Abstract
The Ion AmpliSeq™ HID single nucleotide polymorphism (SNP) panel, a primer pool of 103 autosomal SNPs and 33 Y-SNPs, was evaluated using the Ion 314™ Chip on the Ion PGM™ Sequencer with four DNA samples. The study focused on the sequencing of DNA at three different initial target quantities, related interpretation issues, and concordance of results with another sequencing platform, i.e., Genome Analyzer IIx. With 10 ng of template DNA, all genotypes at the 136 SNPs were detected. With 1 ng of DNA, all SNPs were detected and one SNP locus in one sample showed extreme heterozygote imbalance on allele coverage. With 100 pg of DNA, an average of 1.6 SNP loci were not detected, and an average of 4.3 SNPs showed heterozygote imbalance. The average sequence coverage was 945–600× at autosomal SNPs and 465–209× at Y-SNPs for 10 ng–100 pg of DNA. The average heterozygote allele coverage ratio was 89.6–61.8 % for 10 ng–100 pg of DNA. At 10 ng of DNA, all genotypes of the 95 SNPs shared between the two different sequencing platforms were concordant except for one SNP, rs1029047. The error was due to the misalignment of a flanking homopolymer. Overall, the data support that genotyping a large battery of SNPs is feasible with massively parallel sequencing. With barcode systems, better allele balance, and specifically designed alignment software, a more comprehensive rapid genotyping and more cost-effective results may be obtained from multiple samples in one analysis than are possible with current typing and capillary electrophoresis systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forensic DNA typing can provide useful information on human identification, such as in criminal cases and mass disasters. Short tandem repeats (STRs) are the primary genetic markers used because of their high discrimination power and relatively short amplicon size. However, some evidence samples are highly degraded and may not be characterized well with the current battery of STRs. Although commercial mini-STR typing kits enable generation of amplicons ranging from approximately 70–280 bp, some degraded samples still may produce partial or no STR profiles.
In contrast, single nucleotide polymorphism (SNP) typing may be applied successfully to degraded samples that are not amenable to STR typing. Most SNP containing amplicons can be designed to be smaller than 150 bp and, in theory, as short as 50–60 bp in length. There have been a number of reports on SNP typing methods describing the advantages of short amplicon length and high discrimination power. For example, the SNPforID group developed a multiplex assay with 52 autosomal SNPs with a mean match probability of at least 5.0 × 10−19 in nine different populations [1]. Pakstis et al. [2] reported on a panel of 45 unlinked SNPs providing matching probabilities of less than 1.0 × 10−15 in 44 populations. Considering that the match probability for the 13 CODIS core STR loci is approximately 2.4 × 10−15 in, for example, the US Caucasian population [3], these SNP panels provide discrimination power comparable to that of the STR core loci. Various approaches have been used to analyze SNPs, such as single base extension, chip-based microarrays, allele-specific hybridization assays, and mass spectrometry ([1, 2, 4–6], see refs [7–9] for reviews of technologies]. Each of these methods has some limitations; the most notable is not being able to type a large battery of SNPs in a single analysis.
Next generation sequencing technology, or given its maturity current generation sequencing (CGS), provides a platform for more comprehensive coverage of genetic markers. CGS technologies sequence DNA in a massively parallel fashion with high coverage and high throughput of specified targets. Moreover, with the high-throughput capacity afforded by CGS, many different samples, which can be distinguished by barcoding, may be sequenced simultaneously. In theory, hundreds to thousands of barcodes could be synthesized, but currently 384 different reference samples can be coded at one time [10]. With the economy of scales afforded, sequencing cost and run time of the CGS systems have dropped substantially and now offer a potentially cost-effective approach to genetically characterize samples for human identification purposes. These features make SNP typing on a CGS platform particularly appealing. Given the research that already has identified a number of human identity SNPs, the combination of SNPs and typing by CGS should be seamless. However, there are selection criteria to consider beyond the typical population genetic information of allele frequency, Hardy–Weinberg equilibrium, linkage equilibrium, and population substructure [11]. One of the most important criteria will be compatibility of the SNP with the sequencing chemistry of a CGS system.
In this study, CGS data obtained from four barcoded DNA samples (three males and one female) at 10 ng, 1 ng, and 100 pg of template DNA were described. The results were used to assess the performance of CGS and SNP typing and identify criteria for selecting SNPs for human identification.
Material and methods
Sample preparation
DNA was extracted from whole blood of four volunteers (one female, three males) with informed consent. The QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) was used for DNA extraction. The quantity of extracted DNA was estimated using the Qubit® dsDNA BR Assay Kit (Life Technologies, Foster City, CA) on a Qubit® 2.0 Fluorometer (Life Technologies) and the Quantifiler® Human DNA Quantification Kit (Life Technologies) on an ABI Prism® 7500 Sequence Detection System (Life Technologies).
Human identification SNP primer pool
The 2× Ion AmpliSeq™ HID SNP panel primer pool (panel, HID_SNP_v0.1; Life Technologies) was used for this study. This panel was designed to amplify 103 autosomal SNPs and 33 Y-SNPs. Information on the primer pool is described on Ion Community (http://ioncommunity.lifetechnologies.com/community/applications/hid/snps).
Library preparation
To amplify the targeted 136 SNPs, 10 ng, 1 ng, and 100 pg of genomic DNA were used for each of the four samples. PCRs were prepared using the Ion AmpliSeq™ Library Kit 2.0 and 2× Ion AmpliSeq™ HID SNP panel containing the pool of PCR primers on a GeneAmp® PCR System 9700 (Life Technologies) following the manufacturer’s recommended protocols [12]. The PCR conditions for 10 ng of template DNA were 2 min at 99 °C for polymerase activation and 18 cycles of 15 s at 99 °C for denaturation and 4 min at 60 °C for annealing/extension. For 1 ng of template DNA, the amplification cycles were increased to 22 cycles. For 100 pg of template DNA, the amplification cycles were carried out at 26 and 28 cycles and conducted in duplicate. The resulting amplicons were treated with FuPa Reagent (Life Technologies) to partially digest primers. Amplicons then were ligated to Ion P1 and Ion Xpress™ Barcode adapters (Life Technologies) and purified using Agencourt® AMPure® XP Reagent (Beckman Coulter, Brea, CA). Barcoded libraries were assessed by quantitative PCR with the Ion Library Quantitation Kit (Life Technologies) following the recommended protocol [13] and diluted to ∼20 pM. Equal volumes of the four diluted libraries were combined for the next steps.
Template preparation
The diluted library (20 μl) was used to generate template-positive Ion Sphere™ Particles (ISPs) containing clonally amplified DNA. Emulsion PCR was conducted by using the OneTouch™ 200 Template Kit v2 DL with the Ion OneTouch™ DL configuration (Life Technologies) following the recommended protocol [14]. Template-positive ISPs were enriched with the Ion OneTouch™ ES (Life Technologies). Quality of template-positive ISPs was assessed by using the Ion Sphere™ Quality Control Kit (Life Technologies) on the Qubit® 2.0 Fluorometer.
Sequencing and data analysis
The Ion Torrent Personal Genome Machine (PGM™; Life Technologies), a non-optical sequencing platform that exploits CMOS integrated circuits to detect pH shifts by release of protons during incorporation of nucleotides into growing strands, was the primary CGS instrument used in this analysis. Libraries were sequenced on the Ion 314™ Chip with the Ion PGM™ 200 Sequencing Kit (Life Technologies) following the recommended protocol [15]. The plugin “HID SNP Genotyper” on the Ion Torrent server and Integrative Genomic Viewer (IGV) [16, 17] were used for data analysis. The reference genome was Hg19.
Results and discussion
Library information
For the sequencing data of four samples at 10 ng, 1 ng, 100 pg (26 cycles, twice), and 100 pg (28 cycles, twice) of initial genomic DNA, the number of filtered and trimmed base pairs was 58.29, 50.03, 44.12 ± 3.99 (standard deviation), and 45.17 ± 1.91 Mbp, respectively. Considering a predicted error rate of 1 % (Q20), the total number of bases for 10 ng, 1 ng, 100 pg (26 cycles, twice), and 100 pg (28 cycles, twice) was 51.79, 44.85, 38.37 ± 3.94, and 39.66 ± 1.44 Mbp, respectively.
Allele coverage
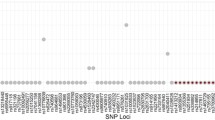
The allele coverage of SNP typing with 10 ng to 100 pg of genomic DNA is presented in Tables S1–6 (in the Electronic supplementary materials, ESM). Allele coverage charts for the 136 SNPs are presented in Figs. S1–6 (ESM). The average coverage of autosomal and Y-SNPs for 10 ng, 1 ng, and 100 pg of template DNA are shown in Figs. 1 and 2.
Results show that at 10 ng of template DNA, there is consistently high coverage with little variation between samples. Genotypes of Y-SNPs were not detected in sample no. 1 (female). However, variation in coverage was observed among the SNPs (Figs. S1–6). Each SNP generally showed similarly high or low coverage across the samples. The lowest coverage was at rs2072422 (a Y-SNP) at 5–9× in the three male DNA samples (Table S1). There were no bases for this SNP with Base Phred Quality (BPQ) of ≤10. The difference in coverage primarily may be related to differences in PCR amplification efficiency. Modifications of the primers and/or primer concentrations could increase amplification and provide more balance and higher yield across the SNPs in the panel.
The average coverage of autosomal SNPs with 1 ng of template DNA was comparable to the 10 ng for the female sample and lower for the male samples (Fig. 1). The average coverage of Y-SNPs for 1 ng samples also was lower than 10 ng samples (Fig. 2). The SNP rs2072422 still displayed the lowest allele coverage of 1–4× in male DNA samples (Table S2). A genotype at the Y-SNP rs891407 was detected with coverage of 2× in the female sample (sample no. 1). This observation could be due to minor contamination (i.e., allele drop-in) with the increased number of PCR cycles or partial homology on the X chromosome. Such low level detection of 2× might be filtered out based on validation studies that establish a minimum coverage level for reliable allele calling. These overall results support that target amplification and detection are feasible with a relative low quantity level of DNA template.
For the average coverage of autosomal and Y-SNPs with 100 pg of DNA for 26 and 28 cycles, overall, observed allele coverage was higher for 26 PCR cycles than for 28 cycles (Figs. 1 and 2). However, this coverage difference did not appear to be directly related to the number of complete locus drop-out and heterozygote imbalance events (see SNP detection section for details).
Allele coverage may depend on the total number of bases obtained from sequencing. However, the number of bases was higher with 100 pg of DNA at 28 cycles than at 26 cycles. Yet 100 pg of DNA with 28 PCR cycles (first run) produced a lower sequencing coverage of 531× (autosomal and Y-SNPs) compared with those using 100 pg of DNA with 26 cycles (611× at second run) and 28 cycles (629× at second run). The total number of bases was 46.52, 41.30, and 43.82 Mbp in 100 pg of DNA with 28 PCR cycles (first run), 26 cycles (second run), and 28 cycles (second run), respectively. Factors such as the number of bases aligned to the reference sequence and base quality might play a role in the minor variation in coverage.
SNP detection
Genotypes at all SNP loci were obtained for all samples with 10 ng of template DNA on the PGM™. True genotypes were based on a combination of the 10 ng results and genotype results obtained from an in-house panel on a different chemistry and platform, i.e., the Genome Analyzer IIx (GAIIx; Illumina, Inc., San Diego, CA) [18] (data not shown). These genotypes were used to determine both locus and allele drop-out at the other template quantities.
Most SNP genotypes were detected correctly at 1 ng of template DNA, except for one SNP in one sample demonstrated extreme heterozygote imbalance on allele coverage (Table 1). At rs13218440 in sample no. 4, the true genotype was AG, and allele coverage of A and G was 452× and 30×, respectively (Table S2). In this study, SNPs that showed allele coverage ratios <20 % were identified. Allele coverage ratio was calculated by dividing the coverage of one allele (showing lower coverage) by the coverage of the other allele (showing higher coverage; ex. 30/452 = 6.6 % at rs13218440 in sample no. 4). The selection of this threshold was arbitrary and further studies will be needed to determine operational thresholds.
SNP typing with 100 pg of template DNA was conducted using 26 PCR cycles and 28 PCR cycles in duplicate (a total of four PCR reactions per sample). Most SNP genotypes were detected. However, 2–6 SNPs and 1–5 SNPs were not detected in sample nos. 3, 4, and 4275 using 26 and 28 PCR cycles, respectively (Table 2). For emulsion PCR, equimolar amounts of each barcoded library were combined and the pooled library was used. Therefore, a portion of autosomal SNPs which is amplified could be higher in the female sample (sample no. 1) than in the male samples (sample nos. 3, 4, and 4275). This coverage difference could be one of the reasons that all genotypes were detected in the female sample. Autosomal SNPs were not detected at rs1554472 in sample no. 3, and rs1355366 and rs727811 in sample no. 4275 (Tables S3–6). A total of 12 sequencing runs were performed at 100 pg for the male samples. In these runs, the following Y-SNPs were not detected: rs2072422 (six occurrences), rs2032673 (five occurrences), rs3900 (two occurrences), rs2032601, rs1800865, rs2075640, rs2267801, rs1865680, rs2032598, rs2032599, rs2032607, and rs2032653. Overall, the number of undetected genotypes was higher for Y-SNPs than autosomal SNPs.
For 26 and 28 PCR cycles, an average of 5.5 SNPs and 3 SNPs showed heterozygote imbalance <20 %, respectively (Table 1). The detailed information on the SNPs with such heterozygote imbalance is shown in Table S7 (ESM). There was no considerable difference of the number of SNPs showing heterozygote imbalance between the two different PCR cycles. No detectable pattern of heterozygote imbalance was observed across the SNP loci, other than those clearly low-performing loci with low coverage at 10 ng of DNA. This phenomenon was more severe in the results from 100 pg of template DNA than those from 1 ng of template DNA. Most discordant genotypes were due to heterozygote imbalance, resulting in changes from heterozygous genotypes to apparent homozygous genotypes. However, one case showed a homozygous genotype that changed to a heterozygous genotype. At rs576261 in sample no. 1, the genotype was designated as AC (true type: CC; Table S4). The number of reads of A and C was 108 and 210, respectively. A possible reason for this observation might be contamination (i.e., allele drop-in), as might be expected with an assay with high sensitivity.
In addition, the number of complete locus drop-out and heterozygote imbalance events was not related to coverage difference. Overall, observed allele coverage was higher for 26 PCR cycles than for 28 cycles. The average number of non-detected genotypes was 1.5 and 1.6 in 26 and 28 cycles, respectively. The average number of SNPs showing heterozygote imbalance was 5.5 and 3 in 26 and 28 cycles, respectively.
Heterozygote allele balance
The coverage ratio of heterozygous alleles was assessed (Fig. 3). Allele coverage ratio was calculated by the aforementioned method on “SNP detection” section. The observed allele coverage ratio with 10 ng, 1 ng, 100 pg (26 cycles), and 100 pg (28 cycles) of template DNA was 89.6 ± 11.3 %, 70.7 ± 18.3 %, 60.4 ± 21.1 %, and 63.2 ± 21.6 %, respectively. This balance remained relatively consistent among samples at each template amount.
In tests with 10 ng of template DNA, SNPs displaying imbalanced allelic coverage ratios of <60 % were investigated further (Table S1). In sample no. 1, rs1029047 showed T(35.3 %)/A(64.7 %) and rs4530059 showed G(25.2 %)/A(74.8 %), which are equivalent to allele coverage ratios of 54.6 and 33.7 %, respectively. In sample no. 3, rs4530059 showed G(23.8 %)/A(76.2 %) and rs576261 showed A(63.5 %)/C(36.5 %), which are equivalent to allele coverage ratios of 31.2 and 57.4 %, respectively. In sample no. 4, rs4530059 showed G(24.4 %)/A(75.6 %), which is equivalent to an allele coverage ratio of 32.2 %. In sample no. 4275, rs576261 showed A(65.5 %)/C(34.5 %), which is equivalent to an allele coverage ratio of 52.7 %. The heterozygote allelic imbalance may be attributed to a primer mismatch causing PCR bias or may just be a stochastic effect that is greater at some loci than others. Further work will be performed by typing more samples to determine if a trend persists. If such a trend continues for certain loci presenting a high allelic imbalance, then primers should be modified or the SNP could be replaced with a better performing SNP.
Problematic SNP
The high throughput of CGS makes it difficult to verify typing results with standard capillary electrophoresis-based methods, as the latter method does not have sufficient throughput. Concordance typing is more efficient for determining correct typing results using two CGS systems based on different chemistries. Of the 103 autosomal SNPs, there were 95 SNPs in common with an in-house panel of SNPs (data not shown). Data from this panel were generated using the GAIIx [18]. The SNPs rs10495407, rs10768550, rs901398, rs2175957, rs4789798, rs689512, rs2292972, and rs9606186 were not part of the in-house panel run on the GAIIx. All SNP typing results using 10 ng of template DNA were concordant for the SNPs in common between the two platforms, except for the SNP rs1029047. This SNP is flanked by homopolymeric stretches, and the SNP states are the same as the homopolymer regions [TTT(T/A)AAAAAAAAA]. A priori this SNP was suspected of posing a potential typing problem because of the continuum of flanking homopolymers. The homopolymer reads interfered with the alignment of bases to the Hg19 genome reference. In sample no. 1, the locus appeared to be heterozygous; a mixture of T and A was observed with IGV (Fig. 4a). However, 44.4 % of bases showed BPQ of ≤10 at the locus when bases with BPQ of ≥4 were counted. IGV aligned bases to the reference genome based on a 3′ end strategy. Therefore, misalignment could have occurred at bases at the 5′ end of the homopolymers when homopolymer length was not correctly determined, e.g., the alignment at the first T position at the T stretch (TTTT). The first T was incorrectly designated as a deletion in 22.6 % of the reads when bases with BPQ of ≥4 were counted; in these reads, the T bases were shifted to other T positions. This observation indicates a high probability that the SNP genotype was incorrect with the PGM™ data. The overall data indicated that the true genotype of rs1029047 for sample no. 1 is AA. The in-house GAIIx panel yielded an AA type for this SNP.
The rs1029047 SNP was examined in the other samples. In sample nos. 3 and 4, 99.0 % of the bases were detected as T (Fig. 4b, c). The TT genotype was correctly called and was concordant with GAIIx-generated results. However, the A deletion and A calls with low BPQ scores were still observed in the homoploymeric A stretch. The insertion of A, AA or TA between A and T stretches also was observed. In sample no. 4275, the portion of reads calling T and A was 67.0 % and 33.0 %, respectively, and the TT genotype was determined using the HID SNP Genotyper plugin (Fig. 4d). When the TTT(T/A)AAAAAAAAA region flanking of SNP rs1029047 was examined using IGV, 14.9 % of the first T in the T stretch was incorrectly designated as a deletion due to a shift of bases when bases with BPQ of ≥4 were counted. After correcting the alignment problem, 46.0 % of bases showed A and 54.0 % of bases showed T at the locus. This observation indicated that the true SNP type was an A/T heterozygote. It also indicated that even with flanking homopolymers it may be possible to overcome incorrect calls with software that uses a specifically designed algorithm for alignment.
Based on the chemistry and detection system of the PGM™, the intensity of the electronic signal due to pH change increases proportionally with the number of incorporated bases added [19]. In theory, a homopolymer with ten residues should produce twice the signal of homopolymer with five residues. However, operationally, signals generated from homopolymers with the PGM™ system are not entirely linear [20], and each locus with adjacent homopolymers needs to be evaluated and tested. Overall, the PGM™ chemistry was quite successful in typing SNPs and showed concordance with the GAIIx data, except for the one SNP. Chemistry enhancements and/or software may reduce the typing difficulty of SNP rs1029047. The GAIIx, because of a different chemistry approach, did not experience the same degree of sequencing problems with homopolymers surrounding SNP rs1029047. For genotyping with the GAIIx, misalignment of bases due to homopolymers was less likely, and an overall error of genotype designation was not observed at SNP rs1029047. At sample no. 1, only 5.5 % of GAIIx reads showed T and 94.5 % of reads showed A (and the genotype was determined as AA) using IGV. At sample no. 4275, 45.1 % of reads showed A and 54.9 % of reads showed T (determined as the AT genotype). These results indicated that the GAIIx was more accurate at sequencing homopolymeric stretches compared with the PGM™. While the low percentage of misalignments with the GAIIx data had no impact on typing single source samples, there may be some limitations with mixture analyses. The 5.5 % T calls might be reduced with better-directed alignment software. In total, an incorrect estimation of homopolymer length and a concomitant alignment issue contributed to the typing problem of the SNP. The alignment issue due to homopolymeric stretches was more severe in samples (nos. 1 and 4275) with at least one A allele at SNP rs1029047 than in samples (nos. 3 and 4) with only a T allele at this position. The length of the A homopolymer was 10 in the former samples and 9 in latter samples. If the SNP is selected for a human identification panel, then special attention will be required to designate heterozygous genotypes compared with homozygous genotypes (see. Fig. 4a and d, samples no. 1 and 4275) by correction of misaligned SNPs and calculation of allele ratios.
Table S8 (ESM) lists those SNPs within the Ion AmpliSeq™ HID SNP panel that were proximal to homopolymers of three bases or more. Only SNP rs1029047 has such an extremely long homopolymer immediately flanking the site. All others demonstrated no genotyping errors, i.e., complete concordance among the two CGS platforms. As an example, a TT genotype [TTT(C/T)T stretch], was determined with a T called in 94.1 % of the reads at SNP rs430046 (sample no. 4). It has been reported that for homopolymers of length 5, the per-base accuracy is 97 % on the PGM™ [19]. Therefore, designating homopolymeric stretches may be correct up to (or slightly beyond) 5 residues but may not be relevant if alignments can anchor on a base adjacent to the SNP. Immediate flanking SNPs that differ from the known allelic state of a SNP could anchor alignments, further reducing error in allele calls. Consider, for example, SNP rs10092491, where a G residue lies immediately 5′ to the C/T SNP. Even if an incorrect estimation of the number of homopolymer bases were to arise, the alignment could anchor on the G residue and reduce the chance of mistyping. Although no typing errors were observed with 94 of the 95 SNPs on the PGM™, it would be beneficial to review sequences around all SNPs for potential homopolymer and alignment issues.
The SNPs in the Ion AmpliSeq™ HID SNP that were not in common with our in-house Genome Analyzer panel were reviewed for adjacent homopolymers, as described. The data support that the calls were correctly obtained. Larger samplings are needed to verify if there are primer binding variants in the population that could impact typing success.
Conclusion
In this study, the Ion AmpliSeq™ HID SNP panel typed on the PGM™ was evaluated. Genotypes were obtained for all 136 SNPs for the three male samples and 103 SNPs for the female sample with 10 ng of initial template DNA. With 1 ng of DNA, most SNPs were detected and typed correctly; there was one example of extreme heterozygote imbalance across the four samples. With 100 pg of DNA, an average of 1.6 SNP loci was undetected and an average of 4.3 SNPs showed heterozygote imbalance <20 % across the samples. All barcoded samples showed high autosomal SNP allele coverage averaging 945× with 10 ng of template DNA, 792× with 1 ng of DNA and 689× with 100 pg of DNA. For Y-SNPs, the samples showed high average allele coverage of 465× with 10 ng of template DNA, 350× with 1 ng of DNA and 257× with 100 pg of DNA. Average heterozygote allele coverage ratios were 89.6, 70.7, and 61.8 % with 10 ng, 1 ng, and 100 pg of template DNA, respectively. Successful and accurate typing with approximately 1 ng of initial template DNA is promising and indicates that the sensitivity of detection of CGS technology may reach the sensitivity of detection of current forensic DNA typing methods.
In the analyses using the standard 10 ng of template DNA, all genotypes were the same as those obtained from an in-house GAIIx SNP panel, except for SNP rs1029047, where a typing error was induced by an adjacent homopolymer. SNPs residing adjacent to homopolymers may require further scrutiny before placing them in panels and, even more so, before selecting them as core markers. One approach would be not to select such SNPs as universal forensic identity markers since they would not work on all known current platforms. In contrast, this approach could be limiting for implementation with new technologies that are certain to be developed. If SNPs are selected based on compatibility with the current platforms and the SNPs become standards, then future technologies could be locked out even if their sequencing capabilities were superior. Another approach may be to let different technologies incorporate whatever panels are accurate and reliable, as long as there is substantial overlap among panels. Given that all but one of the SNPs tested were concordant between CGS systems and that hundreds of SNPs may be typable, panels with substantial SNP overlap are possible and the very few SNPs that may not be amenable to one particular platform should not impact community compatibility. Other factors such as simplicity of library preparation, cost, labor, coverage, accuracy, and robustness may be more meaningful for selecting a particular platform.
This study, although limited to four samples, indicated that typing of samples for a large battery of SNPs is feasible and that CGS technology may be a reality for characterizing at least reference samples for national DNA databases in the near term. In addition, applications of massive SNP typing in forensic genetics go beyond databasing and make feasible testing such as distant familial relationships [21]. Further efforts will focus on increasing sample sizes, simplifying library preparation, balancing the amplicon yield to obtain similar coverage across more of the SNPs in a panel, selection of additional SNPs, elucidating the contributing factors of allele drop-out, and alignment and interpretation guidelines for single-source and mixture samples. Although not the focus of this study, the combination of SNPs from the Kidd-panel and SNPforID 52plex would yield much greater discrimination powers beyond that of either panel. As CGS provides the capability of multiplexes larger than previously conceived, population genetics studies will need to be carried to assess the effects of linkage and linkage disequilibrium that will inevitably be encountered. Such studies will be carried out in the near future to support application of large multiplex analyses.
References
Sanchez JJ, Phillips C, Børsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N (2009) A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 27:1713–1724
Pakstis AJ, Speed WC, Fang R, Hyland FC, Furtado MR, Kidd JR, Kidd KK (2010) SNPs for a universal individual identification panel. Hum Genet 127:315–324
Ge J, Eisenberg A, Budowle B (2012) Developing criteria and data to determine best options for expanding the core CODIS loci. Investig Genet 3:1
Tomas C, Axler-DiPerte G, Budimlija ZM, Børsting C, Coble MD, Decker AE, Eisenberg A, Fang R, Fondevila M, Fredslund SF, Gonzalez S, Hansen AJ, Hoff-Olsen P, Haas C, Kohler P, Kriegel AK, Lindblom B, Manohar F, Maroñas O, Mogensen HS, Neureuther K, Nilsson H, Scheible MK, Schneider PM, Sonntag ML, Stangegaard M, Syndercombe-Court D, Thacker CR, Vallone PM, Westen AA, Morling N (2011) Autosomal SNP typing of forensic samples with the GenPlex™ HID System: results of a collaborative study. Forensic Sci Int Genet 5:369–375
Børsting C, Sanchez JJ, Morling N (2005) SNP typing on the NanoChip electronic microarray. Methods Mol Biol 297:155–168
Mengel-Jørgensen J, Sanchez JJ, Børsting C, Kirpekar F, Morling N (2005) Typing of multiple single-nucleotide polymorphisms using ribonuclease cleavage of DNA/RNA chimeric single-base extension primers and detection by MALDI-TOF mass spectrometry. Anal Chem 77:5229–5235
Budowle B, Planz J, Campbell R, Eisenberg A (2004) SNPs and microarray technology in forensic genetics: development and application to mitochondrial DNA. Forens Sci Rev 16:22–36
Budowle B (2004) SNP typing strategies. Forensic Sci Int 146(Suppl):S139–S142
Sobrino B, Brión M, Carracedo A (2005) SNPs in forensic genetics: a review on SNP typing methodologies. Forensic Sci Int 154:181–194
NuGEN (2013) Encore™ 384 Multiplex System. NuGEN. http://www.nugeninc.com/nugen/index.cfm/products/pl/library-preparation/encore-384-multiplex-system/. Accessed 30 January 2013
Wei YL, Li CX, Jia J, Hu L, Liu Y (2012) Forensic identification using a multiplex assay of 47 SNPs. J Forensic Sci 57:1448–1156
Life Technologies (2012) Ion AmpliSeq™ Library Preparation User Guide. Life Technologies, Foster City (CA)
Life Technologies (2011) Ion Library Quantitation Kit User Guide. Life Technologies, Foster City (CA)
Life Technologies (2011) Ion OneTouch™ 200 Template Kit v2 DL. Life Technologies, Foster City (CA)
Life Technologies (2011) Ion PGM™ 200 Sequencing Kit. Life Technologies, Foster City (CA)
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2012) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. doi:10.1093/bib/bbs017
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Davis C, Warshauer, D, Budowle B (2012) DNA profiling of database reference samples using second generation sequencing. 23rd International symposium on human identification, Nashville (TN)
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352
Voelkerding KV, Dames SA, Durtschi JD (2009) Next-generation sequencing: from basic research to diagnostics. Clin Chem 55:641–658
Keating B, Bansal AT, Walsh S, Millman J, Newman J, Kidd K, Budowle B, Eisenberg A, Donfack J, Gasparini P, Budimlija Z, Henders AK, Chandrupatla H, Duffy DL, Gordon SD, Hysi P, Liu F, Medland SE, Rubin L, Martin NG, Spector TD, Kayser M (2013) First all-in-one inference tool for DNA forensics: parallel genome-wide inference of bio-geographic ancestry, appearance, relatedness and gender with Identitas forensic chip. Int J Leg Med 127:559–572
Acknowledgments
This project was supported in part by Award No. 2012-DN-BX-K033, awarded by the National Institute of Justice, Office of Justice Programs, US Department of Justice. The opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect those of the US Department of Justice. We would like to thank Life Technologies for providing its technical expertise and contributing to the sample preparation and sequencing chemistries used in this study. In addition, we would like to thank Illumina Inc for its contributions for generating the in-house SNP panel.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Seo, S.B., King, J.L., Warshauer, D.H. et al. Single nucleotide polymorphism typing with massively parallel sequencing for human identification. Int J Legal Med 127, 1079–1086 (2013). https://doi.org/10.1007/s00414-013-0879-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-013-0879-7