Abstract
The ability to provide accurate DNA-based forensic intelligence requires analysis of multiple DNA markers to predict the biogeographical ancestry (BGA) and externally visible characteristics (EVCs) of the donor of biological evidence. Massively parallel sequencing (MPS) enables the analysis of hundreds of DNA markers in multiple samples simultaneously, increasing the value of the intelligence provided to forensic investigators while reducing the depletion of evidential material resulting from multiple analyses. The Precision ID Ancestry Panel (formerly the HID Ion AmpliSeq™ Ancestry Panel) (Thermo Fisher Scientific) (TFS)) consists of 165 autosomal SNPs selected to infer BGA. Forensic validation criteria were applied to 95 samples using this panel to assess sensitivity (1 ng-15 pg), reproducibility (inter- and intra-run variability) and effects of compromised and forensic casework type samples (artificially degraded and inhibited, mixed source and aged blood and bone samples). BGA prediction accuracy was assessed using samples from individuals who self-declared their ancestry as being from single populations of origin (n = 36) or from multiple populations of origin (n = 14). Sequencing was conducted on Ion 318™ chips (TFS) on the Ion PGM™ System (TFS). HID SNP Genotyper v4.3.1 software (TFS) was used to perform BGA predictions based on admixture proportions (continental level) and likelihood estimates (sub-population level). BGA prediction was accurate at DNA template amounts of 125pg and 30pg using 21 and 25 PCR cycles respectively. HID SNP Genotyper continental level BGA assignments were concordant with BGAs for self-declared East Asian, African, European and South Asian individuals. Compromised, mixed source and admixed samples, in addition to sub-population level prediction, requires more extensive analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biogeographical ancestry (BGA) prediction, as a form of forensic DNA phenotyping (FDP), is the process of assigning the unknown donors of DNA samples to specific ancestral genetic origins. BGA prediction can provide valuable intelligence from evidential DNA samples when conventional DNA profiling fails to achieve an identification [1, 2].
Ancestry informative single nucleotide polymorphisms (AISNPs) are the genetic markers of choice for BGA prediction due to their low mutation rates and abundance in the genome. [3,4,5,6,7,8]. The Precision ID Ancestry panel (formerly the HID Ion AmpliSeq™ Ancestry Panel) is a collection of 165 autosomal AISNPs commercially available from Thermo Fisher Scientific (TFS) for BGA prediction. The marker set is combination of the entire Kidd AISNP panel [6] and 123 AISNPs from the Seldin panel [7, 8] with 13 SNPs overlapping between the two constituent panels [9]. It is reported to have global coverage for major populations including Africa, Europe, Southwest Asia, South Asia, East Asia, Oceania and the Americas.
The Precision ID Ancestry panel employs massively parallel sequencing (MPS), specifically the Ion PGM™ or Ion S5 (TFS), based on semiconductor sequencing technology. All 165 SNPs are genotyped in a single reaction, and the use of unique oligonucleotide barcode sequences (between six and ten base pairs (bps) in length) enables multiple samples to be sequenced simultaneously. The average amplicon size of the Kidd markers (incorporating primer binding sites and conjoining sequences) is 130 bps while that of the Seldin panel is 122 bps [9]. These characteristics provide a cost-effective approach, minimise the depletion of evidentiary material and confer suitability for analysis of compromised forensic samples. The HID SNP Genotyper v4.3.1 software (TFS) is used to produce two forms of ancestry prediction: admixture proportion (at the continental level) and population likelihood estimates (at the sub-population level). Three varieties of Ion PGM™ semiconductor chips (Ion 314™ v2, 316™ v2 and 318™ v2 chips (Ion Torrent)) can be used to genotype 6, 30 and 59 samples, respectively, when using the Precision ID Ancestry panel, depending on the coverage required [9].
In this study, forensic validation criteria were applied to 95 samples genotyped using the Precision ID Ancestry panel. These included sensitivity (1 ng–15 pg), reproducibility (inter- and intra-run variability), genotype concordance, variation in coverage, analysis of casework-type samples (mixed source, artificially degraded, artificially inhibited and aged blood and bone) and BGA prediction accuracy. The latter was assessed using samples from individuals who self-declared their populations of origin over three generations (n = 50). Every sample was sequenced on at least two Ion 318™ v2 chips on the Ion PGM™, with a total of 48 samples on each chip. HID SNP Genotyper v4.3.1 software was used to perform BGA predictions based on admixture proportions (at the continental level) and maximum likelihood estimates (at the sub-population level).
Methods
Samples
A total of 95 samples were included in this project (Table 1), 50 of which were collected from individuals who self-declared their BGA over three generations where each grandparent was assumed to contribute 25% of their DNA to each individual. Ethics approval for this study was granted by the University of Canberra Committee for Ethics in Human Research (project number 11-119 and its extension, 15-64). The 50 samples were divided into individuals who self-declared their ancestry over three generations as being from a single population of origin (n = 36) or from multiple populations of origin (n = 14). Three standard reference material DNA templates with a 100% European BGA were included. These were the 2800M male human genomic control DNA (Promega), the female human cell line control DNA 9947A (sourced from the AmpFlSTR® Identifiler® Kit: Applied Biosystems) and the male human cell line control DNA 007 (sourced from the AmpFlSTR® Yfiler® Kit: Applied Biosystems). Five replicates of 2800M male human genomic control DNA at 1 ng template input amount were included to assess the reproducibility of genotypes produced by the Precision ID Ancestry panel as well as ancestry proportions reported by HID SNP Genotyper. Sensitivity, genotype concordance and intra-run variability were assessed using 2800M serially diluted from 1 to 0.015 ng and run in duplicate (2800A and 2800B) (n = 2 × 7 = 14). Genotype concordance was assessed using 2800M, 9947A and 007 at 1 ng. The effect of increased PCR cycle number was compared at 21 and 25 cycles using the 2800M sensitivity dilution series.
Samples were analysed in two runs of 48 samples. The first run was performed using 21 PCR cycles and sequenced in duplicate on two Ion 318™ v2 chips (chips 1 and 2) (Table 1). The second run contained lower template and casework-type samples which were amplified using 25 cycles (based on manufacturer’s recommendations for low quality/quantity templates). These consisted of mixed source samples, artificially degraded and artificially inhibited DNA as well as aged bone and blood samples and were also sequenced in duplicate on two Ion 318™ v2 chips (chip 3 and 4) (Table 1). The same library was run on each duplicate chip. The viability of the pooled library (generated from the first 48 samples) after 3 months at 4 °C was assessed by repeating the template preparation using the Ion OneTouch™ 2 System (Ion Torrent) and then sequencing on a fifth Ion 318™ v2 chip according to the manufacturer’s protocol.
Seven mixed source samples were prepared by combining one self-declared European sample with one self-declared African sample at various ratios: 1:10, 1:4, 1:2, 1:1, 2:1, 4:1 and 10:1. Two samples with self-declared European ancestry were artificially degraded by exposing to ultra-violet (UV) light for 30 and 60 min intervals. The light was generated from a 10 W source (Sankyo Denki) at a distance of 13 cm. PCR amplicons were generated from 1 μL of each irradiated sample (1 ng DNA template) for each time interval. Two samples from the same donors (1 ng DNA template) were spiked with 75 and 100 ng humic acid, respectively. Lastly, six samples from unknown donors provided by the NSW Forensic and Analytical Science Service (FASS), NSW, Australia, were also included, five of which were bone samples (unknown age) and the other a blood sample (stored at room temperature for 45 years).
Laboratory analysis
DNA extraction was performed on a BIOMEK® NXp Workstation (Promega) using the DNA IQ™ System (Promega), according to the manufacturer’s protocol. DNA extracts were quantified by real-time PCR analysis in an ABI PRISM® 7500 Real-Time PCR System (Applied Biosystems) using the Quantifiler® Human DNA Quantification Kit (Applied Biosystems), according to the manufacturer’s protocol.
The PCRs were prepared using the Precision ID Ancestry panel according to the manufacturer’s protocol in a total volume of 20 μL. DNA amplification was performed in a Veriti® Thermal Cycler (Applied Biosystems), according to the manufacturer’s protocol, at 21 cycles of amplification. Lower template (down to 0.015 ng), mixed source and compromised/degraded samples were also subjected to 25 cycles of amplification (according to the manufacturer’s recommendation). Library preparation was performed using the Ion AmpliSeq™ Library Kit 2.0 (TFS) for all samples. Template preparation was performed on the Ion Chef™ System (Ion Torrent) using the Ion PGM™ Hi-Q™ Chef Kit (Ion Torrent) according to the manufacturer’s protocol. Pooled libraries were loaded on Ion 318™ v2 Chips (Ion Torrent), and sequencing was performed by the Ion PGM™ System using the Ion PGM™ Hi- Q™ Sequencing Kit (TFS) in accordance with the manufacturer’s protocol. Signal processing, base calling and the generation of unmapped and mapped BAM files (hg19 reference genomic sequence) were performed using Torrent Suite software (version 4.6, TFS). Variant calling was performed with the Torrent Variant Caller Plugin (v4.6.0.7). Default somatic low stringency analysis parameters were applied for the Variant Caller plugin. These included a minimum coverage threshold of 100×, minimum allele frequency of 2% and heterozygote allele frequencies range 35–65%. Default HID SNP Genotyper parameters were also applied, including a minimum coverage of 6 reads and a minimum allele frequency of 10% (implying a minimum homozygote allele frequency of 90%).
Statistical tests
Non-parametric statistical tests using the IBM SPSS package (v. 21) were applied to the data due to the skewed (non-normal) amplicon coverage distribution. A Mann-Whitney U test was performed to test the null hypothesis that there was no difference in coverage between duplicate chips. A Kruskal-Wallis test was employed to test the null hypothesis that there was no difference in coverage between amplicons.
HID SNP Genotyper ancestry assignment
The HID SNP Genotyper v4.3.1 software was used to perform BGA predictions in real time based on likelihood estimates. This software produces two forms of ancestry prediction: admixture proportions (at the continental level) and population likelihood estimates (at the sub-population level). The output produced by HID SNP Genotyper includes a heat map indicating the likely geographical origin of the DNA donor, confidence levels (high or low), admixture proportions of the individual at the continental level (with reference to seven root populations: African, American, East Asian, European, South Asian, Southwest Asian (Middle East) and Oceanian) and log likelihood values of possible sub-population assignments. The root population ‘American’ refers to sub-populations in the ALFRED database (https://alfred.med.yale.edu) which are all derived from Central and South America. Therefore, ‘American’ ancestry, as inferred by HID SNP Genotyper, is indigenous Central/South American. For comparison purposes, the self-declared ancestries of the individuals in this study were aligned with the seven root populations as reported by HID SNP Genotyper. This software references only 151 of the 165 SNPs assayed based on population data available to Thermo Fisher Scientific (Sharon Wootton, Thermo Fisher Scientific, personal communication).
Results
Coverage
The mean depth of coverage for chip 1 and chip 2 (343× and 334×) was higher than the median depth of coverage for both chips (249× and 253×) respectively, indicating a non-normal distribution skewed towards lower coverage in each case. Coverage ranged from zero for both chips to many thousands (9621× for chip 1 and 7777× for chip 2).
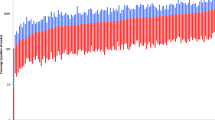
The coverage distribution of SNP amplicons ranked from lowest to highest coverage was consistent between chips (Fig. 1). For example, rs1296819 (left most SNP in Fig. 1) received the lowest coverage on both chips 1 and 2 (medians of 39× and 43×) while rs3793451 (right most SNP in Fig. 1) received the highest coverage (medians of 878× and 901×). A Kruskal-Wallis test indicated a significant difference in coverage between amplicons (p = 0.000) while a two-tailed Mann-Whitney U test indicated no significant difference in amplicon coverage between the two chips (p = 0.715). Mean, median, minimum and maximum coverage for each SNP are shown in Supplementary Table 1.
Coverage distribution across the 165 amplicons in the Precision ID Ancestry panel for 50 samples from individuals who self-declared their ancestry. SNP amplicons (horizontal axis) are ordered from low overall median coverage (left) to high overall median coverage (right) for chip 1 (top) and chip 2 (bottom). Blue bars indicate the range below the median, and red bars indicate the range above the median. A Kruskal-Wallis test indicated significant difference in coverage distribution between amplicons (p = 0.000). A two-tailed Mann-Whitney U test indicated no significant difference in amplicon coverage between the two chips (p = 0.715)
Complete SNP profiles of between 98 and 100% were obtained for all individuals across duplicate runs except for one admixed individual and three individuals (two from Southwest Asia and one from America) which exhibited partial SNP profiles of 92–94%, 92–93%, 79–81% and 82–84%, respectively, across both chips. The partial SNP profiles were characterised by missing genotypes (between 10 and 34 per profile) and a number of SNP loci with low coverage (between 39 and 76 loci per profile).
Reproducibility and genotype concordance
The depth of coverage of the five replicates of the reference DNA (2800M) at 1 ng was used to assess the reproducibility of the Precision ID Ancestry panel across two chips (Fig. 2). Genotyper predicted 100% European BGA for all five replicates across both chips. Four replicates (predicted with high confidence by Genotyper) consisted of full SNP profiles while the fifth replicate (predicted with low confidence by Genotyper) was 98% complete (162/165 loci). Three SNPs had missing data (‘NN’ genotypes) with low coverage; thus, non-concordant genotypes were not observed. Therefore, of the genotypes present, 100% SNP genotype concordance was observed among the five replicates. A Kruskal-Wallis test indicated a significant difference in the total coverage distribution across the five replicates (p = 0.000) while a two-tailed Mann-Whitney U test showed a significant difference in coverage distribution between the two chips (p = 0.026).
Depth of coverage for five replicates of 2800M at 1 ng to assess the reproducibility of the Precision ID Ancestry panel across two chips. Four replicates displayed 100% complete SNP profiles while the fifth was 98% complete (162/165 loci). Boxes represent the interquartile range (25–50%). Whiskers represent 1.5 × interquartile range. Circles (○) represent data that lie outside 1.5 × interquartile range
Genotypes were 100% concordant for 007 on chips 1 and 2 and compared to the expected genotype obtained from the manufacturer. Comparison of 9947A genotypes from chips 1 and 2 revealed 98% concordance due only to missing genotypes for two SNPs (rs12498138 and rs4833103).
Reproducibility of admixture proportions for 50 individuals was assessed by analysing them in duplicate (on chips 1 and 2 or on chips 3 and 4). A two-tailed Mann-Whitney U test indicated no significant difference in admixture proportions between the duplicates (p = 0.872). The admixture proportions were identical for all seven Genotyper BGAs except for five individuals who differed by no more than 5% in no more than two BGAs (Supplementary Table 2).
Sensitivity
Full SNP profiles were generated using 1 ng of DNA template. A decrease in template DNA to 0.5, 0.25 and 0.125 ng resulted in partial SNP profiles of 96–99%, 89–96% and 79–85% respectively across duplicate runs. Genotyper indicated ‘high’ confidence only with template input amounts of 1 ng with ‘low’ confidence calls for all other template input amounts at 21 PCR cycles. However, the correct BGA assignment for 2800M (100% European) was observed down to 125 pg (Fig. 3). Inconsistent BGA assignments, either in the assigned populations or proportions, were observed between chips for template input amounts of less than 125 pg and input amounts of less than 0.06 ng indicated less than 50% concordance. As expected, the depth of coverage decreased as the amount of DNA input decreased with partial SNP profiles of 70% or less from template input amounts of less than 125 pg. A two-tailed Mann-Whitney U test indicated no significant difference in coverage between the two chips (p = 0.270). Figure 4 shows that the ranking of SNP amplicons from low to high genotype concordance in the dilution series was consistent between chips. For example, rs12498138 (left most SNP in Fig. 4) was least concordant on both chips 1 and 2 (in only 2 and 3 samples, respectively).
Genotype concordance and Genotyper biogeographical ancestry assignments of the 2800M sensitivity dilution series on two chips at 21 PCR cycles as a function of DNA template input amount (horizontal axes). Non-concordance was a result of either one allele dropping out or one allele dropping in. Excluded SNPs included those with a quality score of zero (Q = 0) and those that were missing
Genotype concordance for all 14 samples in the 2800M sensitivity dilution series (1, 0.5, 0.25, 0.125, 0.06, 0.03 and 0.015 ng, all in duplicate). SNP amplicons (horizontal axis) are ordered from low overall concordance (left) to high overall concordance (right) for chip 1 (top) and chip 2 (bottom). Non-concordance was a result of either one allele dropping out or one allele dropping in. Excluded SNPs included those with a quality score of zero (Q = 0) and those that were missing
To assess intra-run variability with respect to coverage, each serial dilution series was run in duplicate (2800A and 2800B) on each chip. A Kruskal-Wallis test indicated a significant difference in coverage between the dilution series run on the same chip (p = 0.000).
Increased PCR cycles
The effect of increased PCR cycles on amplicon sequence coverage, completeness of SNP profiles and the accuracy of ancestry prediction was assessed by comparing 21 (recommended for 1 ng template input: Sensitivity) and 25 PCR cycles (recommended for template input of less than 1 ng) using the sensitivity series (serially diluted 2800M 1–0.015 ng). The median value of coverage for 25 cycles was approximately five times higher than for 21 cycles. The mean depth of coverage for both cycle numbers was higher than the median depth of coverage, indicating a distribution skewed towards lower coverage in each case. However, the difference between the mean and median for 25 cycles was higher. The depth of coverage for samples amplified with 21 cycles decreased as the DNA template input decreased. In contrast, the depth of coverage peaked at 0.25-ng DNA template amount for 25 cycles (Fig. 5). At 21 cycles of amplification, a complete SNP profile was only observed for a DNA template input of 1 ng and less DNA template resulted in partial SNP profiles of as low as 4%. In comparison, at 25 cycles, all DNA template inputs generated full, concordant SNP profiles, except for the DNA template input of 0.015 ng which had an 86% partial SNP profile. At 21 amplification cycles, the admixture proportions predicted for all input amounts indicated 100% European BGA down to 125 pg. In contrast to HID SNP Genotyper only indicating ‘high’ confidence using 1 ng at 21 cycles, amplification at 25 cycles achieved ‘high’ confidence for all input amounts except for 0.015 ng. In addition, 100% European BGA was indicated for all template input amounts at 25 cycles.
Boxplots showing the depth of coverage (log scale) for two replicates of seven serially diluted samples ranging between 1 and 0.015 ng at different amplification cycles (21 and 25). Boxes represent the interquartile range (25–50%). Whiskers represent 1.5 × interquartile range. Circles (○) represent data that lie outside 1.5 × interquartile range
Ancestry prediction at the continental level
The accuracy of HID SNP Genotyper ancestry assignments at the continental level was assessed by comparing the predictions to the self-declared populations of origin of 50 individuals from single or multiple populations over three generations (as previously defined). At the continental level, HID SNP Genotyper assigns admixture proportions to the seven root populations. The prediction accuracy was assessed based on the major assigned admixture proportion compared to the self-declared ancestral populations. Figure 6 shows the admixture proportions for individuals self-declared from a single population of origin (n = 36: left) and individuals self-declared from multiple populations of origin (n = 14: right) compared to HID SNP Genotyper assignments. Figure 7 demonstrates that ancestry assignment for individuals self-declared from a single population of origin at the continental level was most accurate for East Asians, followed by Africans, Europeans and South Asians.
HID SNP Genotyper prediction for East Asian individuals was highly concordant with self-declared population of origin at the continental level. Minor contributions from the African, Southwest Asian, South Asian or Oceanian populations were observed in half of the self-declared East Asian individuals (Fig. 6). With the exception of a self-declared Eritrean individual assigned as 55% African/45% Southwest Asian, no admixture was assigned in three self-declared African individuals. South Asian admixture (approx. 30%) and Southwest Asian admixture (15%) was assigned by Genotyper for three self-declared European individuals. Minor African and Oceanian contributions were also assigned with no admixture assigned in only two self-declared Europeans.
Of the self-declared South Asian individuals, minor (< 15%) admixture proportions from the Southwest Asian, African, American, East Asian and Oceanian populations were assigned. However, one self-declared Nepalese individual was assigned a Southwest Asian component (> 25%) with minor European and African components. A self-declared Tamil individual was assigned an African component (approximately 20%) with minor American and European proportions.
HID SNP Genotyper predictions for two of the five Southwest Asian individuals were concordant with self-declared population of origin (with minor East Asian or African admixture) while high European and South Asian admixture (> 40%) was predicted in three individuals (2 × self-declared Kurdish, 1 × self-declared Turkish).
Genotyper was unable to differentiate self-declared Oceanian individuals from self-declared East and South Asians with high admixture (> 40%) observed in the three self-declared Oceanian individuals. Interestingly, one self-declared Fijian individual was assigned greater than 80% South Asian admixture (consistent with Indian migration to Fiji). The major admixture component of the two self-declared American individuals was European indicating only 5 to 10% American BGA. Therefore, HID SNP Genotyper predictions were not concordant with the self-declared, continental level populations of origin of the American and Oceanian individuals but were consistent with the colonial histories of these regions.
HID SNP Genotyper predictions for 14 individuals self-declared from multiple populations of origin were also assessed. With the exception of the three individuals with (indigenous) Australian Aboriginal ancestry (which is not represented in reference data sets), and excluding minor (< 15%) undeclared admixture contributions, HID SNP Genotyper prediction was concordant with self-declared populations of origin for one-third of these individuals while the remaining two-thirds were assigned more admixed ancestry than was declared.
HID SNP Genotyper BGA assignments were reported with high confidence on both chips for 86% (31/36) of the individuals self-declared from a single population of origin. Of the five low confidence predictions, four of these had major BGA proportions consistent with self-declared populations of origin. Low confidence predictions were observed for 50% of the individuals self-declared from multiple populations of origin.
Ancestry prediction at the sub-population level
Genotyper sub-population BGA assignments (population likelihood estimates) are only appropriate for individuals from a single population of origin as they do not include admixture proportions and only represent relative membership likelihoods. This is consistent with the manufacturer’s recommendations for the use of population likelihood estimates. Table 2 compares self-declared population of origin with the five highest likelihood estimates predicted by Genotyper for the 36 individuals self-declared from single populations of origin. Sub-population predictions consistent with self-declared population of origin are shown in bold. For the samples where none of the five highest likelihood estimates are consistent with self-declared population of origin, prediction of a sub-population which is located in close geographic proximity, or a possibly related sub-population/common language group, is indicated in normal text. The remainder are indicated in italics. The five highest likelihood estimates for each sample, corresponding with Table 2, are shown in Supplementary Table 3.
Sub-populations consistent with self-declared populations of origin were predicted by Genotyper in the five highest likelihood estimates for approximately 60% of the samples. Sub-population prediction was accurate for the majority of the self-declared East Asian individuals with specific sub-populations such as ‘Lao Loum’ predicted for self-declared Lao individuals. Of the self-declared African individuals, ‘Hausa’ (language spoken in Ghana) was a consistent sub-population prediction for the Ghanaian individual; however, only related, or geographically close, sub-populations were predicted for the remaining African individuals. The self-declared European individuals were considered to be consistent with the broad groupings ‘European Americans’ or ‘Europeans-HapMap’ but not generally consistent with the alternative sub-populations predicted. However, Genotyper correctly assigned two self-declared Greek individuals.
Sub-population predictions for the self-declared South Asian individuals were consistent with sub-populations such as Keralite, Kachari and Thoti with the exception of the Nepalese individual. Keralite and Pashtun were consistent sub-population predictions for an individual self-declared as 50% Indian (parent from Southern India) and 50% Pakistani. Of the self-declared Southwest Asian individuals, sub-population predictions were only consistent with a self-declared Yemeni and self-declared Kurdish individual. Sub-population assignments for self-declared Oceanian and American individuals were not consistent with self-declared BGA, as observed for continental level ancestry prediction.
Casework-type samples
Mixed source samples were prepared by combining a European sample (both self-declared and assigned) with an African sample (both self-declared and assigned) at various ratios (1:10, 1:4, 1:2, 1:1, 2:1, 4:1 and 10:1). HID SNP Genotyper predicted BGA was concordant with the population of origin of the major contributor (with high confidence) in the high mixture ratios (i.e. 1:10 and 10:1) (Fig. 8). Large discrepancies in Genotyper BGA prediction (low confidence) was observed for the remainder of the mixture ratios. SNP genotype concordance between the mixed source samples and the major contributor decreased as the proportion of the minor contributor increased. However, without the known genotypes of the contributors, and without characterising the expected coverage of each allele at each locus, it may not have been possible to determine the major or minor contributions to the mixture. Therefore, mixture deconvolution is improbable for the Precision ID Ancestry panel, as is the case for biallelic SNP genotypes in general [10].
Artificially degraded samples had, on average (between chips 1 and 2), 34 and 4% complete profiles for 30- and 60-min UV exposure respectively. The inhibited samples indicated an average of 81 and 54% complete profiles for 75 and 100 ng humic acid (HA) addition respectively. Approximately 95 and 100% genotype concordance was observed for the genotypes which were present in artificially degraded samples and 97 and 100% for artificially inhibited samples. Ancestry predicted by HID SNP Genotyper correctly indicated high European BGA (with low confidence). South and/or Southwest Asian admixture was assigned in all samples except for 60-min exposure which may be due to the low coverage in this sample. Figure 9 shows a decrease in coverage with respect to amplicon length in the artificially inhibited and degraded samples.
Partial profiles were observed for the blood (35%, average between chips 1 and 2) and bone (68, 56, 18, 26 and 31%) samples. Both sample types had non-detectable quantities of DNA; therefore, less than 1 ng of DNA was added to the PCR reaction. BGA prediction indicated high European admixture proportions. However, all Genotyper predictions were low confidence. Known genotypes and self-declared populations of origin were not available for these samples.
Assessment of the pooled library after storage
After 3 months of storage at 4 °C, the pooled library from the 48 samples at 21 cycles underwent template preparation using the Ion OneTouch™ 2 System and was then sequenced to assess the viability of the stored library after short-term storage. The mean depths of coverage for the Ion OneTouch™ 2 System and Ion Chef™ System (chip 1 only) (467× and 254×) were higher than the median depths of coverage for both systems (357× and 186×) respectively, again indicating a distribution skewed towards lower coverage in both cases. SNP profile completeness varied between 98 and 100% across both methods. Admixture proportions predicted for all individuals, regardless of the degree of admixture, were concordant between both template preparation methods. A two-tailed Mann-Whitney U test indicated no significant difference in admixture proportions between the two methods (p = 0.954).
Discussion
Forensic validation criteria were applied to 95 samples using the Precision ID Ancestry panel. Coverage analysis indicated a skewed distribution towards shorter amplicons which is consistent with MPS coverage reported in other studies [11, 12]. Statistical tests confirmed that there was a significant difference between amplicon coverage within and between samples indicating that some amplicons received more coverage than others. rs1296819 had consistently poor coverage across all samples. Pereira et al. [13] also reported non-concordant genotypes for this SNP in 2 of 317 population samples genotyped using the Precision ID Ancestry panel, but only from individuals with Danish, Somali and Greenlandic ancestries. In contrast, non-concordant genotypes for rs7251928 and rs7722456 with 13 and 10 non-concordant genotypes respectively were not observed in this study [13]. Low coverage may be the result of less efficient primer binding, or lower amplification efficiencies during PCR [14], a finding consistent with Seo et al. who observed that rs2072422 displayed low coverage across multiple samples in their study [11].
The panel is optimised for a DNA input amount of 1 ng amplified with 21 PCR cycles. Partial SNP profiles and low confidence HID SNP Genotyper predictions were observed for all template input amounts of less than 1 ng. Amplification using 25 cycles greatly increased the sensitivity of the panel with full SNP profiles and high confidence HID SNP Genotyper predictions down to 30 pg. This provides a useful option for analysing low template and degraded DNA which are often encountered in forensic casework [13]. High reproducibility was observed within and between chips with 98% genotype concordance. The 2% deficit was explained by missing data rather than non-concordant genotypes. While this study demonstrated high genotype concordance at low DNA template input amounts using a low stringency (default) coverage threshold (6 reads), further investigation may assist in determining optimal minimum coverage thresholds as well as noise ratios and allelic balance.
Non-admixed individuals are most rigidly defined as those with no less than a 100% contribution from a single BGA while admixed individuals are those who have at least a second measurable contribution, regardless of the size of the proportion. However, in practice, individuals who self-declare as from a single population of origin may have a minor BGA contribution from another population. This is not surprising given the superposition of the large-scale recent human migrations facilitated by modern travel upon the ancient human migrations from Africa, into Europe, across Asia and into the Americas [15,16,17]. At the continental level, BGA assignment for individuals with self-declared single populations of origin indicated a high level of consistency with self-declared populations for East Asian, African, European and South Asian individuals, based on the major admixture proportion as predicted by HID SNP Genotyper. Assignments for East Asian individuals were highly concordant with self-declared population of origin at the continental level (but not necessarily the sub-population level).
High European admixture assigned in self-declared South West Asian individuals may be related to the relatively recent gene flow between southern European countries and the Southwest Asian region [18, 19]. HID SNP Genotyper also assigned a majority European ancestry with only 5 to 10% American admixture for two self-declared Americans (self-declared Colombian and Brazilian individuals). This is indicative of the inherent genetic admixture present as a result of the colonial history of the Americas. Mathias et al. showed that populations from Cartagena (Colombia) and Conde (Brazil) have significant genetic admixture from Europe and Africa as well as an indigenous American ancestral population [20]. Therefore, due to the admixed history of this geographic region, identifying the European and Amerindian components of Americans requires appropriate reference populations for each.
Admixture can be the result of parentage from different ancestral populations; therefore, BGA assignment for individuals from multiple populations of origin is a more complex task than for individuals from a single population of origin. In addition, with respect to BGA assignment, individuals may appear to be admixed as a result of belonging to a BGA which is not represented by a reference population or as a result of a lack of suitable markers to differentiate between the contributing populations. In both cases, admixture proportions of less than 100% will be predicted which is not necessarily reflective of the individual’s actual ancestry.
Sub-population level assignment was accurate for over half of the 36 individuals self-declared as being from single populations of origin; however, the sub-population most consistent with the self-declared population did not always have the highest likelihood estimate. More extensive assessment of sub-population level prediction is required to accurately interpret the likelihood estimates. In addition, this finding suggests that continental-level BGA assignments and sub-population-level likelihood estimates should be examined together to gain a more comprehensive understanding of an individual’s BGA. Sub-population level assignments should not be used as the sole form of classification.
A larger sample set may provide a more accurate representation of the prediction accuracies for the populations assessed in this study. This is especially so for the individuals representing Southwest Asian (n = 5), American (n = 2) and Oceanian (n = 3) populations.
There are a number of important considerations for the assessment of an ancestry-informative assay. The prediction of BGA is heavily reliant, firstly, on the selection of genetic markers and their ability to distinguish between populations. This may be influenced by the genetic markers available (i.e. markers whose genotype frequencies have been characterised in various reference populations). HID SNP Genotyper predictions for self-declared Oceanian individuals indicated limitations in differentiating between East Asian and Oceanian ancestral contributions. Greater resolution may be achieved through the selection of markers that can improve differentiation between these two populations; however, this may be limited due to the paucity of reference individuals [21].
Secondly, BGA prediction accuracy is dependent on the availability and composition of reference populations which are used to train the ancestry prediction algorithms. Some ancestral populations can be scattered across multiple regions (such as the Kurds in the Middle East) while some countries can be included in multiple continental groupings such as those in the Middle East that could be associated with Africa, Europe and/or South Asia. The Southwest Asian (Middle Eastern) region, for example, has been defined in various ways [22, 23]. Phillips et al. highlighted difficulties in assigning (with confidence) individuals from southern Europe (e.g. Spain) and North Africa (e.g. Morocco and Algeria) due to the small genetic distance between these populations [19]. Likewise, the greater-than-expected admixture prediction for a self-declared Eritrean individual in this study could be due to the lack of sufficient markers for this population or the limited number of Eritrean samples in the reference population used. Additionally, Eritrea is close in geographical proximity to Southwest Asia (Middle East); thus, admixture may be expected.
Lastly, BGA assignment is dependent on the assignment algorithm employed. HID SNP Genotyper finds the most likely combination of seven root populations that best explains the genotypes observed in the sample of interest [24]. This is achieved by multiple simulations of various ancestry proportions from members of the root populations with the final proportion being the one that most often resembles the observed genotype. It can also calculate a likelihood estimate for various sub-populations by calculating the relative expected frequency of the observed genotype in reference populations, assuming independence between loci. As such, it is a Bayesian classifier, in common with Structure [25]. These types of Bayesian classifiers have been shown to be more accurate than other classifiers (multinomial logistic regression, genetic distance) for non-admixed individuals [26].
The apparent accuracy of BGA assignment is often reliant on the accuracy of self-declared population of origin. Individuals in this study declared their ancestry over three generations. However, some individuals may declare their affiliation with a particular nationality or ethnic group rather than their genetic ancestry. In this study, a self-declared Fijian individual was predicted by HID SNP Genotyper (with high confidence) to be Southern Indian. Given the large ethnic Indian population in Fiji, it is possible that this individual declared their nationality rather than their BGA. Additionally, Genotyper utilises, and reports, sub-populations with religious or ethnic affiliations such as Yemenite Jews, Ashkenazi Jews, Sephardic Jews and Ethiopian Jews. A non-Jewish individual from Yemen was assigned to the Yemenite Jews sub-population. Although it is known that some of these sub-populations may be genetically distinct from others, as a forensic intelligence tool, it is more appropriate to group individuals based on their BGA rather than ethnic or religious affiliations to avoid problematic and inaccurate ancestry predictions. Genetic determination of BGA will only ever reveal the ancestral origins of an individual, not their current geographical location. This should always be emphasised in any reporting of BGA for forensic intelligence purposes.
Application of the Precision ID Ancestry panel to casework-type samples indicated greater performance for artificially inhibited samples compared to artificially degraded samples. Humic acid spiking was not as detrimental as exposure to UV light. This may be due to the panel being designed and optimised to minimise the effect of PCR inhibitors including humic acid. BGA predictions consistent with self-declared European population of origin were generated by Genotyper (low confidence) but with unexpected South and/or Southwest Asian admixture, possibly due to the high degree of partial profiles generated. Therefore, SNP coverage should be considered in conjunction with BGA prediction, particularly for compromised and degraded samples flagged with low confidence HID SNP Genotyper assignments.
Given the low polymorphic nature of biallelic SNPs, it is difficult to resolve mixtures using this panel. BGA predictions from Genotyper were both reported at high confidence and consistent with declared population of origin only for high mixture ratios (10:1). Genotypes consistent with major contributors were present at high mixture ratios; however, this conclusion was only possible as the genotypes and mixture ratios were known. Further work is required to develop interpretation guidelines for mixture detection including characterisation of expected coverage of each allele at a SNP locus which could be used to detect mixed source samples.
Conclusion
The Precision ID Ancestry panel has demonstrated high sensitivity and reproducibility for use in forensic casework with high quality, single source DNA samples [27]. This study also confirmed the ability of the panel, used in conjunction with the HID SNP Genotyper plugin, to generate accurate continental-level BGA assignments for self-declared East Asian, African, European and South Asian individuals. However, limitations were observed in its capacity to accurately predict continental and sub-population level BGA for individuals self-declared from multiple populations of origin. Improving the performance of this panel may be achieved by employing larger reference populations from under-represented regions, such as South and Southwest Asia, America and Oceania, as well as the inclusion of more informative SNP loci. Development of interpretation criteria will also assist in the assessment of BGA prediction accuracy.
References
Kayser M, de Knijff P (2011) Improving human forensics through advances in genetics, genomics and molecular biology. Nat Rev Genet 12(3):179–192. https://doi.org/10.1038/nrg2952
Pardo-Seco J, Martinón-Torres F, Salas A (2014) Evaluating the accuracy of AIM panels at quantifying genome ancestry. BMC Genomics 15(1):1–24. https://doi.org/10.1186/1471-2164-15-543
Fondevila M, Phillips C, Santos C, Aradas AF, Vallone P, Butler J, Lareu M, Carracedo A (2013) Revision of the SNPforID 34-plex forensic ancestry test: assay enhancements, standard reference sample genotypes and extended population studies. Forensic Sci Int: Genet 7(1):63–74. https://doi.org/10.1016/j.fsigen.2012.06.007
Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T (2008) A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat 29(5):648–658. https://doi.org/10.1002/humu.20695
Kidd JR, Friedlaender FR, Speed WC, Pakstis AJ, De La Vega FM, Kidd KK (2011) Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig Genet 2(1):1. https://doi.org/10.1186/2041-2223-2-1
Kidd KK, Speed WC, Pakstis AJ, Furtado MR, Fang R, Madbouly A, Maiers M, Middha M, Friedlaender FR, Kidd JR (2014) Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci Int: Genet 10:23–32. https://doi.org/10.1016/j.fsigen.2014.01.002
Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF, och p I f g, farmaceutiska v M o, Medicinska f U u (2009) Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat 30(1):69–78. https://doi.org/10.1002/humu.20822
Nassir R, Kosoy R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF (2009) An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC Genet 10(1):39. https://doi.org/10.1186/1471-2156-10-39
Thermo Fisher Scientific (2015) Ion AmpliSeq™ library preparation for human identification applications. Thermo Fisher Scientific Inc., Carlsbad
Eduardoff M, Santos C, de la Puente M, Gross TE, Fondevila M, Strobl C, Sobrino B, Ballard D, Schneider PM, Carracedo Á, Lareu MV, Parson W, Phillips C (2015) Inter-laboratory evaluation of SNP-based forensic identification by massively parallel sequencing using the Ion PGM™. Forensic Sci Int: Genet 17:110–121. https://doi.org/10.1016/j.fsigen.2015.04.007
Seo SB, King JL, Warshauer DH, Davis CP, Ge J, Budowle B (2013) Single nucleotide polymorphism typing with massively parallel sequencing for human identification. Int J Legal Med 127(6):1079–1086. https://doi.org/10.1007/s00414-013-0879-7
Daniel R, Santos C, Phillips C, Fondevila M, van Oorschot RA, Carracedo A, Lareu MV, McNevin D (2015) A SNaPshot of next generation sequencing for forensic SNP analysis. Forensic Sci Int: Genet 14:50–60. https://doi.org/10.1016/j.fsigen.2014.08.013
Pereira V, Mogensen HS, Borsting C, Morling N (2017) Evaluation of the Precision ID Ancestry Panel for crime case work: a SNP typing assay developed for typing of 165 ancestral informative markers. Forensic Sci Int Genet 28:138–145. https://doi.org/10.1016/j.fsigen.2017.02.013
Churchill JD, Chang J, Ge J, Rajagopalan N, Wootton SC, Chang CW, Lagace R, Liao W, King JL, Budowle B (2015) Blind study evaluation illustrates utility of the Ion PGM system for use in human identity DNA typing. Croat Med J 56(3):218–229. https://doi.org/10.3325/cmj.2015.56.218
Cox MP, Karafet TM, Lansing JS, Sudoyo H, Hammer MF (2010) Autosomal and X-linked single nucleotide polymorphisms reveal a steep Asian–Melanesian ancestry cline in eastern Indonesia and a sex bias in admixture rates. Proc R Soc Lond B: Biol Sci:rspb20092041
Xu S, Pugach I, Stoneking M, Kayser M, Jin L, Consortium HP-AS (2012) Genetic dating indicates that the Asian–Papuan admixture through Eastern Indonesia corresponds to the Austronesian expansion. Proc Natl Acad Sci 109(12):4574–4579
Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3(8):611–621. https://doi.org/10.1038/nrg865
Castel C (2014) Inference of biogeographical ancestry and pigmentation phenotype using single nucleotide polymorphisms. Doctoral dissertation, University of Technology, Sydney, Australia
Phillips C, Prieto L, Fondevila M, Salas A, Gomez-Tato A, Alvarez-Dios J, Alonso A, Blanco-Verea A, Brion M, Montesino M, Carracedo A, Lareu MV (2009) Ancestry analysis in the 11-M Madrid bomb attack investigation. PLoS One 4(8):e6583. https://doi.org/10.1371/journal.pone.0006583
Mathias RA, Taub MA, Gignoux CR, Fu W, Musharoff S, O'Connor TD, Vergara C, Torgerson DG, Pino-Yanes M, Shringarpure SS, Huang L, Rafaels N, Boorgula MP, Johnston HR, Ortega VE, Levin AM, Song W, Torres R, Padhukasahasram B, Eng C, Mejia-Mejia DA, Ferguson T, Qin ZS, Scott AF, Yazdanbakhsh M, Wilson JG, Marrugo J, Lange LA, Kumar R, Avila PC, Williams LK, Watson H, Ware LB, Olopade C, Olopade O, Oliveira R, Ober C, Nicolae DL, Meyers D, Mayorga A, Knight-Madden J, Hartert T, Hansel NN, Foreman MG, Ford JG, Faruque MU, Dunston GM, Caraballo L, Burchard EG, Bleecker E, Araujo MI, Herrera-Paz EF, Gietzen K, Grus WE, Bamshad M, Bustamante CD, Kenny EE, Hernandez RD, Beaty TH, Ruczinski I, Akey J, Barnes KC (2016) A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun 7:12522. https://doi.org/10.1038/ncomms12522
Phillips C, Salas A, Sanchez JJ, Fondevila M, Gomez-Tato A, Alvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, Carracedo A, Consortium SN (2007) Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int: Genet 1(3–4):273–280. https://doi.org/10.1016/j.fsigen.2007.06.008
Portland State University (2012) Geography of the Modern Middle East and North Africa. http://www.middleeastpdx.org/resources/original/geography-of-the-modern-middle-east-and-north-africa/. Accessed 08 Nov 2015
Worldatlas.com (2015) Middle East. http://www.worldatlas.com/webimage/countrys/me.htm. Accessed 08 Nov 2015
Thermo Fisher Scientific (2016) HID SNP Genotyper Plugin USER GUIDE v4.3.2 (TFS), publication number MAN0010641 Revision C.0 (2016)
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Cheung EYY, Gahan ME, McNevin D (2017) Prediction of biogeographical ancestry from genotype: a comparison of classifiers. Int J Legal Med 131(4):901–912. https://doi.org/10.1007/s00414-016-1504-3
Hollard C, Keyser C, Delabarde T, Gonzalez A, Vilela Lamego C, Zvenigorosky V, Ludes B (2017) Case report: on the use of the HID-Ion AmpliSeq Ancestry Panel in a real forensic case. Int J Legal Med 131(2):351–358. https://doi.org/10.1007/s00414-016-1425-1
Acknowledgements
The authors acknowledge Dr. Roland van Oorschot (Office of the Chief Forensic Scientist, Victoria Police Forensic Science Centre) for his assistance with sample collection. They would also like to thank the NSW Forensic and Analytical Science Service (FASS) for providing the bone and blood samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Al-Asfi, M., McNevin, D., Mehta, B. et al. Assessment of the Precision ID Ancestry panel. Int J Legal Med 132, 1581–1594 (2018). https://doi.org/10.1007/s00414-018-1785-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-018-1785-9