Abstract
The effects of protists on an indigenous soil bacterial community, putative bacterial genes involved in N-cycling, and the rice plant growth were studied in poultry litter biochar (PL) and rice husk biochar (RH) amended (with two application doses: 2% and 4% w/w) paddy field soil. The bacterial community composition, which was evaluated using 16S rRNA gene amplicon sequencing, was significantly and differentially affected by the protists, the PL and the RH. The effects of protists on the bacterial community composition were decreased by the RH and the PL treatments. The number of protist-affected bioindicator bacterial taxa was decreased from 90 to 46, 29, 43, and 21 in the 2% RH-, 4% RH-, 2% PL-, and 4% PL-treated soils, respectively. The presence of the protist significantly increased the abundance of the putative bacterial genes involved in mineralisation, dissimilatory nitrate reduction to ammonium (DNRA), and NO3- assimilation, and the same occurred with PL treatments. The rice plant growth and N uptake were always higher in the presence of protists and PL amendments. Overall our results suggest a new insight into the effects of biochar on the bacterial community via altering the trophic interactions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biochar amendment is often considered as an important solution for global warming (Herdler et al. 2008; Woolf et al. 2010), and, thus, it is one of the most important practices in sustainable agriculture. Biochar has positive effects on carbon sequestration, greenhouse gas emission, soil fertility, and agricultural productivity (Lehmann et al. 2011; Rondon et al. 2007). It is well documented that the biochar amendments shape microbial communities and functioning (D’Hose et al. 2018; Geisseler and Scow 2014; Gul et al. 2015; Lehmann et al. 2011). The effects of the biochar amendments on the microbial communities are often associated with the changes in soil chemical (i.e., pH and nutrient contents) (Dai et al. 2018; Hale et al. 2015) and physical (i.e., bulk density, porosity) (Gul et al. 2015) properties. Furthermore, biochar pores provide a specific habitat to the soil microorganisms within itself depending on the surface physicochemical properties of the biochar such as porosity, surface area, and nutrient contents (Quilliam et al. 2013). Despite the great body of work conducted to understand the effects of biochar on the microbiome composition and functionality, less is known about its effects on interactions among microorganisms, especially between the prey and predator.
The prey-predator interactions are a central topic in microbial ecology and are among the key factors shaping microbiome composition and functionality (Chow et al. 2014; Fox 2007). The trophic interactions between phagotrophic protists (hereafter referred to as ‘protists’ for simplicity) and bacteria are the most commonly found and well-studied pair as protists are the major bacterial predators (Crotty et al. 2012; Trap et al. 2016). The role of protists as bacterial predators makes them an integral part of the soil microbiome (Bonkowski 2004; Clarholm 1985). The protist predation alters the bacterial community composition (Flues et al. 2017; Kreuzer et al. 2006; Rønn et al. 2002), stimulates bacterial activities (Bonkowski and Brandt 2002; Hünninghaus et al. 2017; Murase et al. 2006) and increases the plant growth (Asiloglu et al. 2020b; Herdler et al. 2008; Kreuzer et al. 2006). Due to selective feeding of protists on bacteria at the genus (Singh 1941, 1942) and even at the species level (Murase and Frenzel 2008), the number of the bacteria susceptible to protist predation markedly decreases (Saleem et al. 2012). Although the protist predation is often considered as a limiting factor for bacterial growth, the effect of protists on bacteria is not always negative. The bacterial species that can avoid predation or the non-target species may benefit from the protist predation (Jousset et al. 2008; Matz and Kjelleberg 2005; Salcher et al. 2005) by the increased availability of nutrients released from the protist-preyed microbial biomass (Bonkowski et al. 2000; Griffiths 1994) and/or by gaining a competitive advantage as a consequence of the protist predation on their strong competitors (Bell et al. 2010; Flues et al. 2017; Jousset et al. 2008; Saleem et al. 2012).
The protist predation and its effects on bacteria vary depending on the physicochemical properties of soil, such as pH, texture, available pore space, and water (Adl 2007; England et al. 1993; Rutherford and Juma 1992). Considering the effects of biochar on the soil physicochemical properties (Gul et al. 2015), the biochar amendment is likely to affect the protist predation and its outcomes on the prey’s community composition. Furthermore, although no quantitative evidence is available, it has been long hypothesized that biochar protects bacteria from predators by allowing them to explore the micro-pore habitats of itself (Lehmann et al. 2011; Quilliam et al. 2013), which is too small for predators. This hypothesis was further supported by a decreased population of relatively big-sized predators, bacterivorous nematodes (Kamau et al. 2019; Liu et al. 2020) and amoeba (Liu et al. 2020) in biochar amendment soils. Taken together, here we hypothesized that the biochar amendment decreases the effects of protist predation and alters the effect of protists on overall bacterial community composition. Since the effects of biochar on protists vary depending on its physicochemical properties (Asiloglu et al. 2020a; Kamau et al. 2019; Liu et al. 2020), we assumed that protists-bacteria interactions might be affected by the physicochemical properties of biochar. Therefore, we used two biochars with different physicochemical properties: rice husk biochar (RH), which has relatively low nutrient content and high surface area, and poultry litter biochar (PL), which has relatively high nutrient content and low surface area. In this study, we aimed to evaluate the effects of protists on an indigenous bacterial community composition in a rice rhizosphere under RH and PL amendments. A 16S rRNA gene amplicon sequencing method, which efficiently reveals the predatory effects of protists on bacterial communities (Asiloglu et al. 2020b; Flues et al. 2017; Kurm et al. 2019), was used to evaluate the effects of four common soil protists isolated from a rice rhizosphere (Asiloglu et al. 2020b) on an indigenous bacterial community that was obtained from a paddy field soil. Previous studies showed that not only bacterial community composition but also bacterial activities, especially on N turnover, were affected by the protists (Hervey and Greaves 1941; Murase et al. 2006) and biochar amendments (Anderson et al. 2014; Wu et al. 2016), which consequently enhance the plant growth (Bonkowski 2004; Lehmann et al. 2011). Therefore, the effects of protists on putative bacterial genes involved in N-cycling and on the rice plant growth in the biochar amended soils were also analysed.
Material and methods
Biochar production, soil samples, microorganisms and rice seedlings
The biochars were produced as described previously (Asiloglu et al. 2020a). Briefly, the raw materials were dried at 105 °C for 24 h, sieved through 2-mm mesh and stored at room temperature (25oC) until pyrolysis. The raw materials were subjected to pyrolysis in an anaerobic pyrolysis chamber (Ankara University, Ankara, Turkey) at 300 °C with a heating rate of 10 °C/min for 120 min to obtain the biochar. The collection of soil samples was performed as described by Asiloglu et al. (2020b). The soil sample had the following characteristics: sand, 33.6%; silt, 47.0%; clay, 19.4%; TC, 1.6%; TN, 0.2%; pH, 5.0 (H2O); and CEC, 15 meq 100 g-1. Prior to the experiment, the soil and the biochars were sterilized by autoclaving at 121 °C for 30 min. This procedure was repeated 3 times in 3 consecutive days.
We used a mixture of four protists previously isolated from a rice rhizosphere (Asiloglu et al. 2020b): Vermamoeba vermiformis (formerly known as Hartmannella vermiformis, Amoebozoa; Tubulinea) (~20 μm), Naegleria sp. (Excavata, Heterolobosea) (~25 μm), Colpoda steinii (Alveolata; Ciliophara) (~30 μm) and Heteromita globosa (Rhizaria; Cercozoa) (~10 μm). The preparation of sterile rice (Oryza sativa L. Nipponbare) seedlings was done as described previously (Asiloglu et al. 2020b). The protist-free indigenous bacterial community was obtained from the non-sterile paddy field soil by a filtering method (0.8 μm pore size mixed cellulose ester membrane filters [Advantec, Tokyo, Japan]) as previously described (Asiloglu et al. 2020b).
Experimental set-up and sampling
Sterile centrifugation tubes (50 mL) were filled with 40 g of the sterile soil. The paddy field soil used in this study was rich in nutrients, especially N, and our previous studies showed that the nutrient content of the 40 g of soil is enough to support the rice plant growth for the early growth period (Asiloglu et al. 2020a, 2020b). A protist-free indigenous bacterial mixture of 4 mL was inoculated to each microcosm. The microcosms (n = 30) were pre-incubated for 1 week under submerged conditions to obtain a stable bacterial community before the addition of protists and/or the biochars. To analyse the effect of the biochar amendments on protist-bacteria interaction, we set up 5 groups of microcosms with 6 replicates: Ctrl group, no biochar addition; RH2 group, 2% (w/w) of rice husk biochar addition; RH4 group, 4% (w/w) of rice husk biochar addition; PL2 group, 2% (w/w) of poultry litter biochar addition; PL4 group, 4% (w/w) of poultry litter biochar addition. Half of the microcosms (n = 3) in each of the 5 groups were inoculated with protists (P+ treatments), while the other half were not inoculated (P- treatments). About 103 protist cells g-1 soil (consisting of a combination of approximately 250 cells of each of the four species) were inoculated into P+ treatments, while the P- treatments received the same amount of sterile water. One rice seedling (26-day old) was transplanted to each microcosm. The microcosms were kept submerged in a growth chamber at 25/30 °C (day/night) with a day length of 16 h (250 μmol m-2 s-1). The microcosms were destructively sampled after 36 days of incubation as previously described (Asiloglu et al. 2020b). Briefly, the surface water of microcosms was removed and the shoots were cut off. Then, the whole root system with the surrounding soil was transferred from the microcosms into a 300-mL sterilized beaker. The non-rhizosphere soil was removed by repeatedly shaking by hand. The rice roots with the surrounding rhizosphere soil carefully selected and placed into a new 300-mL sterilized beaker. Afterwards the rice roots with the surrounding soil were cut (< 5 mm) with sterilized scissors and mixed with the rhizosphere soil. The rice roots were sampled together with the rhizosphere soil in the microcosms (hereafter called rhizosphere). Immediately after mixing, 0.5 g of rhizosphere samples were placed into 2-mL DNA extraction tubes and stored at −80 °C until use for nucleic acid extraction.
Population of protists and physicochemical analyses of biochar and soil
The number of soil protists was estimated using the MPN method (Darbyshire et al. 1974) with slight modifications as previously described (Asiloglu et al. 2020b). The pH of biochars and the soil samples was measured in de-ionised water at a 1:2 (w/w) mass ratio using a pH meter (Mettler Toledo, FP20), and the electrical conductivity (EC) of the biochars was measured using an EC meter (Consort, C3010) as described by Rajkovich et al. (2012). The ash and organic matter contents of the biochar samples were determined by burning at 550 °C for 8 h. The surface area of the biochars was analysed in a surface analyser (Nova, Quantachrome Instruments 77 K, USA) with N2 and was calculated by the Brunauer-Emmett-Teller (BET) equation (Brunauer et al. 1938). The biochar samples were degassed at 100 °C for 18 h prior to N2 physisorption. The total C and N contents (TC and TN, respectively) in the biochar, soil, and plant shoot samples were analysed after drying at 105 °C for 24 h using an MT-700 Mark 2 CN analyser (Yanaco, Kyoto, Japan). The available P was extracted from the soil using 0.002 N H2SO4 and measured by the colourimetric method using a spectrophotometer (Shimadzu, UV-160A, Kyoto, Japan) with the Truog method (Truog 1930). Exchangeable forms of Ca, Mg, K and Na in the biochars and soil samples were extracted according to Pansu and Gautheyrou (2006) with neutral 1 M ammonium acetate and measured using the polarized Zeeman atomic absorption spectrophotometer (Za3300, Hitachi Ltd., Tokyo).
Illumina library preparation and bioinformatics
The DNA extraction, PCR, Illumina Miseq sequencing, primary analysis of raw FASTQ data, taxonomy assignment and the alpha diversity indexes were performed as described previously (Asiloglu et al. 2020b). A negative control was used in all steps from the DNA extraction to the PCR analyses to make sure contamination did not occur. The statistical differences in the community composition among the treatments were estimated as follows: the samples that were rarefied to 30,000 sequences were used to generate the dissimilarity matrices based on the Bray–Curtis distances using the phyloseq package in the R program version 3.6.1. The matrices were then ordinated using the adonis function to calculate the permutational multivariate analysis of variance (PERMANOVA). A non-metric multidimensional scaling (NMDS) analysis was performed based on the Bray–Curtis dissimilarity index at the genus level using the envfit function in the vegan package of the R program version 3.6.1 to evaluate the significant correlations between bacterial community composition and the environmental variables. We used the NMDS to visualize the effect of treatments on overall bacterial community composition in each sample and to visualize significantly important correlations among the bacterial communities and the environmental variables. The raw FASTQ files obtained in this study for the Miseq libraries have been deposited to the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA622382. All other datasets including codes for statistical analysis used during the current study are available from the corresponding author on a reasonable request.
To reveal the effect of protists in the presence and absence of the biochars, we compared the relative abundances of bacteria at multiple taxonomic levels between protist-inoculated and non-inoculated treatments in each group (Ctrl, RH2, RH4, PL2, PL4) using the linear discriminant analysis effect size (LEfSe) (Segata et al. 2011) on Galaxy server (http://huttenhower.sph.harvard.edu/galaxy).
Since protists select their prey mainly at the genus level (Singh 1941, 1942), we further investigate the effect of protists on bacterial genera. The top 100 bacterial taxa at the genus level, which represents over 95% of the total bacterial community, were analysed by comparing P- and P+ treatments for each of the 100 taxa in each of Ctrl, RH2, RH4, PL2, and PL4 treatments. We used 3 categories to show the effect of protists on the bacterial genera: negative effect (significant decrease in the relative abundance of a bacterial genus by the presence of protists), positive effect (significant increase in the relative abundance of a bacterial genus the by the presence of protists) and no effect (no significant difference in the relative abundance of a bacterial genus between protist inoculated and non-inoculated treatments), which were evaluated separately for each treatment. Then the number of genera and their total relative abundances were calculated manually in each category and treatment.
The functional genes putatively involved in specific N-cycling processes were predicted using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software (Langille et al. 2013) version 1.1.3 (normalize_by_copy_number.py). The details of the analysis and the KEGG orthologs for specific N-cycling processes are described elsewhere (Chen et al. 2019). Briefly, metagenome contributions.py script was used to collect the bacterial OTUs with specific N-cycling processes based on Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) database. The gene abundances for specific N-cycling processes were calculated as suggested by the KEGG modules: K00260 + K00261+ K00262 for N mineralization, K10535 for nitrification, (K00370 + K00371 + K00374)/3 + (K02567 + K02568)/2 for NO3- reduction, K00368 for NO2- reduction, (K04561 + K02305)/2 for NO reduction, K00376 for N2O reduction, (K02588 + K02586 + K02591)/3 for N fixation, (K00362+ K00363)/2 + K03383 for dissimilatory nitrate reduction to ammonium (DNRA) and K00366 for NO3 assimilation. The data for each process were normalized to the highest values of individual treatments, which were set to one. The accuracy of PICRUSt predictions for each sample was verified using the weighted NSTI scores, which summarize the extent to which microorganisms in a given sample are related to the sequenced genomes (Langille et al. 2013).
Statistical analysis
The beta diversities were analysed with PERMANOVA with 999 random permutations (p < 0.05), which was obtained based on Bray-Curtis dissimilarities with the adonis function of the vegan package in R version 3.5.2 (https://www.r-project.org/). The homogeneity of multivariate dispersions (PERMDISP) (Anderson 2006) was tested using the ‘betadisper’ function in the vegan package to calculate the significance of the protist effect in each group. Significant biological consistency and effect relevance of the treatment-specific bacterial groups were analysed by LEfSe as follows: Firstly, the non-parametric factorial Kruskal-Wallis sum-rank test (p < 0.05) was conducted to detect features with significant differential abundances. After this step, linear discriminant analysis (LDA), in which the logarithmic score was set to 2.0, was conducted to estimate the significant effect size of each differentially abundant feature.
Differences in the number of protists, soil physicochemical properties and plant growth parameters were analysed by one-way analysis of variance (ANOVA) using R version 3.5.2 (https://www.r-project.org/). Comparison between means was performed with Tukey’s honestly significant difference (HSD) test at 0.05 level, which was carried out using R with ‘glht’ function in the ‘multcomp’ package (Hothorn et al. 2008). To evaluate the effect of protists on a bacterial genus in each group, ANOVA tests were carried out for P- and P+ treatments of each group for each bacterial genus for the top 100 genera.
Results
Growth of protists and the physicochemical properties of the soil
At the end of the experiment, the introduced protists (1 × 103 cells g-1 soil) successfully populated the microcosms (> 6 × 104 MPN g-1 soil) in all of the protist-inoculated microcosms, while the no-protist microcosms did not contain detectable levels of protists (Supplementary Fig. S1). The RH or PL amendments did not significantly affect the total number of protist isolates (p < 0.05).
The physicochemical properties of the RH and PL were reported previously (Asiloglu et al. 2020a). Briefly, the nutrient contents, especially the C, N, P and K contents, of PL were relatively higher than those of RH. The pH and EC were higher in PL, while RH was characterized with a higher C/N ratio and low bulk density. Biochar amendments for 36 days significantly affected the soil physicochemical properties (p < 0.05), while treatment with protists did not affect the tested soil physicochemical properties at the end of the experiment (Table 1). The RH and PL groups at both low (2%) and high (4%) doses significantly affected the pH, total C content and C/N ratio (p < 0.05). The PL treatment significantly increased the total N, K, Mg and Ca contents (p < 0.05). Only the PL4 treatment significantly increased available P and Mg (p < 0.05).
Bacterial community composition
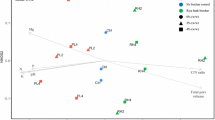
Treatment with the protists (PERMANOVA, R2 = 0.173, p < 0.001), the PL (PERMANOVA, R2 = 0.178, p < 0.001), and the RH (PERMANOVA, R2 = 0.059, p < 0.01) significantly affected the bacterial community composition (Table 2). There was no significant effect of the applied biochar doses (PERMANOVA, R2 = 0.036, p > 0.1) on the bacterial community composition (Table 2). The NMDS analysis revealed that the bacterial communities were affected by the presence of the protists, as well as by the PL and RH amendments (Fig. 1). The shifts in the bacterial community composition due to the presence of protists did not correlate with the soil chemical properties (Fig 1). The effect of PL was associated with the changes in pH (p < 0.001), total N (p < 0.001), available P (p < 0.001), Mg (p < 0.001), K (p < 0.001), Ca (p < 0.001) and Na (p < 0.005), while that of RH was only associated with increased C/N ratio (p < 0.001).
Comparison of the bacterial community composition between the treatments with non-metric multidimensional scaling (NMDS) plot based on Bray–Curtis dissimilarity index at the genus level. The arrows show significant (p < 0.05) correlation between bacterial community composition and environmental properties. Green colour, protist treatments; red colour, no-protist treatments; circle, controls with no biochar addition; triangle, rice husk biochar; square, poultry litter biochar. Ctrl, control with no biochar addition; RH2 and RH4, 2% and % (w/w) rice husk biochar; PL2 and PL4, 2% and 4 (w/w) poultry litter biochar
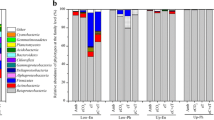
The effect of protists on the bacterial community composition in the absence of biochar (Ctrl group) as expressed in PERMDISP was F = 87.3 (p = 8.5 x 10-10), and this was decreased to F = 15.1 (p = 6.2 x 10-5) and F = 30.9 (p = 1.7 x 10-7) in the RH and PL treatments, respectively. The decreased effect of protists on the bacterial community was confirmed by the LEfSe analysis, which revealed the protist-affected bioindicator groups of the bacterial communities at multiple taxonomic levels in the absence and presence of biochar amendments (Fig. 2 and Supplementary Fig. S4). In the control group, 90 bacterial groups including members of Firmicutes, Cyanobacteria, Chloroflexi, Actinobacteria, Acidobacteria and several bacteria from Alphaproteobacteria were detected by the LEfSe as bioindicator taxa that were affected by the protists in the absence of the biochars (Fig. 2a and Supplementary Fig. S4A). The LEfSe revealed that the addition of PL and RH at 2 and 4% doses decreased the number of protist-affected bioindicator bacteria at multiple taxonomic levels (Figs. 2b, c, d, e, and Supplementary Figs. S4B, C, D, E). The number of bioindicator bacterial taxa was decreased from 90 to 46, 29, 43 and 21 in the RH2, RH4, PL2 and PL4 groups, respectively. Members of Armatimonadetes and Alphaproteobacteria in the RH group and members of Clostridia, Alphaproteobacteria and Bacteroidia in the PL group were detected as protist-affected bioindicators.
The bacterial taxa affected by protists in the presence and absence of the biochars. A linear discriminant analysis effect size (LEfSe) analysis identifies the significantly different (p < 0.05, Kruskal-Wallis test, LDA score > 2.0) bacteria at multiple taxonomic levels. Effect of protists on bacteria was determined by comparing the community composition of protist treatments (P+) with no-protist treatments (P-) in control group a, RH2 group b, RH4 group c, PL2 group d and PL4 group e. The taxa with significantly different abundances between treatments are represented by coloured dots, and from the centre outward, they represent the kingdom, phylum, class, order, family and genus levels. The coloured shadows represent trends of the significantly differed taxa in the absence (red colour) or presence (green colour) of protists. The LDA scores are available in Supplementary Fig. S4
The significant effects of protists on bacteria at the genus level (p < 0.05) are shown in Supplementary Fig. S5 for the top 100 genera, which covers over 95% of the total community. From the top 100 bacterial genera, 74 of them were either negatively (36) or positively (38) affected by the protists in the no-biochar control group (Supplementary Fig. S5A). This represents 72.8% (the relative abundance) of the total bacterial community that was negatively (34.5%) or positively (38.3%) affected by protists (Supplementary Fig. S5B). The enormous effects of the protists on bacteria were decreased in the presence of both biochars at both doses (Supplementary Fig. S5). Averagely in all bacterial groups of the biochar amended treatments, a total of 56 bacterial genera (49.4% by relative abundance) were affected by protists, which represents 24.6% (number of genera) and 32.1% (relative abundance) decrease in protist-affected bacterial taxa by the biochar amendments. In the presence of biochar, bacterial genera that were not affected by protists were increased by 69.2% in the total number of genera and by 104.5% in the relative abundance (Supplementary Fig. S5).
Bacterial genes putatively involved in N-cycling
We used the PICRUSt analysis to predict the abundance of genes involved in specific N-cycling processes. The reliability of PICRUSt predictions for each sample was checked using the nearest sequenced taxon index (NSTI) scores. The reliability of predictions increases with a lower NSTI score, and NSTI score < 0.170 is usually considered to have reliable predictions for soil samples (Langille et al. 2013). The NSTI scores of our samples were 0.102 ± 0.008 and ranged from 0.092 to 0.119 (Supplementary Table S3), which indicated the accuracy of predictions. Treatment with protists in the absence of biochar significantly (p < 0.05) increased the abundance of bacterial genes involved in N mineralization, NO3- reduction, DNRA and NO3- assimilation (Fig. 3). The effect of RH was different from that of PL. Both RH and PL treatments exhibited significantly increased abundance of bacterial genes involved in nitrification. Only PL treatment resulted in a positive effect on the abundance of bacterial genes involved in mineralization, DNRA and NO3- assimilation. The effects of protists on the bacterial genes were not significant for DNRA, NO3- assimilation and NO3- reductions in the presence of biochar.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) predicted bacterial 16S rRNA gene abundances involved in specific N-cycling processes in the rhizosphere. All data were normalized to the highest values of individual processes, which were set to one. Error bars represent standard deviation. Different letters indicate significant differences (p < 0.05). Blue bars, control with no biochar addition (Ctrl); green bars, rice husk biochar (RH); orange bars, poultry litter biochar (PL); horizontal striped bars, absence of protists (P-), filled bars, presence of protists (P+)
Plant growth and N uptake
The rice plants exhibited healthy growth in all microcosms. Comparison of P- and P+ treatments showed that the presence of protists significantly (p < 0.05) enhanced the shoot biomass in the absence or presence of the biochars except for the PL2. The amount of the total N in the plants significantly increased in the presence of protists in all groups (Fig. 4). The shoot biomass and the total N in the PL treatment group were higher than those in the control treatment group, while the RH had no significant effect. There was no significant difference between the applied doses of biochars on the shoot biomass. However, the total N in the rice plants was affected by the applied dose of PL both in the presence and absence of the protists.
Plant growth parameters on day 36. a Shoot biomass. b total N uptake. Ctrl, control with no biochar addition; RH2 and RH4, 2 and 4% (w/w) rice husk biochar; PL2 and PL4, 2 and 4% (w/w) poultry litter biochar; P˗, no-protist treatments; P+, protist treatments. Error bars represent standard deviation. Different letters indicate significant differences (p < 0.05)
Discussion
The microbial prey-predator interactions are among the main factors shaping the microbiome composition (Bouvy et al. 2011; Chow et al. 2014). Here, we showed that the biochar amendment affected the trophic interaction between protists and bacteria, as well as its outcomes for the prey’s community composition, while the effect of protists on the rice plant growth and N uptake was consistent. To the best of our knowledge, this is the first reference to show the effects of the biochar on the trophic interactions. The effects of protists on overall bacterial community composition were decreased by biochar amendment, which corroborates our hypothesis. Both of the biochars with different physicochemical properties significantly affected the trophic interactions, and we did not observe a correlation between the effect of biochar on trophic interaction and its physicochemical properties such as the size of the micro-pores and nutrient content. This suggests that the effect of biochar on prey-predator interactions is likely to be more complex than previously assumed.
Prey-predator interaction in the biochar amended soil
Compared with the no-biochar control treatment, the effect of protists on the bacteria was significantly reduced in the biochar-amended treatments, suggesting that biochar is likely to protect bacteria from protist predation. Small pores or channels in soil particles may create a protected habitat for bacteria that have relatively small body sizes than their predators (Ekelund and Rønn 1994; England et al. 1993; Wright et al. 1995). This supports the long-existing hypothesis that micro-pores of biochar physically protect bacteria from predation (Lehmann et al. 2011; Quilliam et al. 2013). However, contrary to the hypothesis, protists can pass through channels smaller than their body size (Wang et al. 2005) and micro-pores as small as 2 μm are accessible to many small soil flagellates and amoeba (Edwards 2003). Although bacteria inhabiting the micro-pores of biochar may be protected from predation by relatively big protists, ciliates, for instance, the small soil protists may still be able to feed on them. This is supported with previous findings, where biochar amendment caused a decrease in the number of big-sized bacterivorous predators, nematodes and big soil amoeba (Kamau et al. 2019; Liu et al. 2020) and an increase in the number of small-sized predators, flagellates (Liu et al. 2020). This balance between big-sized and small-sized predators may explain why the total population of protists did not differ between the presence and absence of the biochars in our study. We suggest that the effect of biochar on bacterivorous predators may vary depending on the body size of the predators and accessibility of the biochar micro-pores to the predators is most likely a critical factor controlling the microbial food web.
The bacterial predation by protists reduces the dominance of strong competitors (Flues et al. 2017); hence, formerly subordinate bacterial species gain a competitive advantage (Jousset et al. 2008). Biochar amendments decrease the positively affected bacterial taxa by protists. The total relative abundance of Clostridia and Bacteroidia, which were among the top 3 most abundant classes in the no-protist control treatment (Ctrl[P-]), decreased from 43.4 to 18.4% upon treatment with protists (Ctrl[P+]). This decrease was reflected as an increase in less abundant bacterial classes. For instance, the relative abundances of Saprospirae were increased from 1.5 to 10.5%, and that of Actinobacteria was increased from 1.6 to 7.7%. However, such patterns were not observed in the biochar-amended treatments. Therefore, it is likely that the less abundant bacterial taxa may not be able to benefit from the advantage of protist predation in the biochar amended soils through the decreased effect of protist predation on their competitive bacteria by biochar amendments.
The biochar-induced changes in the physicochemical properties of soil could influence the behaviour of protists. For instance, the pH was significantly increased by both biochar applications, which should also change the status of chemical compounds like organic acids. The soil pH affects the abundance and feeding behaviour of protists (Ekelund and Rønn 1994; Hansen et al. 2019). Homma and Cook (1985) showed that the feeding efficiency of a vampyrellid amoeba, which was maximum at neutral pH, was decreased under increased or decreased pH conditions. The protist isolates used in this study are native to the experimental soil. Elevated pH conditions by the biochars may affect their comfort zone and, therefore may decrease their predation efficiency. Additionally, the biochar-induced changes in the physicochemical properties of soil affected the bacterial community composition. The shift in the bacterial food sources of the protists may also alter their effect on the overall community composition.
Bacterial genes involved in the N-cycling and the plant growth
Positive effects of protist predation (Asiloglu et al. 2020b; Herdler et al. 2008; Kreuzer et al. 2006) and the biochar amendments (Bhattacharjya et al. 2016) on the rice plant growth are well documented. In line with the previous work, in this study, rice plant growth and N uptake were significantly higher in P+ treatments than the P- treatments. The positive effects of protists on plant growth is generally explained by increased nutrient turnover; for instance, plants can utilize considerable amounts of NH4+ released from consumed bacterial biomasses as a result of protist predation (Bonkowski et al. 2000; Griffiths 1994). However, not all N comes from the preyed bacterial biomass. Protists influence bacterial activities of N mineralization to NH4+ in soil (Zahn et al. 2016), including paddy soil (Murase et al. 2006). The PICRUSt analysis predicted the high abundance of bacterial genes involved in N mineralization, DNRA, and NO3- assimilation in the presence of protists. This suggested that the bacterial communities regulated by the protists may promote plant growth through increased bacterial mineralization of organic matter to NH4+ and through prevention of NH4+ loss by DNRA and NO3- assimilation.
Depending on the chemical properties, biochar alters the microbially mediated transformation of nutrients, especially N (Lehmann et al. 2011). Previously, Anderson et al. (2014) showed that the increased bacterial activities on nitrification, denitrification and NO3- assimilation were linked to the high N input by the biochar amendment. Similar results also obtained by Wu et al. (2016), who showed that a biochar with low-N content had no effect on the bacterial marker genes involved in N-cycling, while pre-composted biochar with rice straw enhanced the gene copy numbers due to higher amount of N input. This is in line with our results, where the high-N content biochar (PL) treatment increased the bacterial genes involved in mineralisation, nitrification, DNRA and NO3- assimilation, while the low-N content biochar (RH) had no effect on them. Besides the obvious chemical impact of biochar on plant growth, we suggest that the effects of biochar on the plant growth could also be linked to changes in bacterial community composition and functionality, which was also previously reported (Kolton et al. 2017).
Conclusion
Here we studied the prey-predator interaction between indigenous soil bacteria and the protists in a submerged rice rhizosphere as affected by the biochar amendments. Our results showed that the response of the bacterial community composition was distinct to the presence of protists and the biochar amendments. The effects of protists on the overall community composition of bacteria were decreased by the biochar amendments, which corroborates our hypotheses. The rice plant growth and N uptake were always higher in the presence of protists and PL treatments, which is likely to be linked with the enhanced expression of bacterial genes involved in the N mineralisation, dissimilatory nitrate reduction to ammonium (DNRA) and NO3- assimilation by the protists and the PL treatments. Overall, our results suggest that the interfered trophic interaction by the biochar amendments is most likely to affect bacterial community dynamics. Considering the importance of prey-predator interaction for the bacterial community, functioning and plant growth, our results might help future efforts to better understand the effects of biochar amendments on the soil bacterial community. Future studies should focus on revealing the mechanism behind the decreased prey-predator interaction in the biochar amended soils.
References
Adl SM (2007) Motility and migration rate of protozoa in soil columns. Soil Biol Biochem 39:700–703. https://doi.org/10.1016/j.soilbio.2006.09.008
Anderson CR, Hamonts K, Clough TJ, Condron LM (2014) Biochar does not affect soil N-transformations or microbial community structure under ruminant urine patches but does alter relative proportions of nitrogen cycling bacteria. Agric Ecosyst Environ 191:63–72. https://doi.org/10.1016/j.agee.2014.02.021
Anderson MJ (2006) Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253. https://doi.org/10.1111/j.1541-0420.2005.00440.x
Asiloglu R, Samuel SO, Sevilir B, Akca MO, Acar Bozkurt P, Suzuki K, Murase J, Turgay OC, Harada N (2020a) Biochar affects taxonomic and functional community composition of protists. Biol Fertil Soils:1–15. https://doi.org/10.1007/s00374-020-01502-8
Asiloglu R, Shiroishi K, Suzuki K, Turgay OC, Murase J, Harada N (2020b) Protist-enhanced survival of a plant growth promoting rhizobacteria, Azospirillum sp. B510, and the growth of rice (Oryza sativa L.) plants. Appl Soil Ecol 154:103599. https://doi.org/10.1016/j.apsoil.2020.103599
Bell T, Bonsall MB, Buckling A, Whiteley AS, Goodall T, Griffiths RI (2010) Protists have divergent effects on bacterial diversity along a productivity gradient. Biol Lett 6:639–642. https://doi.org/10.1098/rsbl.2010.0027
Bhattacharjya S, Chandra R, Pareek N, Raverkar KP (2016) Biochar and crop residue application to soil: effect on soil biochemical properties, nutrient availability and yield of rice ( Oryza sativa L.) and wheat ( Triticum aestivum L.). Arch Agron Soil Sci 62:1095–1108. https://doi.org/10.1080/03650340.2015.1118760
Bonkowski M (2004) Protozoa and plant growth: the microbial loop in soil revisited. New Phytol 162:617–631. https://doi.org/10.1111/j.1469-8137.2004.01066.x
Bonkowski M, Brandt F (2002) Do soil protozoa enhance plant growth by hormonal effects? Soil Biol Biochem 34:1709–1715. https://doi.org/10.1016/S0038-0717(02)00157-8
Bonkowski M, Griffiths B, Scrimgeour C (2000) Substrate heterogeneity and microfauna in soil organic ‘hotspots’ as determinants of nitrogen capture and growth of ryegrass. Appl Soil Ecol 14:37–53. https://doi.org/10.1016/S0929-1393(99)00047-5
Bouvy M, Bettarel Y, Bouvier C, Domaizon I, Jacquet S, Le Floc’h E, Montanié H, Mostajir B, Sime-Ngando T, Torréton JP, Vidussi F, Bouvier T (2011) Trophic interactions between viruses, bacteria and nanoflagellates under various nutrient conditions and simulated climate change. Environ Microbiol 13:1842–1857. https://doi.org/10.1111/j.1462-2920.2011.02498.x
Brunauer S, Emmett PH, Teller E (1938) Adsorption of gases in multimolecular layers. J Am Chem Soc 60:309–319. https://doi.org/10.1021/ja01269a023
Chen H, Xia Q, Yang T, Bowman D, Shi W (2019) The soil microbial community of turf: linear and nonlinear changes of taxa and N-cycling gene abundances over a century-long turf development. FEMS Microbiol Ecol 95:1–14. https://doi.org/10.1093/femsec/fiy224
Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA (2014) Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8:816–829. https://doi.org/10.1038/ismej.2013.199
Clarholm M (1985) Interactions of bacteria, protozoa and plants leading to mineralization of soil nitrogen. Soil Biol Biochem 17:181–187. https://doi.org/10.1016/0038-0717(85)90113-0
Crotty FV, Adl SM, Blackshaw RP, Murray PJ (2012) Using stable isotopes to differentiate trophic feeding channels within soil food webs. J Eukaryot Microbiol 59:520–526. https://doi.org/10.1111/j.1550-7408.2011.00608.x
D’Hose T, Molendijk L, Van Vooren L, van den Berg W, Hoek H, Runia W, van Evert F, ten Berge H, Spiegel H, Sandèn T, Grignani C, Ruysschaert G (2018) Responses of soil biota to non-inversion tillage and organic amendments: An analysis on European multiyear field experiments. Pedobiologia (Jena) 66:18–28. https://doi.org/10.1016/j.pedobi.2017.12.003
Dai Z, Enders A, Rodrigues JLM, Hanley KL, Brookes PC, Xu J, Lehmann J (2018) Soil fungal taxonomic and functional community composition as affected by biochar properties. Soil Biol Biochem 126:159–167. https://doi.org/10.1016/j.soilbio.2018.09.001
Darbyshire JF, Wheatley RE, Greaves MP, Inkson RHE (1974) A rapid micromethod for estimating bacterial and protozoan populations in soil. Rev Écol Biol Sol 11:465–475
Edwards CA (2003) Encyclopedia of Soil Science. Appl Soil Ecol 23:279. https://doi.org/10.1016/S0929-1393(03)00051-9
Ekelund F, Rønn R (1994) Notes on protozoa in agricultural soil with emphasis on heterotrophic flagellates and naked amoebae and their ecology. FEMS Microbiol Rev 15:321–353. https://doi.org/10.1111/j.1574-6976.1994.tb00144.x
England LS, Lee H, Trevors JT (1993) Bacterial survival in soil: Effect of clays and protozoa. Soil Biol Biochem 25:525–531. https://doi.org/10.1016/0038-0717(93)90189-I
Flues S, Bass D, Bonkowski M (2017) Grazing of leaf-associated Cercomonads (Protists: Rhizaria: Cercozoa) structures bacterial community composition and function. Environ Microbiol 19:3297–3309. https://doi.org/10.1111/1462-2920.13824
Fox JW (2007) The dynamics of top-down and bottom-up effects in food webs of varying prey diversity, composition, and productivity. Oikos 116:189–200. https://doi.org/10.1111/j.0030-1299.2007.15280.x
Geisseler D, Scow KM (2014) Long-term effects of mineral fertilizers on soil microorganisms – A review. Soil Biol Biochem 75:54–63. https://doi.org/10.1016/j.soilbio.2014.03.023
Griffiths BS (1994) Microbial-feeding nematodes and protozoa in soil: Their effectson microbial activity and nitrogen mineralization in decomposition hotspots and the rhizosphere. Plant Soil 164:25–33. https://doi.org/10.1007/BF00010107
Gul S, Whalen JK, Thomas BW, Sachdeva V, Deng H (2015) Physico-chemical properties and microbial responses in biochar-amended soils: Mechanisms and future directions. Agric Ecosyst Environ 206:46–59. https://doi.org/10.1016/j.agee.2015.03.015
Hale L, Luth M, Crowley D (2015) Biochar characteristics relate to its utility as an alternative soil inoculum carrier to peat and vermiculite. Soil Biol Biochem 81:228–235. https://doi.org/10.1016/j.soilbio.2014.11.023
Hansen BW, Andersen CMB, Hansen PJ, Nielsen TG, Vismann B, Tiselius P (2019) In situ and experimental evidence for effects of elevated pH on protistan and metazoan grazers. J Plankton Res 41:257–271. https://doi.org/10.1093/plankt/fbz020
Herdler S, Kreuzer K, Scheu S, Bonkowski M (2008) Interactions between arbuscular mycorrhizal fungi (Glomus intraradices, Glomeromycota) and amoebae (Acanthamoeba castellanii, Protozoa) in the rhizosphere of rice (Oryza sativa). Soil Biol Biochem 40:660–668. https://doi.org/10.1016/j.soilbio.2007.09.026
Hervey RJ, Greaves JE (1941) Nitrogen fixation by Azotobacter chroococcum in the presence of soil protozoa. Soil Sci 51:85–100
Homma Y, Cook RJ (1985) Influence of matric and osmotic water potentials and soil pH on the activity of giant vampyrellid amoebae. Phytopathology 75:243. https://doi.org/10.1094/Phyto-75-243
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50:346–363. https://doi.org/10.1002/bimj.200810425
Hünninghaus M, Koller R, Kramer S, Marhan S, Kandeler E, Bonkowski M (2017) Changes in bacterial community composition and soil respiration indicate rapid successions of protist grazers during mineralization of maize crop residues. Pedobiologia (Jena) 62:1–8. https://doi.org/10.1016/j.pedobi.2017.03.002
Jousset A, Scheu S, Bonkowski M (2008) Secondary metabolite production facilitates establishment of rhizobacteria by reducing both protozoan predation and the competitive effects of indigenous bacteria. Funct Ecol 22:714–719. https://doi.org/10.1111/j.1365-2435.2008.01411.x
Kamau S, Karanja NK, Ayuke FO, Lehmann J (2019) Short-term influence of biochar and fertilizer-biochar blends on soil nutrients, fauna and maize growth. Biol Fertil Soils 55:661–673. https://doi.org/10.1007/s00374-019-01381-8
Kolton M, Graber ER, Tsehansky L, Elad Y, Cytryn E (2017) Biochar-stimulated plant performance is strongly linked to microbial diversity and metabolic potential in the rhizosphere. New Phytol 213:1393–1404. https://doi.org/10.1111/nph.14253
Kreuzer K, Adamczyk J, Iijima M, Wagner M, Scheu S, Bonkowski M (2006) Grazing of a common species of soil protozoa (Acanthamoeba castellanii) affects rhizosphere bacterial community composition and root architecture of rice (Oryza sativa L.). Soil Biol Biochem 38:1665–1672. https://doi.org/10.1016/j.soilbio.2005.11.027
Kurm V, van der Putten WH, Weidner S, Geisen S, Snoek BL, Bakx T, Hol WHG (2019) Competition and predation as possible causes of bacterial rarity. Environ Microbiol 21:1356–1368. https://doi.org/10.1111/1462-2920.14569
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Lehmann J, Rillig MC, Thies J, Masiello CA, Hockaday WC, Crowley D (2011) Biochar effects on soil biota – A review. Soil Biol Biochem 43:1812–1836. https://doi.org/10.1016/j.soilbio.2011.04.022
Liu T, Yang L, Hu Z, Xue J, Lu Y, Chen X, Griffiths BS, Whalen JK, Liu M (2020) Biochar exerts negative effects on soil fauna across multiple trophic levels in a cultivated acidic soil. Biol Fertil Soils 56:597–606. https://doi.org/10.1007/s00374-020-01436-1
Matz C, Kjelleberg S (2005) Off the hook – how bacteria survive protozoan grazing. Trends Microbiol 13:302–307. https://doi.org/10.1016/j.tim.2005.05.009
Murase J, Frenzel P (2008) Selective grazing of methanotrophs by protozoa in a rice field soil. FEMS Microbiol Ecol 65:408–414. https://doi.org/10.1111/j.1574-6941.2008.00511.x
Murase J, Noll M, Frenzel P (2006) Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl Environ Microbiol 72:5436–5444. https://doi.org/10.1128/AEM.00207-06
Pansu M, Gautheyrou J (2006) Exchangeable Cations. In: Handbook of soil analysis. Springer, Berlin, pp 667–676
Quilliam RS, Glanville HC, Wade SC, Jones DL (2013) Life in the ‘charosphere’ – Does biochar in agricultural soil provide a significant habitat for microorganisms? Soil Biol Biochem 65:287–293. https://doi.org/10.1016/j.soilbio.2013.06.004
Rajkovich S, Enders A, Hanley K, Hyland C, Zimmerman AR, Lehmann J (2012) Corn growth and nitrogen nutrition after additions of biochars with varying properties to a temperate soil. Biol Fertil Soils 48:271–284. https://doi.org/10.1007/s00374-011-0624-7
Rondon MA, Lehmann J, Ramírez J, Hurtado M (2007) Biological nitrogen fixation by common beans (Phaseolus vulgaris L.) increases with bio-char additions. Biol Fertil Soils 43:699–708. https://doi.org/10.1007/s00374-006-0152-z
Rønn R, McCaig AE, Griffiths BS, Prosser JI (2002) Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl Environ Microbiol 68:6094–6105. https://doi.org/10.1128/AEM.68.12.6094-6105.2002
Rutherford PM, Juma NG (1992) Influence of texture on habitable pore space and bacterial-protozoan populations in soil. Biol Fertil Soils 12:221–227. https://doi.org/10.1007/BF00336036
Salcher M, Pernthaler J, Psenner R, Posch T (2005) Succession of bacterial grazing defense mechanisms against protistan predators in an experimental microbial community. Aquat Microb Ecol 38:215–229. https://doi.org/10.3354/ame038215
Saleem M, Fetzer I, Dormann CF, Harms H, Chatzinotas A (2012) Predator richness increases the effect of prey diversity on prey yield. Nat Commun 3:1305. https://doi.org/10.1038/ncomms2287
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. https://doi.org/10.1186/gb-2011-12-6-r60
Singh BN (1941) Selectivity in bacterial food by soil amoebae in pure mixed culture and in sterilized soil. Ann Appl Biol 28:52–64. https://doi.org/10.1111/j.1744-7348.1941.tb07536.x
Singh BN (1942) Selection of bacterial food by soil flagellates and amoebae. Ann Appl Biol 29:18–22. https://doi.org/10.1111/j.1744-7348.1942.tb06917.x
Trap J, Bonkowski M, Plassard C, Villenave C, Blanchart E (2016) Ecological importance of soil bacterivores for ecosystem functions. Plant Soil 398:1–24. https://doi.org/10.1007/s11104-015-2671-6
Truog E (1930) The determination of the readily available phosphorus of soils. Agron J 22:874–882. https://doi.org/10.2134/agronj1930.00021962002200100008x
Wang W, Shor LM, LeBoeuf EJ, Wikswo JP, Kosson DS (2005) Mobility of protozoa through narrow channels. Appl Environ Microbiol 71:4628–4637. https://doi.org/10.1128/AEM.71.8.4628-4637.2005
Woolf D, Amonette JE, Street-Perrott FA, Lehmann J, Joseph S (2010) Sustainable biochar to mitigate global climate change. Nat Commun 1:1–9. https://doi.org/10.1038/ncomms1053
Wright DA, Killham K, Glover LA, Prosser JI (1995) Role of pore size location in determining bacterial activity during predation by protozoa in soil. Appl Environ Microbiol 61:3537–3543. https://doi.org/10.1128/AEM.61.10.3537-3543.1995
Wu H, Zeng G, Liang J, Chen J, Xu J, Dai J, Li X, Chen M, Xu P, Zhou Y, Li F, Hu L, Wan J (2016) Responses of bacterial community and functional marker genes of nitrogen cycling to biochar, compost and combined amendments in soil. Appl Microbiol Biotechnol 100:8583–8591. https://doi.org/10.1007/s00253-016-7614-5
Zahn G, Wagai R, Yonemura S (2016) The effects of amoebal bacterivory on carbon and nitrogen dynamics depend on temperature and soil structure interactions. Soil Biol Biochem 94:133–137. https://doi.org/10.1016/j.soilbio.2015.11.021
Acknowledgements
This research was supported by The Yanmar Environmental Sustainability Support Association (K30059) and by a Kakenhi grant (19H00305H) from the Japan Society for the Promotion of Science to R.A.
Author information
Authors and Affiliations
Contributions
RA conceived and designed the study, analysed the sequence data, performed bioinformatic and statistical analyses and prepared the manuscript. RA and JM interpreted the data. RA, BS, SOS, MA, MOA and OCT performed the laboratory works. MA, KS, OCT, JM and NH provided feedback and comments. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 994 kb)
Rights and permissions
About this article
Cite this article
Asiloglu, R., Sevilir, B., Samuel, S.O. et al. Effect of protists on rhizobacterial community composition and rice plant growth in a biochar amended soil. Biol Fertil Soils 57, 293–304 (2021). https://doi.org/10.1007/s00374-020-01525-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-020-01525-1