Exp Transl Clin. https://doi.org/10.1177/20552173211065731
Fuh-Ngwa V, Zhou Y, Charlesworth JC et al (2021) Developing a clinical-environmental-genotypic prognostic index for relapsing-onset multiple sclerosis and clinically isolated syndrome. Brain Commun. https://doi.org/10.1093/braincomms/fcab288
Johnson KB, Wei WQ, Weeraratne D et al (2021) Precision medicine, AI, and the future of personalized health care. Clin Transl Sci 14:86–93. https://doi.org/10.1111/cts.12884
Naji Y, Mahdaoui M, Klevor R, Kissani N (2023) Artificial intelligence and multiple sclerosis: up-to-date review. Cureus 14: 86–93. https://doi.org/10.7759/cureus.45412
Cortese R, Collorone S, Ciccarelli O, Toosy AT (2019) Advances in brain imaging in multiple sclerosis. Ther Adv Neurol Disord 12:1756286419859722. https://doi.org/10.1177/1756286419859722
Hemond CC, Bakshi R (2018) Magnetic resonance imaging in multiple sclerosis. Cold Spring Harb Perspect Med 8:a028969. https://doi.org/10.1101/cshperspect.a028969
Shoeibi A, Khodatars M, Jafari M et al (2021) Applications of deep learning techniques for automated multiple sclerosis detection using magnetic resonance imaging: a review. Comput Biol Med 136:104697. https://doi.org/10.1016/j.compbiomed.2021.104697
Lapucci C, Schiavi S, Signori A et al (2022) The role of disconnection in explaining disability in multiple sclerosis. Eur Radiol Exp 6: 23. https://doi.org/10.1186/s41747-022-00277-x
Biggio M, Caligiore D, D’Antoni F et al (2022) Machine learning for exploring neurophysiological functionality in multiple sclerosis based on trigeminal and hand blink reflexes. Sci Rep 12: 21078. https://doi.org/10.1038/s41598-022-24720-6
Alharthi HM, Almurdi MM (2023) Association between cognitive impairment and motor dysfunction among patients with multiple sclerosis: a cross-sectional study. Eur J Med Res 28: 110. https://doi.org/10.1186/s40001-023-01079-6
Meyer-Moock S, Feng YS, Maeurer M et al (2014) Systematic literature review and validity evaluation of the Expanded Disability Status Scale (EDSS) and the Multiple Sclerosis Functional Composite (MSFC) in patients with multiple sclerosis. BMC Neurol 14:1–10. https://doi.org/10.1186/1471-2377-14-58
Cohen M, Bresch S, Thommel Rocchi O et al (2021) Should we still only rely on EDSS to evaluate disability in multiple sclerosis patients? A study of inter and intra rater reliability. Mult Scler Relat Disord 54: 103144. https://doi.org/10.1016/j.msard.2021.103144
Barzegar M, Najdaghi S, Afshari-Safavi A et al (2021) Early predictors of conversion to secondary progressive multiple sclerosis. Mult Scler Relat Disord 54: 103115. https://doi.org/10.1016/j.msard.2021.103115
Moazami F, Lefevre-Utile A, Papaloukas C, Soumelis V (2021) Machine learning approaches in study of multiple sclerosis disease through magnetic resonance images. Front Immunol. https://doi.org/10.3389/fimmu.2021.700582
Aslam N, Khan IU, Bashamakh A et al (2022) Multiple sclerosis diagnosis using machine learning and deep learning: challenges and opportunities. Sensors 22: 7856. https://doi.org/10.3390/s22207856
Seccia R, Romano S, Salvetti M et al (2021) Machine learning use for prognostic purposes in multiple sclerosis. Life 11:1–18. https://doi.org/10.3390/life11020122
Macaron G, Ontaneda D (2019) Diagnosis and management of progressive multiple sclerosis. Biomedicines 7: 56. https://doi.org/10.3390/biomedicines7030056
Filippi M, Agosta F (2010) Imaging biomarkers in multiple sclerosis. J Magn Reson Imaging 31:770–788. https://doi.org/10.1002/jmri.22102
Truyen L, Van Waesberghe JHTM, Van Walderveen MAA et al (1996) Accumulation of hypointense lesions (“black holes”) on T, spin-echo MRI correlates with disease progression in multiple sclerosis. Neurology 47: 1469–1476. https://doi.org/10.1212/WNL.47.6.1469
Sahraian MA, Radue EW, Haller S, Kappos L (2010) Black holes in multiple sclerosis: Definition, evolution, and clinical correlations. Acta Neurol Scand 122:1–8. https://doi.org/10.1111/j.1600-0404.2009.01221.x
Kostic D, Dincic E, Jovanovski A et al (2023) Evolution of acute “black hole” lesions in patients with relapsing–remitting multiple sclerosis. Acta Neurol Belg 123:831–838. https://doi.org/10.1007/s13760-022-01938-9
Kolb H, Al-Louzi O, Beck ES et al (2022) From pathology to MRI and back: clinically relevant biomarkers of multiple sclerosis lesions. Neuroimage Clin 36:103194. https://doi.org/10.1016/j.nicl.2022.103194
Tiu VE, Enache I, Panea CA et al (2022) Predictive MRI biomarkers in MS—a critical review. Medicina (Kaunas) 58: 377. https://doi.org/10.3390/medicina58030377
Barkhof F (2002) The clinico-radiological paradox in multiple sclerosis revisited. Curr Opin Neurol 15:239–245
Hackmack K, Weygandt M, Wuerfel J et al (2012) Can we overcome the “clinico-radiological paradox” in multiple sclerosis? J Neurol 259:2151–2160. https://doi.org/10.1007/s00415-012-6475-9
Chard D, Trip SA (2017) Resolving the clinico-radiological paradox in multiple sclerosis. F1000Research. https://doi.org/10.12688/f1000research.11932.1
Mollison D, Sellar R, Bastin M et al (2017) The clinico-radiological paradox of cognitive function and MRI burden of white matter lesions in people with multiple sclerosis: a systematic review and meta-analysis. PLoS One 15:e0177727. https://doi.org/10.1371/journal.pone.01777
Kwong KCNK, Mollison D, Meijboom R et al (2021) The prevalence of paramagnetic rim lesions in multiple sclerosis: a systematic review and meta-analysis. PLoS One 16(9):e0256845. https://doi.org/10.1371/journal.pone.0256845
Marcille M, Hurtado Rúa S, Tyshkov C et al (2022) Disease correlates of rim lesions on quantitative susceptibility mapping in multiple sclerosis. Sci Rep 12:4411. https://doi.org/10.1038/s41598-022-08477-6
Castellaro M, Tamanti A, Pisani AI et al (2020) The use of the central vein sign in the diagnosis of multiple sclerosis: a systematic review and meta-analysis. Diagnostics 10:1025. https://doi.org/10.3390/diagnostics10121025
Sati P, Oh J, Todd Constable R et al (2016) The central vein sign and its clinical evaluation for the diagnosis of multiple sclerosis: a consensus statement from the North American Imaging in Multiple Sclerosis Cooperative. Nat Rev Neurol 12:714–722. https://doi.org/10.1038/nrneurol.2016.166
Maggi P, Fartaria MJ, Jorge J et al (2020) CVSnet: a machine learning approach for automated central vein sign assessment in multiple sclerosis. NMR Biomed 33:e4283. https://doi.org/10.1002/nbm.4283
Oh J, Suthiphosuwan S, Sati P et al (2021) Cognitive impairment, the central vein sign, and paramagnetic rim lesions in RIS. Mult Scler J 27:2199–2208. https://doi.org/10.1177/13524585211002097
Klaver R, De Vries HE, Schenk GJ, Geurts JJG (2013) Grey matter damage in multiple sclerosis: A pathology perspective. Prion 7:66–75. https://doi.org/10.4161/pri.23499
Haider L, Simeonidou C, Steinberger G et al (2014) Multiple sclerosis deep grey matter: the relation between demyelination, neurodegeneration, inflammation and iron. J Neurol Neurosurg Psychiatry 85:1386–1395. https://doi.org/10.1136/jnnp-2014-307712
Rothstein TL (2020) Gray matter matters: a longitudinal magnetic resonance voxel-based morphometry study of primary progressive multiple sclerosis. Front Neurol 11: 581537. https://doi.org/10.3389/fneur.2020.581537
Calabrese M, Favaretto A, Martini V, Gallo P (2013) Grey matter lesions in MS from histology to clinical implications. Prion. 7:20–27. https://doi.org/10.4161/pri.22580
Tóth E, Szabó N, Csete G et al (2017) Gray matter atrophy is primarily related to demyelination of lesions in multiple sclerosis: a diffusion tensor imaging MRI study. Front Neuroanat. https://doi.org/10.3389/FNANA.2017.00023
Andica C, Hagiwara A, Kamagata K et al (2019) Gray matter alterations in early and late relapsing-remitting multiple sclerosis evaluated with synthetic quantitative magnetic resonance imaging. Sci Rep 9:8147. https://doi.org/10.1038/s41598-019-44615-3
Honce JM (2013) Gray matter pathology in MS: neuroimaging and clinical correlations. Mult Scler Int 2013:1–16. https://doi.org/10.1155/2013/627870
Higazi MM, El GHSA, Fathy AW et al (2022) Diagnostic accuracy of double inversion recovery (DIR) in detection of cortical gray matter lesions in patients with MS. Egyptian J Radiol Nucl Med 53:13. https://doi.org/10.1186/s43055-021-00668-x
Bouman PM, Steenwijk MD, Pouwels PJW et al (2020) Histopathology-validated recommendations for cortical lesion imaging in multiple sclerosis. Brain 143:2988–2997. https://doi.org/10.1093/brain/awaa233
Bussas M, El Husseini M, Harabacz L et al (2022) Multiple sclerosis lesions and atrophy in the spinal cord: distribution across vertebral levels and correlation with disability. Neuroimage Clin 34:103006. https://doi.org/10.1016/j.nicl.2022.103006
Ontaneda D, Sakaie K, Lin J et al (2017) Measuring brain tissue integrity during 4 years using diffusion tensor imaging. Am J Neuroradiol 38:31–38. https://doi.org/10.3174/ajnr.A4946
Cairns J, Vavasour IM, Traboulsee A et al (2022) Diffusely abnormal white matter in multiple sclerosis. J Neuroimaging 32:5–16
Seewann A, Vrenken H, Van Der Valk P et al (2009) Diffusely abnormal white matter in chronic multiple sclerosis imaging and histopathologic analysis. Arch Neurol 66:601–609
West J, Aalto A, Tisell A et al (2014) Normal appearing and diffusely abnormal white matter in patients with multiple sclerosis assessed with quantitative MR. PLoS One 9:e95161. https://doi.org/10.1371/journal.pone.0095161
Vrenken H, Seewann A, Knol DL et al (2010) Diffusely abnormal white matter in progressive multiple sclerosis: in vivo quantitative MR imaging characterization and comparison between disease types. Am J Neuroradiol 31:541–548. https://doi.org/10.3174/ajnr.A1839
Pareto D, Garcia-Vidal A, Alberich M et al (2020) Ratio of T1-weighted to T2-weighted signal intensity as a measure of tissue integrity: comparison with magnetization transfer ratio in patients with multiple sclerosis. Am J Neuroradiol 41:461–463. https://doi.org/10.3174/AJNR.A6481
Fritz NE, Edwards EM, Keller J et al (2020) Combining magnetization transfer ratio mri and quantitative measures of walking improves the identification of fallers in MS. Brain Sci 10:1–12. https://doi.org/10.3390/brainsci10110822
Tozlu C, Olafson E, Jamison KW et al (2023) The sequence of regional structural disconnectivity due to multiple sclerosis lesions. Brain Commun. https://doi.org/10.1093/braincomms/fcad332
Pagani E, Rocca MA, De Meo E et al (2020) Structural connectivity in multiple sclerosis and modeling of disconnection. Mult Scler J 26:220–232. https://doi.org/10.1177/1352458518820759
Rocca MA, Filippi M (2007) Functional MRI in multiple sclerosis. J Neuroimaging. https://doi.org/10.1111/j.1552-6569.2007.00135.x
Tahedl M, Levine SM, Greenlee MW et al (2018) Functional connectivity in multiple sclerosis: recent findings and future directions. Front Neurol 9:828. https://doi.org/10.3389/fneur.2018.00828
Segato A, Marzullo A, Calimeri F, De Momi E (2020) Artificial intelligence for brain diseases: a systematic review. APL Bioeng 4:041503. https://doi.org/10.1063/5.0011697
Lima AA, Mridha MF, Das SC et al (2022) A comprehensive survey on the detection, classification, and challenges of neurological disorders. Biology (Basel) 11:469. https://doi.org/10.3390/biology11030469
Al-Amyn A, Daniel V, Eric R, Oermann K (2019) Deep learning and neurology: a systematic review. Neurol Ther 8:351–365. https://doi.org/10.6084/m9.figshare.9272951
Habehh H, Gohel S (2021) Machine learning in healthcare. Curr Genomics 22:291–300. https://doi.org/10.2174/1389202922666210705124359
Alanazi A (2022) Using machine learning for healthcare challenges and opportunities. Inform Med Unlocked 30:100924. https://doi.org/10.1016/j.imu.2022.100924
Javaid M, Haleem A, Pratap Singh R et al (2022) Significance of machine learning in healthcare: features, pillars and applications. Int J Intell Networks 3:58–73. https://doi.org/10.1016/j.ijin.2022.05.002
Sarker IH (2021) Machine learning: algorithms, real-world applications and research directions. SN Comput Sci 2:160. https://doi.org/10.1007/s42979-021-00592-x
Alnuaimi AFAH, Albaldawi THK (2024) An overview of machine learning classification techniques. BIO Web Conf EDP Sci 97:00133. https://doi.org/10.1051/bioconf/20249700133
Ying X (2019) An overview of overfitting and its solutions. J Phys Conf Series 1168:022022. https://doi.org/10.1088/1742-6596/1168/2/022022
Yousef LA, Yousef H, Rocha-Meneses L (2023) Artificial intelligence for management of variable renewable energy systems: a review of current status and future directions. Energies (Basel) 16:8057. https://doi.org/10.3390/en16248057
Tufail S, Riggs H, Tariq M, Sarwat AI (2023) Advancements and challenges in machine learning: a comprehensive review of models, libraries, applications, and algorithms. Electronics (Switzerland) 12(8):1789
Boateng EY, Abaye DA (2019) A review of the logistic regression model with emphasis on medical research. J Data Anal Inf Process 07:190–207. https://doi.org/10.4236/jdaip.2019.74012
Cervantes J, Garcia-Lamont F, Rodríguez-Mazahua L, Lopez A (2020) A comprehensive survey on support vector machine classification: applications, challenges and trends. Neurocomputing 408:189–215. https://doi.org/10.1016/j.neucom.2019.10.118
Guido R, Ferrisi S, Lofaro D, Conforti D (2024) An overview on the advancements of support vector machine models in healthcare applications: a review. Information (Switzerland) 15:235. https://doi.org/10.3390/info15040235
Taunk K, De S, Verma S, Swetapadma A (2019) A Brief Review of Nearest Neighbor Algorithm for Learning and Classification. In: Proceedings of the International Conference on Intelligent Computing and Control Systems (ICICCS 2019). IEEE
Wickramasinghe I, Kalutarage H (2021) Naive Bayes: applications, variations and vulnerabilities: a review of literature with code snippets for implementation. Soft comput 25:2277–2293. https://doi.org/10.1007/s00500-020-05297-6
Kingsford C, Salzberg SL (2008) What are decision trees? Nat Biotechnol 26:1011–1012. https://doi.org/10.1038/nbt0908-1011
Kotsiantis SB (2013) Decision trees: a recent overview. Artif Intell Rev 39:261–283. https://doi.org/10.1007/s10462-011-9272-4
Mienye ID, Jere N (2024) A survey of decision trees: concepts, algorithms, and applications. IEEE Access 12:86716–86727. https://doi.org/10.1109/ACCESS.2024.3416838
Mayr A, Binder H, Gefeller O, Schmid M (2014) The evolution of boosting algorithms: from machine learning to statistical modelling. Methods Inf Med 53:419–427. https://doi.org/10.3414/ME13-01-0122
Chen T, Guestrin C (2016) XGBoost: A Scalable Tree Boosting System. In: KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. pp 785–794
Mohammed A, Kora R (2023) A comprehensive review on ensemble deep learning: opportunities and challenges. J King Saud Univ Comput Inf Sci 35:757–774. https://doi.org/10.1016/j.jksuci.2023.01.014
An Q, Rahman S, Zhou J, Kang JJ (2023) A comprehensive review on machine learning in healthcare industry: classification, restrictions opportunities and challenges. Sensors 23:4178. https://doi.org/10.3390/s23094178
Eckhardt CM, Madjarova SJ, Williams RJ et al (2023) Unsupervised machine learning methods and emerging applications in healthcare. Knee Surg Sports Traumatol Arthrosc 31:376–381. https://doi.org/10.1007/s00167-022-07233-7
Dadu A, Satone VK, Kaur R et al (2023) Application of aligned-UMAP to longitudinal biomedical studies. Patterns 4:100741. https://doi.org/10.1016/j.patter.2023.100741
Aggarwal R, Sounderajah V, Martin G et al (2021) Diagnostic accuracy of deep learning in medical imaging: a systematic review and meta-analysis. NPJ Digit Med 4:65. https://doi.org/10.1038/s41746-021-00438-z
Hira ZM, Gillies DF (2015) A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinf 2015:198363. https://doi.org/10.1155/2015/198363
Pudjihartono N, Fadason T, Kempa-Liehr AW, O’Sullivan JM (2022) A review of feature selection methods for machine learning-based disease risk prediction. Frontiers in Bioinf 2:927312. https://doi.org/10.3389/fbinf.2022.927312
Venkatesh B, Anuradha J (2019) A review of feature selection and its methods. Cybern Inf Technol 19:3–26. https://doi.org/10.2478/CAIT-2019-0001
Jović A, Brkić K, Bogunović N (2015) A review of feature selection methods with applications. In: 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics, MIPRO 2015—Proceedings. Institute of Electrical and Electronics Engineers Inc. pp 1200–1205
Suganyadevi S, Seethalakshmi V, Balasamy K (2022) A review on deep learning in medical image analysis. Int J Multimed Inf Retr 11:19–38. https://doi.org/10.1007/s13735-021-00218-1
Chai J, Zeng H, Li A, Ngai EWT (2021) Deep learning in computer vision: a critical review of emerging techniques and application scenarios. Mach Learn Appl. 6:100134. https://doi.org/10.24433/CO.0411648.v1
Sistaninejhad B, Rasi H, Nayeri P (2023) A Review paper about deep learning for medical image analysis. Comput Math Methods Med 2023:7091301. https://doi.org/10.1155/2023/7091301
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) pp 770–778.
Simonyan K, Zisserman A (2014) Very Deep convolutional networks for large-scale image recognition. CoRR. https://api.semanticscholar.org/CorpusID:14124313
Rayed ME, Islam SMS, Niha SI et al (2024) Deep learning for medical image segmentation: State-of-the-art advancements and challenges. Inform Med Unlocked 47:101504. https://doi.org/10.1016/j.imu.2024.101504
Zhang S, Yang J, Zhang Y et al (2023) The combination of a graph neural network technique and brain imaging to diagnose neurological disorders: a review and outlook. Brain Sci 13:1462. https://doi.org/10.3390/brainsci13101462
Meng X, Zou T (2023) Clinical applications of graph neural networks in computational histopathology: a review. Comput Biol Med 164:107201. https://doi.org/10.1016/j.compbiomed.2023.107201
Zhang L, Zhao Y, Che T et al (2023) Graph neural networks for image-guided disease diagnosis: a review. iRADIOLOGY. 1:151–166. https://doi.org/10.1002/ird3.20
Wang J (2023) Review of graph neural networks for medical image. EAI Endorsed Trans e-Learn. https://doi.org/10.4108/eetel.4358
Araf I, Idri A, Chairi I (2024) Cost-sensitive learning for imbalanced medical data: a review. Artif Intell Rev. https://doi.org/10.1007/s10462-023-10652-8
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Band S, S, Yarahmadi A, Hsu CC, et al (2023) Application of explainable artificial intelligence in medical health: A systematic review of interpretability methods. Inform Med Unlocked 40:101286. https://doi.org/10.1016/j.imu.2023.101286
Amann J, Blasimme A, Vayena E et al (2020) Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Med Inform Decis Mak 20:310. https://doi.org/10.1186/s12911-020-01332-6
Zhou SK, Greenspan H, Davatzikos C et al (2020) A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc IEEE 109:820–838. https://doi.org/10.1109/JPROC.2021.3054390
Ponce de Leon-Sanchez ER, Dominguez-Ramirez OA, Herrera-Navarro AM et al (2023) A deep learning approach for predicting multiple sclerosis. Micromachines (Basel) 14:749. https://doi.org/10.3390/mi14040749
Hou Y, Jia Y, Hou J (2018) Natural course of clinically isolated syndrome: a longitudinal analysis using a markov model. Sci Rep 8:1–7. https://doi.org/10.1038/s41598-018-29206-y
Efendi H (2015) Clinically isolated syndromes: clinical characteristics, differential diagnosis, and management. Noropsikiyatri Arsivi 52:S1–S11. https://doi.org/10.5152/npa.2015.12608
Schwenkenbecher P, Wurster U, Konen FF et al (2019) Impact of the McDonald criteria 2017 on early diagnosis of relapsing-remitting multiple sclerosis. Front Neurol 10:188. https://doi.org/10.3389/fneur.2019.00188
Filippi M, Amato MP, Centonze D et al (2022) Early use of high-efficacy disease-modifying therapies makes the difference in people with multiple sclerosis: an expert opinion. J Neurol 269:5382–5394. https://doi.org/10.1007/s00415-022-11193-w
Vollmer BL, Wolf AB, Sillau S et al (2022) Evolution of disease modifying therapy benefits and risks: an argument for de-escalation as a treatment paradigm for patients with multiple sclerosis. Front Neurol 12:799138. https://doi.org/10.3389/fneur.2021.799138
Winkelmann A, Loebermann M, Reisinger EC et al (2016) Disease-modifying therapies and infectious risks in multiple sclerosis. Nat Rev Neurol 12:217–233. https://doi.org/10.1038/nrneurol.2016.21
Fischer S, Proschmann U, Akgün K, Ziemssen T (2021) Lymphocyte counts and multiple sclerosis therapeutics: between mechanisms of action and treatment-limiting side effects. Cells 10:3177. https://doi.org/10.3390/cells10113177
Zhang H, Alberts E, Pongratz V et al (2019) Predicting conversion from clinically isolated syndrome to multiple sclerosis–an imaging-based machine learning approach. Neuroimage Clin 21:101593. https://doi.org/10.1016/j.nicl.2018.11.003
Wottschel V, Alexander DC, Kwok PP et al (2015) Predicting outcome in clinically isolated syndrome using machine learning. Neuroimage Clin 7:281–287. https://doi.org/10.1016/j.nicl.2014.11.021
Bendfeldt K, Taschler B, Gaetano L et al (2019) MRI-based prediction of conversion from clinically isolated syndrome to clinically definite multiple sclerosis using SVM and lesion geometry. Brain Imaging Behav 13:1361–1374. https://doi.org/10.1007/s11682-018-9942-9
Rasouli S, Dakkali MS, Azarbad R et al (2024) Predicting the conversion from clinically isolated syndrome to multiple sclerosis: an explainable machine learning approach. Mult Scler Relat Disord 86:105614. https://doi.org/10.1016/j.msard.2024.105614
Tayyab M, Metz LM, Li DKB et al (2023) Accounting for uncertainty in training data to improve machine learning performance in predicting new disease activity in early multiple sclerosis. Front Neurol 14:1165267. https://doi.org/10.3389/fneur.2023.1165267
Wottschel V, Chard DT, Enzinger C et al (2019) SVM recursive feature elimination analyses of structural brain MRI predicts near-term relapses in patients with clinically isolated syndromes suggestive of multiple sclerosis. Neuroimage Clin 24:102011. https://doi.org/10.1016/j.nicl.2019.102011
Rehan Afzal HM, Lechner-Scott J, Luo S, et al (2018) Automatic prediction of the conversion of clinically isolated syndrome to multiple sclerosis using deep learning. ACM International Conference Proceeding Series. pp 231–235. https://doi.org/10.1145/3301506.3301526
Yoo Y, Tang LYW, Li DKB et al (2019) Deep learning of brain lesion patterns and user-defined clinical and MRI features for predicting conversion to multiple sclerosis from clinically isolated syndrome. Comput Methods Biomech Biomed Eng Imaging Vis 7:250–259. https://doi.org/10.1080/21681163.2017.1356750
Yoo Y, Tang LYW, Brosch T, et al (2016) Deep learning of brain lesion patterns for predicting future disease activity in patients with early symptoms of multiple sclerosis. In: Deep learning and data labeling for medical applications. pp 86–94. https://api.semanticscholar.org/CorpusID:34138081
Pareto D, Garcia-Vidal A, Groppa S et al (2022) Prognosis of a second clinical event from baseline MRI in patients with a CIS: a multicenter study using a machine learning approach. Neuroradiology 64:1383–1390. https://doi.org/10.1007/s00234-021-02885-7
Polman CH, Reingold SC, Edan G et al (2005) Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria.” Ann Neurol 58:840–846. https://doi.org/10.1002/ana.206703
Polman CH, Reingold SC, Banwell B et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302. https://doi.org/10.1002/ana.22366
Oreja-Guevara C, Blanco TA, Ruiz LB et al (2019) Cognitive dysfunctions and assessments in multiple sclerosis. Front Neurol 10:1–9. https://doi.org/10.3389/fneur.2019.00581
Portaccio E, Amato MP (2022) Cognitive impairment in multiple sclerosis: an update on assessment and management. NeuroSci 3:667–676. https://doi.org/10.3390/neurosci3040048
Elshebawy H, Fahmy EM, Elfayoumy NM et al (2021) Clinical predictors to cognitive impairment in multiple sclerosis patients. Egypt J Neurol Psychiatry Neurosurg 57:38. https://doi.org/10.1186/s41983-021-00292-6
Denissen S, Chén OY, De Mey J et al (2021) Towards multimodal machine learning prediction of individual cognitive evolution in multiple sclerosis. J Pers Med 11:1349. https://doi.org/10.3390/jpm11121349
Meca-Lallana V, Gascón-Giménez F, Ginestal-López RC et al (2021) Cognitive impairment in multiple sclerosis: diagnosis and monitoring. Neurol Sci 42:5183–5193. https://doi.org/10.1007/s10072-021-05165-7
Islas MÁM, Ciampi E (2019) Assessment and impact of cognitive impairment in multiple sclerosis: an overview. Biomedicines 7:1–19. https://doi.org/10.3390/biomedicines7010022
Benedict RHB, Amato MP, DeLuca J, Geurts JJG (2020) Cognitive impairment in multiple sclerosis: clinical management, MRI, and therapeutic avenues. Lancet Neurol 19:860–871. https://doi.org/10.1016/S1474-4422(20)30277-5
DeLuca J, Chiaravalloti ND, Sandroff BM (2020) Treatment and management of cognitive dysfunction in patients with multiple sclerosis. Nat Rev Neurol 16:319–332. https://doi.org/10.1038/s41582-020-0355-1
Chen MH, Goverover Y, Genova HM, DeLuca J (2020) Cognitive efficacy of pharmacologic treatments in multiple sclerosis: a systematic review. CNS Drugs 34:599–628. https://doi.org/10.1007/s40263-020-00734-4
Sumowski JF (2015) Cognitive reserve as a useful concept for early intervention research in multiple sclerosis. Front Neurol 6:176. https://doi.org/10.3389/fneur.2015.00176
Sumowski JF, Benedict R, Enzinger C et al (2018) Cognition in multiple sclerosis: state of the field and priorities for the future. Neurology 90:278–288. https://doi.org/10.1212/WNL.0000000000004977
Labiano-Fontcuberta A, Costa-Frossard L, Sainz de la Maza S et al (2023) Predictive models of multiple sclerosis-related cognitive performance using routine clinical practice predictors. Mult Scler Relat Disord 76:104849. https://doi.org/10.1016/j.msard.2023.104849
Nasios G, Bakirtzis C, Messinis L (2020) Cognitive impairment and brain reorganization in MS: underlying mechanisms and the role of neurorehabilitation. Front Neurol 11:1–8. https://doi.org/10.3389/fneur.2020.00147
Dong X, Xu G, Wang J et al (2022) Clinical and MRI predictors of cognitive decline in patients with relapsing-remitting multiple sclerosis: a 2-year longitudinal study. Mult Scler Relat Disord 65:103838. https://doi.org/10.1016/j.msard.2022.103838
Johnen A, Bürkner PC, Landmeyer NC et al (2019) Can we predict cognitive decline after initial diagnosis of multiple sclerosis? Results from the German National early MS cohort (KKNMS). J Neurol 266:386–397. https://doi.org/10.1007/s00415-018-9142-y
Eijlers AJC, Van Geest Q, Dekker I et al (2018) Predicting cognitive decline in multiple sclerosis: a 5-year follow-up study. Brain 141:2605–2618. https://doi.org/10.1093/brain/awy202
Marzi C, d’Ambrosio A, Diciotti S et al (2023) Prediction of the information processing speed performance in multiple sclerosis using a machine learning approach in a large multicenter magnetic resonance imaging data set. Hum Brain Mapp 44:186–202. https://doi.org/10.1002/hbm.26106
Buyukturkoglu K, Zeng D, Bharadwaj S et al (2021) Classifying multiple sclerosis patients on the basis of SDMT performance using machine learning. Mult Scler J 27:107–116. https://doi.org/10.1177/1352458520958362
Brummer T, Muthuraman M, Steffen F et al (2022) Improved prediction of early cognitive impairment in multiple sclerosis combining blood and imaging biomarkers. Brain Commun 4:1–11. https://doi.org/10.1093/braincomms/fcac153
Smith A (1982) Symbol digits modalities test. Western psychological services, Los Angeles. https://doi.org/10.3390/app112110335
Al-iedani O, Lea S, Alshehri A et al (2024) Multi-modal neuroimaging signatures predict cognitive decline in multiple sclerosis: a 5-year longitudinal study. Mult Scler Relat Disord 81:105379. https://doi.org/10.1016/j.msard.2023.105379
Lopez-Soley E, Martinez-Heras E, Andorra M et al (2021) Dynamics and predictors of cognitive impairment along the disease course in multiple sclerosis. J Pers Med 11:1107. https://doi.org/10.3390/jpm11111107
Schmitz-peiffer H, Aust E, Linse K et al (2022) Motor-independent cognitive testing in motor degenerative diseases. J Clin Med 11:814. https://doi.org/10.3390/jcm11030814
Kurtzke JF (1983) Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology. 33:1444–1452. https://doi.org/10.1212/WNL.33.11.1444
Zurawski J, Glanz BI, Chua A et al (2019) Time between expanded disability status scale (EDSS) scores. Mult Scler Relat Disord 30:98–103. https://doi.org/10.1016/j.msard.2019.02.007
Piri Çinar B, Güven Yorgun Y (2018) What we learned from the history of multiple sclerosis measurement: expanded disability status scale. Noropsikiyatri Arsivi. 55:S69–S75. https://doi.org/10.29399/NPA.23343
Demir S (2022) Expanded Disability Status Scale (EDSS) in multiple sclerosis. Cam Sakura Med J 2:82–89. https://doi.org/10.4274/csmedj.galenos.2022.2022-11-11
Dimitrov I, Georgiev R, Kaprelyan A et al (2015) Brain and lesion volumes correlate with edss in relapsing-remitting multiple sclerosis. J IMAB Ann Proc. 21:1015–1018. https://doi.org/10.5272/jimab.2015214.1015
Fisniku LK, Brex PA, Altmann DR et al (2008) Disability and T2 MRI lesions: A 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain 131:808–817. https://doi.org/10.1093/brain/awm329
Minneboo A, Jasperse B, Barkhof F et al (2008) Predicting short-term disability progression in early multiple sclerosis: added value of MRI parameters. J Neurol Neurosurg Psychiatry 79:917–923. https://doi.org/10.1136/jnnp.2007.124123
Gajofatto A, Calabrese M, Benedetti MD, Monaco S (2013) Clinical, MRI, and CSF markers of disability progression in multiple sclerosis. Dis Markers 35:687–699. https://doi.org/10.1155/2013/484959
Sedaghat S, Jang H, Athertya JS et al (2023) The signal intensity variation of multiple sclerosis (MS) lesions on magnetic resonance imaging (MRI) as a potential biomarker for patients’ disability: a feasibility study. Front Neurosci 17:1–8. https://doi.org/10.3389/fnins.2023.1145251
Dwyer MG, Bergsland N, Ramasamy DP et al (2018) Atrophied brain lesion volume: a new imaging biomarker in multiple sclerosis. J Neuroimaging 28:490–495. https://doi.org/10.1111/jon.12527
Tur C, Kanber B, Eshaghi A et al (2020) Clinical relevance of cortical network dynamics in early primary progressive MS. Mult Scler J 26:442–456. https://doi.org/10.1177/1352458519831400
Zhao Y, Healy BC, Rotstein D et al (2017) Exploration of machine learning techniques in predicting multiple sclerosis disease course. PLoS ONE 12:1–13. https://doi.org/10.1371/journal.pone.0174866
Law MTK, Traboulsee AL, Li DKB et al (2019) Machine learning in secondary progressive multiple sclerosis: an improved predictive model for short-term disability progression. Mult Scler J Exp Transl Clin 5:2055217319885983. https://doi.org/10.1177/2055217319885983
Zhao Y, Wang T, Bove R et al (2020) Ensemble learning predicts multiple sclerosis disease course in the SUMMIT study. NPJ Digit Med 3:1–8. https://doi.org/10.1038/s41746-020-00338-8
Roca P, Attye A, Colas L et al (2020) Artificial intelligence to predict clinical disability in patients with multiple sclerosis using FLAIR MRI. Diagn Interv Imaging 101:795–802. https://doi.org/10.1016/j.diii.2020.05.009
Tousignant A, Paul LemaˆıtreLemaˆıtre M, Doina Precup C, Arnold DL (2019) Prediction of disease progression in multiple sclerosis patients using deep learning analysis of MRI data. PMLR 102:483–492. https://api.semanticscholar.org/CorpusID:195785399
Taloni A, Farrelly FA, Pontillo G et al (2022) Evaluation of disability progression in multiple sclerosis via magnetic-resonance-based deep learning techniques. Int J Mol Sci 23:10651. https://doi.org/10.3390/ijms231810651
Pontillo G, Tommasin S, Cuocolo R et al (2021) A combined radiomics and machine learning approach to overcome the clinicoradiologic paradox in multiple sclerosis. Am J Neuroradiol 42:1927–1933. https://doi.org/10.3174/ajnr.A7274
Tommasin S, Cocozza S, Taloni A et al (2021) Machine learning classifier to identify clinical and radiological features relevant to disability progression in multiple sclerosis. J Neurol 268:4834–4845. https://doi.org/10.1007/s00415-021-10605-7
Campanioni S, Veiga C, Prieto-González JM et al (2024) Explainable machine learning on baseline MRI predicts multiple sclerosis trajectory descriptors. PLoS One 19:e0306999. https://doi.org/10.1371/journal.pone.0306999
Dular L, Brecl-Jakob G, Savšek L, et al (2023) Predicting future multiple sclerosis disease progression from MR scans. In: Proc. SPIE 12465, Medical Imaging 2023: Computer-Aided Diagnosis. SPIE-Intl Soc Optical Eng. https://doi.org/10.1117/12.2654416
Zhang K, Lincoln JA, Jiang X et al (2023) Predicting multiple sclerosis severity with multimodal deep neural networks. BMC Med Inform Decis Mak 23:255. https://doi.org/10.1186/s12911-023-02354-6
Ruggieri S, Prosperini L, Petracca M et al (2023) The added value of spinal cord lesions to disability accrual in multiple sclerosis. J Neurol 270:4995–5003. https://doi.org/10.1007/s00415-023-11829-5
Song X, Li D, Qiu Z et al (2020) Correlation between EDSS scores and cervical spinal cord atrophy at 3T MRI in multiple sclerosis: a systematic review and meta-analysis. Mult Scler Relat Disord 37:101426. https://doi.org/10.1016/j.msard.2019.101426
Celik NG, Karabulut AK, Fazliogullari Z et al (2023) Relationship between cervical spinal cord morphometry and clinical disability in patients with multiple sclerosis. Rev Assoc Med Bras 69:e20230949. https://doi.org/10.1590/1806-9282.20230949
Rajavenkatanarayanan A, Kanal V, Tsiakas K et al (2019) A survey of assistive technologies for assessment and rehabilitation of motor impairments in multiple sclerosis. Multimod Technol Interact 3:6. https://doi.org/10.3390/mti3010006
Edwards EM, Kegelmeyer DA, Kloos AD et al (2020) Backward walking and dual-task assessment improve identification of gait impairments and fall risk in individuals with MS. Mult Scler Int 2020:1–10. https://doi.org/10.1155/2020/6707414
Larocca NG (2011) Impact of walking impairment in multiple sclerosis perspectives of patients and care partners. Patient-Patient-Centered-Outcome-Re. 4:189–201. https://doi.org/10.2165/11591150-000000000-00000
Filli L, Sutter T, Easthope CS et al (2018) Profiling walking dysfunction in multiple sclerosis: characterisation, classification and progression over time. Sci Rep 8:4984. https://doi.org/10.1038/s41598-018-22676-0
Caselli S, Sabattini L, Cattaneo D et al (2023) When ‘good’ is not good enough: a retrospective Rasch analysis study of the Berg Balance Scale for persons with Multiple Sclerosis. Front Neurol 14:1171163. https://doi.org/10.3389/fneur.2023.1171163
Rehák Bučková B, Mareš J, Škoch A et al (2023) Multimodal-neuroimaging machine-learning analysis of motor disability in multiple sclerosis. Brain Imaging Behav 17:18–34. https://doi.org/10.1007/s11682-022-00737-3
Pellegrino L, Coscia M, Muller M et al (2018) Evaluating upper limb impairments in multiple sclerosis by exposure to different mechanical environments. Sci Rep 8:2110 https://doi.org/10.1038/s41598-018-20343-y
Ingram LA, Butler AA, Brodie MA et al (2022) Quantifying upper-limb motor impairment in people with multiple sclerosis: a physiological profiling approach. Ann Phys Rehabil Med 65:101625. https://doi.org/10.1016/j.rehab.2021.101625
Koch MW, Repovic P, Mostert J et al (2023) The nine hole peg test as an outcome measure in progressive MS trials. Mult Scler Relat Disord 69:104433. https://doi.org/10.1016/j.msard.2022.104433
Solaro C, Grange E, Di Giovanni R et al (2020) Nine hole peg test asymmetry in refining upper limb assessment in multiple sclerosis. Mult Scler Relat Disord 45:102422. https://doi.org/10.1016/j.msard.2020.102422
Cordani C, Preziosa P, Valsasina P et al (2022) MRI of transcallosal white matter helps to predict motor impairment in multiple sclerosis. Radiology 302:639–649. https://doi.org/10.1148/radiol.2021210922
Mostafa S, Song IHJ, Metwally AA, et al (2021) Predicting upper limb disability progression in primary progressive multiple sclerosis using machine learning and statistical methods. In: Proceedings—2021 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2021. Institute of Electrical and Electronics Engineers Inc. pp 1453–1457. https://doi.ieeecomputersociety.org/10.1109/BIBM52615.2021.9669665
Braune S, Bluemich S, Bruns C et al (2023) The natural history of primary progressive multiple sclerosis: insights from the German NeuroTransData registry. BMC Neurol 23:258. https://doi.org/10.1186/s12883-023-03273-9
VanNostrand M, Sogoloff B, Giroux C et al (2022) Predicting falls in adults with multiple sclerosis using patient-reported measures: are perceptions of dual-tasking missing? Mult Scler Relat Disord 68:104115. https://doi.org/10.1016/j.msard.2022.104115
Coote S, Comber L, Quinn G et al (2020) Falls in people with multiple sclerosis. Int J MS Care 22:247–255. https://doi.org/10.7224/1537-2073.2020-014
Masanneck L, Rolfes L, Regner-Nelke L et al (2022) Detecting ongoing disease activity in mildly affected multiple sclerosis patients under first-line therapies. Mult Scler Relat Disord 63:103927. https://doi.org/10.1016/j.msard.2022.103927
Hegen H, Bsteh G, Berger T (2018) ‘No evidence of disease activity’—is it an appropriate surrogate in multiple sclerosis? Eur J Neurol 25:1107–e101. https://doi.org/10.1111/ene.13669
Rotstein DL, Healy BC, Malik MT et al (2015) Evaluation of no evidence of disease activity in a 7-year longitudinal multiple sclerosis cohort. JAMA Neurol 72:152–158. https://doi.org/10.1001/jamaneurol.2014.3537
Doyle A, Elliott C, Karimaghaloo Z, et al (2018) Lesion detection, segmentation and prediction in multiple sclerosis clinical trials. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer Verlag, New York. pp 15–28
Chien C, Seiler M, Eitel F et al (2022) Prediction of high and low disease activity in early MS patients using multiple kernel learning identifies importance of lateral ventricle intensity. Mult Scler J Exp Transl Clin 8:20552173221109770. https://doi.org/10.1177/20552173221109770
Andorra M, Freire A, Zubizarreta I et al (2023) Predicting disease severity in multiple sclerosis using multimodal data and machine learning. J Neurol 271:1133–1149. https://doi.org/10.1007/s00415-023-12132-z
Du S, Yuan C, Zhou Q et al (2023) Deep learning-based PET/MR radiomics for the classification of annualized relapse rate in multiple sclerosis. Mult Scler Relat Disord 75:104750. https://doi.org/10.1016/j.msard.2023.104750
Basu S, Munafo A, Ben-Amor A-F et al (2022) Predicting disease activity in patients with multiple sclerosis: an explainable machine-learning approach in the Mavenclad trials. CPT Pharmacometrics Syst Pharmacol 11:843–853. https://doi.org/10.1002/psp4.12796
Noteboom S, Seiler M, Chien C et al (2024) Evaluation of machine learning-based classification of clinical impairment and prediction of clinical worsening in multiple sclerosis. J Neurol 271:5577–5589. https://doi.org/10.1007/s00415-024-12507-w
Storelli L, Azzimonti M, Gueye M et al (2022) A deep learning approach to predicting disease progression in multiple sclerosis using magnetic resonance imaging. Adv Artif Intell Mach Learn Res. 57:423–432. https://doi.org/10.1097/RLI.0000000000000854
Oship D, Jakimovski D, Bergsland N et al (2022) Assessment of T2 lesion-based disease activity volume outcomes in predicting disease progression in multiple sclerosis over 10 years. Mult Scler Relat Disord 67:104187. https://doi.org/10.1016/j.msard.2022.104187
Jonkman LE, Soriano AL, Amor S et al (2015) Can MS lesion stages be distinguished with MRI? A postmortem MRI and histopathology study. J Neurol 262:1074–1080. https://doi.org/10.1007/s00415-015-7689-4
Absinta M, Sati P, Masuzzo F et al (2019) Association of chronic active multiple sclerosis lesions with disability in vivo. JAMA Neurol 76:1474–1483. https://doi.org/10.1001/jamaneurol.2019.2399
Luchetti S, Fransen NL, van Eden CG et al (2018) Progressive multiple sclerosis patients show substantial lesion activity that correlates with clinical disease severity and sex: a retrospective autopsy cohort analysis. Acta Neuropathol 135:511–528. https://doi.org/10.1007/s00401-018-1818-y
Saade C, Bou-Fakhredin R, Yousem DM et al (2018) Gadolinium and multiple sclerosis: Vessels, barriers of the brain, and glymphatics. Am J Neuroradiol 39:2168–2176. https://doi.org/10.3174/ajnr.A5773
Martín-Aguilar L, Presas-Rodriguez S et al (2022) Gadolinium-enhanced brain lesions in multiple sclerosis relapse. Neurologia. 37:557–563. https://doi.org/10.1016/j.nrl.2021.10.005
Calvi A, Haider L, Prados F et al (2022) In vivo imaging of chronic active lesions in multiple sclerosis. Mult Scler J 28:683–690. https://doi.org/10.1177/1352458520958589
Pukoli D, Vécsei L (2023) Smouldering lesion in MS: microglia, lymphocytes and pathobiochemical mechanisms. Int J Mol Sci 24:12631. https://doi.org/10.3390/ijms241612631
Todea RA, Lu PJ, Fartaria MJ et al (2020) Evolution of cortical and white matter lesion load in early-stage multiple sclerosis: correlation with neuroaxonal damage and clinical changes. Front Neurol 11:973. https://doi.org/10.3389/fneur.2020.00973
Maggi P, Sati P, Nair G et al (2020) Paramagnetic rim lesions are specific to multiple sclerosis: an international multicenter 3T MRI study. Ann Neurol 88:1034–1042. https://doi.org/10.1002/ana.25877
Hemond CC, Reich DS, Dundamadappa SK (2022) Paramagnetic Rim lesions in multiple sclerosis: comparison of visualization at 1.5-T and 3-T MRI. Am J Roentgenol 219:120–129. https://doi.org/10.2214/AJR.21.26777
Diaz-Hurtado M, Martínez-Heras E, Solana E et al (2022) Recent advances in the longitudinal segmentation of multiple sclerosis lesions on magnetic resonance imaging: a review. Neuroradiology 64:2103–2117. https://doi.org/10.1007/s00234-022-03019-3
Shanmuganathan M, Almutairi S, Aborokbah MM et al (2020) Review of advanced computational approaches on multiple sclerosis segmentation and classification. IET Signal Proc 14:333–341. https://doi.org/10.1049/iet-spr.2019.0543
Zeng C, Gu L, Liu Z, Zhao S (2020) Review of deep learning approaches for the segmentation of multiple sclerosis lesions on brain MRI. Front Neuroinform 14:610967. https://doi.org/10.3389/fninf.2020.610967
Doyle A, Precup D, Arnold DL, Arbel T (2017) Predicting future disease activity and treatment responders for multiple sclerosis patients using a bag-of-lesions brain representation. Int Conf Med Image Comput Comput-Assist Interv. https://doi.org/10.1007/978-3-319-66179-7
Peng Y, Zheng Y, Tan Z et al (2021) Prediction of unenhanced lesion evolution in multiple sclerosis using radiomics-based models: a machine learning approach. Mult Scler Relat Disord 53:102989. https://doi.org/10.1016/j.msard.2021.102989
Sepahvand NM, Hassner T, Arnold DL, Arbel T (2019) CNN prediction of future disease activity for multiple sclerosis patients from baseline MRI and lesion labels. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer Verlag, New York, pp 57–69
Sepahvand NM, Arnold DL, Arbel T (2020) CNN Detection of New and Enlarging Multiple Sclerosis Lesions from Longitudinal Mri Using Subtraction Images. In: Proceedings - International Symposium on Biomedical Imaging. IEEE. pp 127–130
Yılmaz Acar Z, Başçiftçi F, Ekmekci AH (2022) Future activity prediction of multiple sclerosis with 3D MRI using 3D discrete wavelet transform. Biomed Signal Process Control 78:103940. https://doi.org/10.1016/j.bspc.2022.103940
Durso-Finley J, Falet J-PR, Nichyporuk B, et al (2022) Personalized prediction of future lesion activity and treatment effect in multiple sclerosis from baseline MRI. PMLR 172:387–406.
Prabhakar C, Li HB, Paetzold JC et al (2023) Self-pruning graph neural network for predicting inflammatory disease activity in multiple sclerosis from brain MR images. Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. Springer, Cham 226–236. https://doi.org/10.1007/978-3-031-43993-3_22
Chataway J, Williams T, Li V et al (2024) Review clinical trials for progressive multiple sclerosis: progress, new lessons learned, and remaining challenges. Lancet Neurol 23: 277–301. https://doi.org/10.1016/S1474-4422(24)00027-9
Newsome SD, Binns C, Kaunzner UW et al (2023) No evidence of disease activity (NEDA) as a clinical assessment tool for multiple sclerosis: clinician and patient perspectives [narrative review]. Neurol Ther 12:1909–1935. https://doi.org/10.1007/s40120-023-00549-7
Amin M, Hersh CM (2023) Updates and advances in multiple sclerosis neurotherapeutics. Neurodegener Dis Manag 13:47–70. https://doi.org/10.2217/nmt-2021-0058
Salvi M, Loh HW, Seoni S et al (2024) Multi-modality approaches for medical support systems: a systematic review of the last decade. Information Fusion 103:102134. https://doi.org/10.1016/j.inffus.2023.102134
Schoonheim MM, Broeders TAA, Geurts JJG (2022) The network collapse in multiple sclerosis: an overview of novel concepts to address disease dynamics. Neuroimage Clin 35:103108. https://doi.org/10.1016/j.nicl.2022.103108
Bessadok A, Mahjoub MA, Rekik I (2021) Graph neural networks in network neuroscience. IEEE Trans Pattern Anal Mach Intell. 45(5):5833–5848. https://doi.org/10.48550/arXiv.2106.03535
Vrenken H, Jenkinson M, Pham DL et al (2021) Opportunities for understanding MS Mechanisms and progression with MRI using large-scale data sharing and artificial intelligence. Neurology 97:989–999. https://doi.org/10.1212/WNL.0000000000012884
Ramspek CL, Jager KJ, Dekker FW et al (2021) External validation of prognostic models: what, why, how, when and where? Clin Kidney J 14:49–58. https://doi.org/10.1093/ckj/sfaa188
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
H.Y and B.M.T conceived the idea and designed the scope and content. H.Y wrote the first draft. H.Y, B.M.T. and F.C. collaboratively modified the manuscript to reach the final version. All authors approved of the final version.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yousef, H., Malagurski Tortei, B. & Castiglione, F. Predicting multiple sclerosis disease progression and outcomes with machine learning and MRI-based biomarkers: a review. J Neurol (2024). https://doi.org/10.1007/s00415-024-12651-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00415-024-12651-3