Abstract
Extracts of the brown alga, Ascophyllum nodosum, are widely used as plant biostimulants to improve growth and to impart tolerance against abiotic stresses. However, the molecular mechanisms by which A. nodosum extract (ANE) mediates stress tolerance are still largely unknown. The aim of this study was to study selected anti-stress mechanisms at the transcriptome level. We show that methanolic sub-fractions of ANE improved growth of Arabidopsis thaliana under NaCl stress; biomass increased by approximately 50% under 100 mM and 150 mM NaCl, relative to the control. Bioassay-guided fractionation revealed that the ethyl acetate sub-fraction of ANE (EAA) had the majority of stress alleviating, bioactive components. Microarray analysis showed that EAA elicited substantial changes in the global transcriptome on day 1 and day 5, after treatment. On day one, 184 genes were up-regulated while this number increased to 257 genes on day 5. On the other hand, 91 and 262 genes were down-regulated on day 1 and day 5, respectively. On day 1, 2.2% of the genes altered were abiotic stress regulated and this increased to 6% on day 5. EAA modulate the expression of number of the genes involved in stress responses, carbohydrate metabolism, and phenylpropanoid metabolism. Thus, our results suggested that bioactive components in the ethyl acetate fraction of A. nodosum induced salinity tolerance in A. thaliana by modulating the expression of a plethora of stress-responsive genes, providing a better understanding of the mechanisms through which ANE mediates tolerance by plants to salinity stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Global effects such as desertification, soil salinization, atmospheric CO2 enrichment, and nutrient imbalances (including mineral toxicity and deficiency), negatively affect productivity of arable lands (dos Reis et al. 2012). Among environmental stresses, salinity is a major abiotic factor that reduces agricultural productivity (Boyer 1982). Approximately 7% of the world’s land (Szabolcs 1994) and 20% of irrigated agricultural land are affected by soil salinity (Chinnusamy et al. 2005; Al-Maskri et al. 2010). High salinity adversely affects plant growth and development by disrupting intracellular ion homeostasis that results in membrane dysfunction, attenuation of metabolic activity, and secondary effects that inhibit growth and ultimately cell death (Yadav et al. 2012). Salinity stress tolerance is a complex process that involves response at physiological and molecular levels. Salt tolerance mechanism(s) include a complex network of genes involved in signaling pathways, osmolyte production (Delauney and Verma 1993), detoxification of reactive oxygen species (ROS) (Mittler et al. 2004), vacuolar sequestration (Apse et al. 1999) and protection of macromolecules (LEA protein, chaperones). The regulatory proteins and transcription factors such as DREB/CBF, bZIP, MYC, and MYB, control the expression of many downstream genes that play a role in stress tolerance (Agarwal et al. 2013; Shinozaki and Yamaguchi-Shinozaki 2000). The products of these genes converge and interact in different pathways related to abiotic stress conferring stress tolerance (Agarwal et al. 2013).
Plants maintain an ionic balance for cellular metabolism, optimum growth, and productivity. Plants mitigate elevated Na+ concentration by a number of strategies that include (1) activation of Na+ efflux mediated by a plasma membrane Na+/H+ antiporter encoded by SOS1 (salt overly sensitive 1), (2) Na+ sequestration in vacuole mediated by NHX1 (present on the tonoplast), and (3) Na+ influx prevention (Zhu 2003). Due to the complex metabolic pathways involved in salinity tolerance, there has been limited success in generating salinity tolerant crop varieties through genetic engineering. Additionally, transgenic approaches have limitations of meeting regulatory demands and consumer approval. Another approach to improve salinity tolerance in plants is the use of plant biostimulants (Jithesh et al. 2012; Khan et al. 2009). Extracts of brown seaweeds possess plant growth-promoting activity and are widely used in agricultural and horticultural crop production (Battacharyya et al. 2015). These treatments increase plant carbon and nitrogen content, leaf chlorophyll (Blunden et al. 1997), increase water retention capacity, and ameliorate stress (Zhang and Schmidt 1999).
One of the most commonly used biostimulants in agriculture is obtained from the brown seaweed Ascophyllum nodosum (rockweed), widely distributed in the North Atlantic Ocean. Application of A. nodosum extract (ANE) increases plant productivity (Craigie 2011; Khan et al. 2009). ANE promotes root and shoot growth of Arabidopsis by the regulation of phytohormone metabolism (Rayorath et al. 2008; Wally et al. 2013). ANE and its lipophilic fraction enhanced freezing tolerance in Arabidopsis by modulating the expression of freezing responsive stress inducible genes involved in the accumulation of osmo-protectants and of various types of fatty acids and sugar alcohols (Nair et al. 2012; Rayirath et al. 2009). While ANE is known to improve salinity stress tolerance in plants (Jithesh et al. 2012; Van Oosten et al. 2017), the molecular mechanism of ANE-mediated salinity stress tolerance is largely unknown. Further, the nature of chemical components present within ANE which mediate salinity stress tolerance is still elusive. Therefore, the aim of this study was to test the effect of fractions of ANE and to elucidate the mechanism of activity of the most active fraction by studying the differences in the whole-genome transcriptome.
Materials and Methods
Arabidopsis Growth Conditions
Seeds of Arabidopsis thaliana (Col-0) were purchased from Lehle Seeds (Round Rock, TX, USA). Arabidopsis was grown on pre-soaked Jiffy-7 pellets (Jiffy Products, Shippagan, NB, Canada) under greenhouse conditions (temperature 22 ± 2 °C, photoperiod of 16/8 h (day/night) with 100 µmol photons/m/s incident light conditions, and 65% humidity).
Preparation of A. nodosum Extracts and Organic Sub-fractions
Soluble seaweed extract powder (ANE), produced from A. nodosum (Acadian Seaplants Limited, Dartmouth, NS, Canada) was used in the experiment. Methanolic and various organic sub-fractions of ANE were prepared according to Rayirath et al. (2009). 10 g of ANE was dissolved in 40 mL methanol, the supernatant was separated and evaporated to dryness under a stream of nitrogen and resuspended in 10 mL methanol. The resultant solution was filter sterilized using a 0.22-µm syringe filter and stored in sterile centrifuge tubes at 4 °C until further use. For sub-fractionation, the dry solids, after evaporation of the methanol, were resuspended in 50 mL sterile distilled water to give an aqueous suspension. This was sub-fractionated by sequential extraction with three volumes of hexane, three volumes of chloroform, and subsequently with three volumes of ethyl acetate. The sub-fractions were dried under a stream of nitrogen and resuspended in a minimal quantity of methanol (100 µL/g equivalent) and the volume made up with sterile distilled water to adjust the final concentration of methanol in the sub-fractions to 0.1%.
Treatment of Arabidopsis with A. nodosum Extracts and Salt Stress
Solutions of the organic fraction of ANE were prepared in sterile distilled water to a final concentration of 1 g/L equivalent. The salinity stress alleviating effect of the methanolic fraction of ANE was studied on Arabidopsis plants treated with 100 and 150 mM NaCl, while the effects of other organic fractions were tested on the Arabidopsis plants challenged with 150 mM NaCl. 3-week-old Arabidopsis plants were initially exposed to NaCl once for 24 h by placing pellets in a tray containing NaCl solution, 40 mL per pellet. After 24 h, plants were irrigated individually with 20 mL of 1 g/L equivalent of either the methanol extract of A. nodosum (MEA) or the ethyl acetate sub-fraction of A. nodosum (EAA). Control plants were grown under identical conditions and also exposed to 150 mM NaCl for 24 h but were treated with 0.1% aqueous methanol instead of organic fractions of ANE. Additionally, the plants were irrigated on alternate days with distilled water to maintain uniform moisture for optimum growth. Observations on plant height, number of leaves, leaf area, and fresh weight were taken after 4 weeks. The experiment was conducted in a completely randomized block design with 15 plants per treatment (n = 15). These morphological parameters were analyzed using Fisher’s LSD with p ≤ 0.05 using CoStat statistical software (CoHort Software, Monterey, CA).
Arabidopsis Seedling Growth and Root-Bending Assay
Seeds of Arabidopsis were surface-sterilized with 2% sodium hypochlorite solution for 1 min and subsequently rinsed three times in sterile distilled water. The sterilized seeds were germinated in Petri dishes (9 cm diameter) containing half-strength Murashige and Skoog basal medium (Sigma, St. Louis, MO) supplemented with 1% sucrose and 0.8% agar (pH 5.7). Petri dishes were incubated in a low-temperature incubator set at 4 °C in dark for 48 h to facilitate uniform germination. After 2 days, the Petri plates were taken out and staked vertically under a cool fluorescent light (100 µmol photons/m2/s) with a 16/8 h (day/night cycle) at 22 ± 2 °C for 4 days. At the end of 4 days, the plants were about 2 cm long with well-differentiated roots and shoots; uniform plants were then selected and used for the root-bending assay. The root-bending phenotypic screen for NaCl tolerance of salt-tolerant mutants has been described previously (Verslues et al. 2006). Seedlings were then transferred to MS basal medium supplemented with 100 mM NaCl in square plates, and phenotypic analysis characterized by observing the extent of root bending. Salt overly sensitive mutants (sos1, sos2, sos3) were purchased from the Arabidopsis Biological Resource Center (ABRC), Ohio State University (Columbus, OH).
RNA Extraction, cRNA Preparation, Labeling, Hybridization, and Signal Acquisition
Arabidopsis plants were initially exposed to 150 mM NaCl for 24 h, after which plants were treated with either the methanol extract of A. nodosum (MEA) or the ethyl acetate sub-fraction of A. nodosum (EAA). Control plants were also exposed to 150 mM NaCl but were treated with 0.1% aqueous methanol. Total RNA was isolated from Arabidopsis plants after day 1 and 5 of extract application using an RNAqueous Plant RNA isolation kit (Ambion Inc. Austin, TX). The RNA samples were quantified using Nanodrop (Thermo Scientific, Beverly, MA) and the integrity was checked on an Agilent Bioanalyzer 2100W (Agilent Technologies Canada Inc. Mississauga, ON). Copy RNA was prepared using an Illumina Total Prep RNA Amplification kit according to the manufacturer’s protocol. The synthesized cRNA was later purified and quantitated.
Labeling, hybridization, and detection were performed at the McGill University and Genome Quebec Innovation Centre (http://genomequebec.mcgill.ca). Single-stranded, then double-stranded cDNA was synthesized from the poly (A) + mRNA present in the isolated total RNA (10 µg total RNA starting material each sample reaction) using the SuperScript double-stranded cDNA synthesis kit (Invitrogen) and poly (T)-nucleotide primers that contained a sequence recognized by T7 RNA polymerase. A portion of the resulting double-stranded cDNA was used as a template to generate biotin-tagged cRNA from an in vitro transcription reaction, using the Affymetrix GeneChip IVT labeling kit. The resulting biotin-tagged cRNA was fragmented in to strands of 35–200 bases in length following prescribed protocols (Affymetrix GeneChip Expression Analysis Technical Manual). Subsequently, the fragmented target cRNA was hybridized to probe sets present on an Affymetrix Arabidopsis genome ATH1 array. Details of the microarray design and hybridization data have been deposited at NCBI GEO repository with accession number GSE86841.
Statistical Analysis of Microarray Data
Hybridization data were analyzed using Gene Chip Operating Software. Image files were analyzed to generate raw data files that were saved as CEL files. The expression values were log2 transformed after calculating the expression index. The raw MAS 5.0 data files obtained from scanned array images were then imported into Gene Spring 7.3.1 (Silicon Genetics). Only genes with Present (P) calls were included in the analysis. Raw signals of each gene were normalized using RMA with the median of all measurements on the chip. The average normalized value of the signal intensity for each gene in three replicate hybridization experiments was adopted as the expression value of the gene. Expression data were analyzed using one-way ANOVA to identify differentially regulated transcripts. False discovery rate multiple-testing corrections (Benjamini and Hochberg 1995) were calculated based on the P-value generated from the one-way ANOVA using the T-statistic test (Wright and Simon 2003). The T-statistic is calculated by the software to estimate the P-value. Using a false discovery rate of 5% that corresponded to P-value = 0.05, we selected only statistically significant genes that were regarded as up-regulated only if their fold change was ≥ 1.5, and down-regulated only if their fold change was ≤ 0.67. Comparative analysis of samples from ethyl acetate and methanol extract-treated and control salt stress-treated plants were performed using Flex array software (McGill University, Montreal, Canada). Pathways affected by treatments were performed using tools available at TAIR, including AraCyc (http://arabidopsis.org).
Functional Classification of Differentially Expressed Genes
The differentially expressed group of genes were distributed into functional categories using the web-based tools, Classification Super-Viewer (http://bbc.botany.utoronto.ca/ntools/cgi-in/ntools_classification_superviewer.cgi). The genes were classified as detailed in the Munich Information Center for Protein Sequences (MIPS) database. The broad spectrum of gene functions is similar to that predicted for the entire Arabidopsis genome (The Arabidopsis Genome Initiative, 2000). The genes were classified either as “stress,” “cellular organization and biogenesis,” “other metabolic process,” “transcription,” “cell wall,” “transporter activity,” “electron transport or energy pathways,” or “signal transduction,” Additionally, genes that have known abiotic or biotic stress related function or fall in other sub-categories were classified separately. IDs falling into classification categories other than unclassified or un-clear classifications were removed from these categories.
Validation of Microarray Using Real-Time PCR
Real-time expression analysis was used for confirmation of microarray results by using primers listed in Table S1. 2.5 µg RNA of Arabidopsis treated with ethyl acetate fraction of ANE in the presence of 150 mM NaCl, after day 1 and 5 of extract application was treated with DNase I (Promega) followed by first-strand cDNA synthesis using a high-capacity cDNA reverse transcription kit (Ambion, USA). Plants treated with 0 and 150 mM NaCl without ANE served as control. Real-time PCR was done using cDNA by Step One™ Real-Time PCR system, Applied Biosystems. Actin was used as an internal control gene. The specificity of PCR amplification was checked at the end of the PCR cycles, by melt-curve analysis. Each reaction was replicated three times and relative fold expression was determined using the Livak method (Livak and Schmittgen 2001).
Characterization of T-DNA Lines and Phenotypic Screen
T4 segregating T-DNA mutants of wall-associated kinase 1 (WAK1) (SALK_107175, SALK_137260); RNA-binding protein, putative 247575_at (SALK_007455, SALK_007592, SALK_098675, SALK_078241, SALK_078146, SALK_078245); glycine-rich RNA-binding protein 8 (GRBP8) (CCR1), 252885_at (SALK_114101, SALK_149544, CS803581 SALK_047963C) cellulose synthase family protein 260592_at (SALK_072106, CS25016); cellulose synthase family protein 254189_at (SALK_008597, SALK_123441); invertase/pectin methylesterase inhibitor family protein 262640_at (SALK_072421, SALK_084836C, SALK_007858C) were obtained from the Arabidopsis Biological Resource Center (ABRC), Ohio State University. T-DNA insertion mutants were screened for NaCl sensitivity by root gravitropic bending, phenotypic screening, as described above. Briefly, Arabidopsis seeds were sown on Murashige and Skoog (MS) agar medium (Murashige and Skoog 1962), stratified for 1 day and then incubated at 22 °C for 4 days. Seedlings were transferred to MS basal medium supplemented with 125 mM NaCl in square plates, and phenotypic analysis was performed. Seedlings grown on plates were placed in a controlled environment with 100 µmol photons/m/s incident light and photoperiod of 16 h of light and 8 h of darkness at 22 °C.
Results
ANE Confers Salinity Tolerance in Arabidopsis
The salt stress alleviating effect of methanolic fraction of ANE (MEA) was studied by irrigating 2-week-old A. thaliana plants with 100 and 150 mM NaCl for 24 h and subsequently treating with 1 g/L equivalent of methanolic fraction of ANE. After 3 weeks, plant height, fresh weight (FW), leaf number, and leaf area were recorded. Plants treated with MEA showed an increase in leaf area in NaCl stress, as compared to untreated controls. At 150 mM NaCl stress, plants treated with MEA showed a 37.30% increase in leaf area, as compared to NaCl stressed, but not treated with the extract (Fig. S1A). Similarly, there was 29 and 33% increase in plant height and 26 and 33% increase in the number of leaves at 100 mM and 150 mM NaCl, respectively (Fig. S1B, C). MEA treatment showed higher biomass under NaCl stress, the shoot fresh weight increased by 46 and 57% at 100 and 150 mM NaCl, respectively (Fig. S1D). Similarly, MEA treatment showed a higher number of leaves and fresh weight in plants even without the NaCl stress (Fig. S1). We further sub-fractionated the methanolic fraction into water-soluble, chloroform, and ethyl acetate sub-fractions and all fractions were examined for their salt stress alleviating effects. In the presence of the 150 mM NaCl, organic fractions (chloroform and ethyl acetate) showed increased plant growth (Fig. S2). The plants treated with 1 g/L equivalent of ethyl acetate and chloroform fraction showed 48 and 58% increases in plant height, as compared to control in the presence of salinity stress. (Fig. S2A). Plants treated with a 1 g/L equivalent of ethyl acetate fraction showed 52% higher fresh weight than control plants, while treatment with the chloroform fraction resulted in a 36% increase in fresh weight, as compared to control, in the presence of 150 mM NaCl (Fig. S2B). Treatment with the ethyl acetate fraction resulted in 45% increase in the number of leaves while chloroform and water-soluble fractions resulted in 15 and 11% increase in the number of leaves, respectively (Fig. S2C). Further, the ethyl acetate fraction reversed the negative effect of NaCl on leaf area more effectively than the chloroform fraction. Plants treated with ethyl acetate fractions resulted in a 62% increase in leaf area while chloroform caused only a 33% increase (Fig. S2D). Overall, the ethyl acetate fraction was found to mitigate NaCl injury more effectively than chloroform, or the water-soluble extract and was therefore tested in further experiments (Fig. S2).
Ethyl Acetate Fraction of A. nodosum Improved Plant and Root Growth
The ethyl acetate fraction of A. nodosum (EAA) improved plant growth in the presence of salinity stress (Fig. 1). The root-bend assay was a convenient and rapid screening method for testing improved root growth under salt stress conditions. We used this method to test the bioactivity of the ethyl acetate fraction. NaCl (150 mM) decreased root elongation by ~ 50%, as compared to a water control. EAA concentrations of 0.5 and 1.0 g/L improved root growth in the presence of 150 mM NaCl, however, a slightly higher concentration (2.0 g/L) of the EAA was inhibitory to root growth (Fig. S3).
Compounds present in EAA did not complement catalase deficiency and SOS phenotype. Plants deficient in catalase are hypersensitive to NaCl stress. To test if the salt stress alleviating effect of EAA was independent of the catalase mediated pathway, we used two transgenic lines of Arabidopsis (CAT2HP1 and CAT2HP2) that had 20 and 7% residual catalase activity, respectively. Since these genotypes were not tested for NaCl sensitivity, we initially compared root growth in media containing 75 mM NaCl (moderate salt stress) (Fig. S4). In WT (wild-type) plants, root growth decreased by about 20% in WT in media containing NaCl. On the other hand, the effect of low-level catalase in plants was evident in both CAT2HP1 and CAT2HP2, which showed a 40% decrease in growth. Addition of EAA in NaCl containing medium did not result in improved root growth of the CAT2HP1 line (Fig. S4). Surprisingly, EAA potentiated the lethality of CAT2HP2 plants. However, root growth in media supplemented with an ethyl acetate fraction was comparable to CAT2HP1 indicating that the compounds present in EAA did not complement catalase deficiency in Arabidopsis.
Since the SOS pathway functions in Na+ homeostasis, we tested the effect of EAA on the phenotype of sos mutants under salt stress. The root growth and ability of WT and sos mutants for continued root bending were compared in a medium containing 75 mM NaCl, with or without EAA. At 0 mM NaCl, there were no differences in root bending and root growth between WT and sos mutants (Fig. S4). A concentration of 75 mM NaCl did not affect the root growth of WT; however, as expected, root growth of sos1, sos2, and sos3 mutants was as affected (Fig. S4). Thus, from these results, we concluded that the addition of EAA in 75 mM NaCl containing media did not cause a reversal of the sos mutant phenotype.
EAA Elicits Global Transcriptome Changes in Arabidopsis Leaves
To study the molecular mechanisms of A. nodosum extract-elicited salt tolerance in Arabidopsis, we performed a global gene expression profiling on the ATH1 GeneChip platform. The ATH1 GeneChip consists of over 22500-probe sets representing nearly 90% of the Arabidopsis genome, thus providing a means to ascertain global transcriptional changes elicited by organic sub-fractions of A. nodosum during extract treatment.
Global Expression Profile
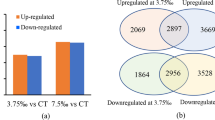
The changes in global transcriptome elicited by extract are depicted in Fig. 2. MEA and EAA caused significant changes in the expression of a small sub-set of the genes, although most transcripts remain unchanged by the treatments (Fig. 2). EAA treatments elicited a stronger response and affected a larger number of genes as compared to MEA treatments as evident in the volcano plot (Fig. S5). There was little difference in the gene expression profile between the replicates within a treatment resulting in a low P-value. Therefore, we used 1.5-fold change as the cut-off in our analysis. In the EAA treatment, 184 and 257 genes were up-regulated in day 1 and day 5, respectively. Six genes were common in day 1 and 5. On the other hand, 91 and 262 genes were down-regulated on day 1 and day 5 by this treatment as illustrated in the Venn diagrams (Fig. 3). A smaller number of genes (34 and 86) were induced by the methanolic fraction sub-fraction treatment on day 1 and day 5, respectively, while 115 and 173 genes were repressed by MEA treatment (data not shown). It is evident from the heat maps that EAA perturbed greater changes to the transcriptome, as compared to MEA (Figs. S6, S7). Annotations were made based on MIPS Functional category classifications and listed (Fig. S8). To study EAA-mediated regulation of gene expression, only changes in mRNA abundance more than, or equal to, 1.5 fold are presented in Tables S3–S6. This threshold cut-off was lower than previously reported microarray studies (Reymond et al. 2000; Seki et al. 2002; Schenk et al. 2000), and has also been validated in other differential expression studies to represent significant changes (Swidzinski et al. 2002; Kumari et al. 2008; Urano et al. 2009; Abercrombie et al. 2008). The genes that were modulated by the EAA fraction of ANE were divided into four categories.
Scatter plots of Arabidopsis ATH1 GeneChip data with extract treatments. Normalized signal intensities are plotted for comparison of a NaCl and EAA treatment samples after day 1; b NaCl and EAA treatment samples after day 5; c comparison of NaCl and MEA treatment samples after day 1 and d NaCl and MEA treatment samples after day 5. Signal intensities are the average of three biological replicates for all experiments. Arabidopsis plants were given NaCl treatment (150 mM) for 24 h and then treated with MEA or EAA at 1 g/L equivalents for 1 day or 5 days
Category 1: Up-Regulated Genes in EAA Treatment on Day 1
Table S3 lists the genes that were up-regulated on day 1 of EAA treatment under 150 mM NaCl stress. Of the 184 genes that showed changes, the largest groups were annotated as involved in metabolism (27%) and 19.8% as “unknown,” 16% as predicted to be involved in regulating gene expression i.e., transcription factors, 2.2% function in abiotic stress response group, and 7.2% in cellular defense (Table S3). Among all of the gene responses, the transcripts for late embryogenesis abundant 3 family protein/LEA3 family protein (At1g02820) and myb-related transcription factor (CCA1; At2g46830) were observed as the most strongly induced (i.e., 2.73 times for LEA3; and 3.6 times for CCA1). In the abiotic stress group, the genes that showed differential expression included late embryogenesis abundant protein LEA group 1 (At5g06760) and LEA 3 family (At1g02820); drought-responsive protein (At4g15910) and HVA 22d genes (At4g24960).
Genes involved in cellular organization and biogenesis such as the lipid transfer family protein LTP6 (At3g08770), endo-1,4-beta-glucanase, and putative/cellulases (At1g64390) were induced by this treatment. In the metabolism group, EAA treatment activated increased levels of myoinositol-1-phosphate synthase 2 (At2g22240) transcripts. Galactinol synthetase genes ATGOLS3 (At1g09350); ATGOLS2 (At1g56600) were also up-regulated. Thus, two families of genes that function in the biosynthesis of raffinose oligosaccharide (myoinositol and galactinol synthetases) were up-regulated by the A. nodosum extract. Furthermore, raffinose synthase (At5g40390) was also up-regulated (Table S3).
The transcription factors up-regulated in this category were mainly zinc finger, myb transcription factors, and AP2 domain containing transcription factor. Transcription factors DRE-binding protein (DREB1A)/CRT/DRE-binding factor 3 (CBF3) and DRE-binding protein (DREB1C)/CRT/DRE-binding factor 2 (CBF2) were significantly induced by EAA. The DREB/CBF pathway has been established to be the converging point of NaCl, drought, and freezing stress signaling. In the cellular rescue and defense group, glutathione S-transferase (At5g172200) was shown to be up-regulated (Table S3).
Category 2: Up-Regulated Genes in EAA Treatment on Day 5
Category 2 included 257 genes that were up-regulated on day 5 of EAA treatment. Interestingly, on day 5 of the treatment, the proportion of abiotic stress-regulated genes increased to 6.0%. On the other hand, the percentage of genes under transcription factors group decreased on day 5 of the treatment (6.5%) in comparison to day 1 of the treatment. Genes that were up-regulated on day 5 are listed in Table S4, including several group 1 LEA and group 2 LEA proteins (dehydrins). An abscisic acid (ABA)-induced stress regulation gene, AtHVA22b (At5g62490), was induced by EAA. Similarly, Di21 (At4g15910) was also up-regulated. Overall, these genes were clearly involved in abiotic stress and have been characterized in the ABA-dependent signaling pathways induced abiotic stress response. The expression study also revealed a higher expression of genes involved in signal transduction such as SnRK2 (At4g40010; 4.47 fold), CIPK25 (At5g25110; 2.60 fold), and phospholipase D delta (At4g35790; 1.81 fold).
Many of the genes involved in lipid transport, mainly lipid transfer proteins (LTPs) were induced. LTPs are small, abundant basic proteins in higher plants. EAA treatment induced transcription of a number of LTPs (At4g33550; At4g15910; At5g59310.1At1g62510.1; At3g18280; At5g59320). LTPs function by binding fatty acids and by transferring phospholipids between membranes in vitro. Other genes that were induced included glutathione S-transferases (At5g62480), Annexin (At1g35720), several transcription factors like RING zinc finger proteins and MYB (Table S4).
Category 3: Down-Regulated Genes in EAA Treatment on Day 1
Genes that were down-regulated by the EAA treatment on day 1 are listed in Table S5. This category consisted of genes involved in cellular organization and biogenesis genes like cellulose synthase family protein (At1g55850) and xyloglucan endotransglucosylase 3 (At5g57550). The plant cell wall polysaccharide cellulose is synthesized in the plasma membrane by cellulose synthase (CesA) proteins. The CesA genes encode polytopic membrane proteins belonging to the glycosyltransferase family 2 (At1g55850; At4g24000). Besides, the auxin-responsive gene (At2g23170), several heat shock proteins, that is, the senescence-associated gene 12 (At5g45890), and an invertase/pectin methylesterase inhibitor family protein (At1g62760) were down-regulated.
Category 4: Down-Regulated Genes in the EAA Treatment on Day 5
Table S6 lists the genes that were down-regulated by EAA treatment on day 5. In the cell wall group, wall-associated kinase (WAK1) and a few pectin-esterases (At1g53840; At3g10720) were inhibited. Transcripts of RNA-binding proteins (At5g61030 and At4g39260) were also reduced. PDH (At3g30775) was also down-regulated in the presence of EAA. It has been shown that reciprocal regulation of P5CS and PDH genes appeared to regulate the concentration of proline under osmotic stress (Peng et al. 1996).
Validation of Microarrays Using Real-Time qPCR
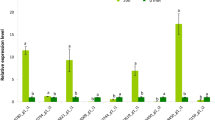
To confirm the validity of microarray data, we conducted Real-time expression analysis of selected genes that showed differential expression upon treatment with EAA in the presence of 0 and 150 mM NaCl. As seen in the microarray data, the Real-time expression analysis also confirmed that AtLEA, AtRD29B, AtDi2l, AtRAB18, AtLTP4, AtNIP6-1, AtGLOS, AtGBSS, AtCHI, and AtDFR expression levels were induced upon treatment with EAA (Fig. 4).
Several Genes Down-Regulated in Treatments with EAA Showed a Salt Tolerance Phenotype
Several genes were significantly repressed by the EAA treatment, as compared to the 150 mM NaCl treatment. We hypothesized that Arabidopsis mutants with reduced expression of these genes may be tolerant to NaCl or negative regulators of the NaCl stress response. Therefore, T-DNA insertion lines for WAK1, RNA-binding protein, glycine-rich RNA-binding protein, cellulose synthetase, and invertase genes were tested for NaCl tolerance. Under ambient growth conditions (without NaCl), no difference was observed between growth of the SALK mutants and the wild-type Col-0 plants. However, under 125 mM NaCl stress, some of the mutants [wall-associated kinase 1-SALK_107175 (Exon), glycine-rich RNA-binding protein 8-SALK_114101 (promoter), Invertase/pectin-SALK_072421 (promoter), SALK_007858C (promoter), RNA-binding protein 3-SALK_078146 (intron) and SALK_007455 (promoter)] grew better and therefore were tolerant to NaCl stress conditions. For instance, 71% of the invertase, SALK_072421 plants survived on 125 mM NaCl medium while only 20% of Col-0 survived (Table S2).
Discussion
Environmental stresses reduce agricultural productivity. Earlier studies from our laboratory showed that bioactive compounds present in A. nodosum extract improved plant growth under stressful conditions (Nair et al. 2012; Shukla et al. 2018). A. nodosum extracts also imparted enhanced tolerance to drought stress (Goni et al. 2018; Shukla et al. 2018; Zhang and Ervin 2004), and improved photosynthetic efficiency and performance of Paspalum vaginatum, exposed to prolonged water stress and salinity shock (Elansary et al. 2017). In the present study, we tested the salinity-alleviating effect of organic sub-fractions of A. nodosum extract (ANE) and explored possible mechanisms of action by studying its effect on the global transcriptome. Among the different organic sub-fractions of ANE, EAA imparted a greater salinity tolerance in Arabidopsis. The root-bend assay showed similar results confirming that compounds in the EAA improved root growth under NaCl stress. EAA consists of lipophilic components such as fatty acids and sterols that improved freezing tolerance in A. thaliana (Nair et al. 2012; Rayirath et al. 2009). Similarly, Eckol, a plant growth stimulant prepared from the ethyl acetate extract of Ecklonia maxima significantly improved growth of maize seedlings (Rengasamy et al. 2015). Therefore, to better understand the mechanism of action of EAA, whole-genome transcriptome profile was carried out.
LEA proteins are highly diverse, and according to their primary sequences were grouped into eight families (that is, dehydrins, LEA_1, LEA_2, LEA_3, LEA_4, LEA_5, LEA_6, and seed maturation protein) (Candat et al. 2014). The previously published reports showed that the expression of LEA genes was increased by cold, drought, and salinity (Candat et al. 2014; Hundertmark and Hincha 2008). EAA treatment induced the expression of several stress-regulated genes on day 1 and day 5 post-treatment. LEA group 3 (At1g02820), LEA group 1 (At5g06760), Di 21 (At4g15910), and HVA 22d (At4g24960) were identified as stress-regulated genes induced by EAA fractions on day 1 in response to salinity stress in Arabidopsis. Some of the abiotic stress-regulated genes induced on day 5 of the treatment were common to day 1 including LEA group 1 (At5g06760) and Di 21 (At4g15910). Apart from these, other LEA isoforms such as LEA group 4 (At3g17520, At1g52690), RAB18 (At5g66400), and LEA group 1 (At2g35300) were also highly induced on day 5. Similarly, Goni et al. (2016) compared the transcriptome of two biostimulants prepared from Ascophyllum nodosum using different extraction methods. The results presented in this study also showed up-regulation of different LEA isoforms in response to the ANE application. LEAs protect cellular structures from water loss by acting as a hydration buffer, by sequestering ions, by direct protection of other proteins, or by re-naturing unfolded proteins (Goyal et al. 2005; Wise and Tunnacliffe 2004). Several reports suggested the positive correlation between LEA expression and stress tolerance (Babu et al. 2004). Interestingly, Group 2 LEA proteins and dehydrins (RAB 18) were specifically up-regulated in day 5 after treatment. The accumulation of dehydrins is one of the prominent components of plant adaptation to extreme environmental conditions. Goni et al. (2018) reported the role of different ANEs prepared by distinct procedures in alleviating drought tolerance in tomato plants by regulating the expression of dehydrins. Thus, the tolerance to salt stress conditions mediated by EAA may be, in part, mediated by an increase in the expression of LEAs genes. RAB 18 encodes a serine and lysine-rich protein and its transcripts accumulate in response to ABA (Lang and Palva 1992). RD29B is induced by NaCl and drought conditions and has an ABA-dependent mode of expression. RD29B contains two ABREs (abscisic acid response elements) (Yamaguchi-shinozaki and Shinozaki 1994). Many ABA-dependent genes were up-regulated by the EAA treatment. Among others like RAB 18, LTP3, and LTP4, Di-21, we also observed HVA homologues, HVA 22d and HVA 22b were also induced on day 1 and day 5, respectively, of extract treatment. Chen et al. (2002) reported that among the Arabidopsis HVA22a, b, c, d, and e homologues, HVA 22d and HVA22b were tightly regulated ABA-inducible genes in Arabidopsis (Chen et al. 2002).
Plant responses to abiotic and biotic stresses are mediated by signal molecules such as ABA, jasmonic acid (JA), and salicylic acid (SA). SnRK2, serine/threonine kinases play a pivotal role in stomatal movement, ABA-inducible gene expression, and stress responses in plants (Hirayama and Shinozaki 2007). SnRK2 are activated in response to saline stress by the phosphorylation of a specific serine residue (Coello et al. 2011; Boudsocq et al. 2007). SnRK2 was induced 4.5 fold in the EAA-treated Arabidopsis exposed to salinity. The C-terminal of SnRK2 was involved in activation of the ABA-signaling network (Coello et al. 2011), suggesting the role of EAA in regulating the stress-induced signal transduction. Phospholipid metabolism has been suggested to play an important role in signal transduction pathways of plants and animals (Wang 2005). Incidentally, we also observed increased expression of phospholipase D delta/PLD delta by salinity stress in A. thaliana by the EAA treatment at day 5 of the treatment. PLD 1-derived phosphatidic acid interacted with ABI1 phosphatase 2C and promoted abscisic acid signaling in plants (Christmann et al. 2006). Seo et al. (2001) demonstrated that S-adenosyl-l-methionine: jasmonic acid carboxyl methyltransferase (JMT) (expressed on day 1 of treatment) was a key enzyme for the jasmonate-regulated plant responses. JMT catalyzes the methylation of JA to form MeJA. MeJA and its free acid jasmonic acid (JA), collectively referred to as jasmonates induced plant defense responses against a group of pathogens and mechanical or herbivorous insect-driven wounding (Seo et al. 2001). MeJA by itself might have a role in jasmonate-regulated signaling process. The SA and JA signaling pathways are mutually antagonistic (Kunkel and Brooks 2002). It is interesting to note from our studies conducted using these extracts, pathogenesis-related (PR) proteins, that were known SA markers were not up-regulated when challenged with Pseudomonas (Subramanian et al. 2011). Thus, from our studies, it might be concluded that JA signaling might be up-regulated in EAA-treated plants and play an essential role in defense response.
A number of transcription factors (TFs) play an important role in conferring stress tolerance in plants (Agarwal and Jha 2010). The TFs interact with cis-elements in the promoter regions of various downstream genes and regulate their expression (Agarwal et al. 2013). Transcription factors DRE-binding protein (DREB1A)/CRT/DRE-binding factor 3 (CBF3), DRE-binding protein (DREB1C)/CRT/DRE-binding factor 2 (CBF2) which play an important role in abiotic stress tolerance were activated on day 1 of the EAA treatment. It is well known that DREB/CBF pathway is the central point on which signals from several abiotic stressors converge (Agarwal et al. 2017). In Arabidopsis, CBF3 is induced by low temperature, but not affected by drought or high salinity. However, over-expression of the Arabidopsis CBF3 in transgenic rice increased tolerances to drought and high salinity (Oh et al. 2005). The application of EAA conferred freezing tolerance in Arabidopsis by up-regulation of hyperosmotic salinity-responsive gene, AZF2 (Nair et al. 2012). The transcription factors COR47, COR15A, AGF2 were also up-regulated in response to an A. nodosum extract application (Goni et al. 2018). Environmental stresses modulate the plant circadian clock, which disrupts co-ordination between the plant metabolic processes and the external environment (Lai et al. 2012; Hotta et al. 2007). The ethyl extract fraction of A. nodosum induced the expression of circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY1) on day 1, which affected the transcriptional regulation of genes involved in ROS homeostasis and tolerance to oxidative stress (Lai et al. 2012). EAA-induced expression of CCA1 and LHY1 also regulated the expression of the cold-regulated gene 15A (COR15A), COR47, and COR78 (Dong et al. 2011). These results suggested that the bioactive components present in the ethyl acetate fraction of A. nodosum extract regulated the circadian clock by maneuvering the expression of DREB1C/CBF2 involved in cross-talk between signaling pathways (Shinozaki and Shinozaki 2000; Dong et al. 2011). The heat shock transcription factor (AtHsfA6a, At5g43840) was highly induced in the presence of ABA, NaCl, and drought, and transcriptionally regulated by ABA-responsive elements (Hwang et al. 2014). EAA induced the expression of HsfA6a on day 5 of treatment, in the presence of salinity. In addition to this, an EAA application, in the presence of NaCl, showed higher transcript accumulation of nuclear factor-YA that binds to the CCAAT-box known as the CCAAT-binding transcription factor. Leyva-González et al. (2012), showed that the nuclear factor-YA (NF-YA) was strongly induced in the presence of salinity and ABA via transcriptional and miR169-related post-translational mechanisms. Thus, these results showed that EAA regulated the convergence and interaction of various transcription factors for better growth of plants in the presence of salinity.
The gene-encoding inositol-3-phosphate synthase (isozyme 2) (IPS 2) was induced on day 1 of the extract treatment. The transcript level of IPS 2 was up-regulated and levels of free myoinositol accumulated during NaCl stress (Ishitani et al. 1996). However, Arabidopsis does not accumulate inositol or increased levels of IPS 2 RNA during salt stress. Taji et al. (2004) hypothesized that raffinose family oligosaccharides were synthesized from either inositol-1-phosphate synthetase or from galactinol synthetase. Because the galactinol synthetase genes ATGOLS3, ATGOLS2, and raffinose synthase were up-regulated on day 1 of the EAA application in the presence of salinity, it was possible that these pathways were activated by the extract treatment. Thus, the higher biosynthesis of galactinol and raffinose, in the presence of EAA + NaCl, acted as osmo-protectants and provided an adaptation to salinity stress (Valliyodan and Nguyen 2006). Interestingly, the salt-tolerant Arabidopsis relative, Thellungiella halophila, showed up-regulation of all these genes and it is hypothesized that they accumulated raffinose (Taji et al. 2004).
Salinity stress leads to the reduction in of stomatal apertures in plants, and hence reduces the rate of photosynthesis (Negrão et al. 2017). EAA induces the expression of light harvesting complex photosystem II (LHCB4 3, At2g40100) and glucose-6-phosphate transport (At1g61800) involved in photosynthesis. The gene-encoding enzymes involved in the biosynthesis of carbohydrates (for example, starch, sucrose, raffinose), amino acids (for example, proline, isoleucine), and sugar alcohol (for example, inositol, trehalose) were found to be up-regulated in EAA-supplemented Arabidopsis, in the presence of NaCl. The enhanced biosynthesis of these osmolytes, in the presence of EAA, improved the growth of the plants treated with NaCl. Sugar and sugar alcohols regulate plant growth and development in the presence of abiotic stresses by acting as osmolytes (Gupta and Kaur 2005). Among them, starch is a key polysaccharide involved in plant responses to abiotic stresses (Thalmann and Santelia 2017). EAA was found to induce the expression of starch synthase (At1g32900) after day 1 of application in the presence of salinity. Increased starch biosynthesis in the EAA-treated plants provided enough carbon and energy as required by the plants during salinity stress (Garg et al. 2016). Plant use sucrose and hexose sugars as a substrate for cellular respiration, or as osmolytes to maintain cellular homeostasis, in response to salinity stress (Rosa et al. 2009). In the present study, the application of EAA induced the expression of sucrose phosphate synthase (At4g10120) (day 1) and sucrose synthase (At4g02280) involved in sucrose biosynthesis. In addition to this, EAA induced the expression of glycerol-3-phosphate transporter (At3g47420) and phosphoglycerate involved in carbohydrate transport (At3g01550) on day 1 of application. Plants strictly regulate their vacuolar storage and transport of sugar to cope with different environmental stresses (Chandran 2015). On day 5 of the treatment, the EAA application showed a higher expression of bi-directional sugar transporters such as SWEET 15 (At5g13170), SWEET 4 (At3g28007), and sugar transporter family protein (At1g73220), whereas the expression of SWEET 13 (At5g50800) was reduced on Day 5 of treatment. Seo et al. (2011) showed that the expression of SWEET15 was highly induced in the presence of different abiotic stresses and modulated cell viability during abiotic stresses. Thus, the evidence presented revealed that EAA improved sugar fluctuations during salinity stress by regulating carbohydrate metabolism and transport in Arabidopsis.
Sugars and phytohormones relay messages to form an intricate network of signaling which regulates plant growth and development (Loreti et al. 2008). In this study, EAA induced the expression of gibberellin-regulated protein 3 (GASA3, At4g09600) and gibberellin 3-beta-dioxygenase (GA3, At1g15550) which is known to be involved in gibberellin biosynthesis in plants grown in the presence of salinity. Similarly, Goni et al. (2018) showed higher expression of GASA1 in the plants treated with the extracts of Ascophyllum nodosum. Gibberellin accumulation during abiotic stress regulated the sugar signaling and biosynthesis of antioxidative enzymes (Fahad et al. 2015). EAA-induced accumulation of gibberellic acid (GA) promoted the activity of sucrose phosphate synthase (At4g10120), and stimulated phloem-loading. Thus, application of the EAA modulated the source-sink metabolism by regulating the cross-talk between gibberellin and sucrose biosynthesis.
EAA applications induced the accumulation of transcripts of xyloglucan: xyloglucosyl transferase (At3g44990) and UDP-glucoronosyl/UDP-glucosyl transferase (At2g16890) involved in cell wall biosynthesis. In addition to this, our microarray analyses also revealed many genes that were down-regulated by extracts on days 1 and 5 of treatment. These included cell-wall-related genes and others involved in signal transduction such as WAK 1 that might play a negative role under salt stress (Tables S5, S6). Plant cell walls consist of a complex matrix of cellulose, non-cellulosic polysaccharides, proteins, and lignin. Plant cell walls determine cell shape and size, providing strength and flexibility during stress and also have a role to defend plants against pathogens (Le Gall et al. 2015). We observed that cellulose synthetases, invertase/pectin methylesterase inhibitor family protein, and pectin-esterases were down-regulated by EAA applications. Pectin methylesterases (PME, EC. 3.1.1.11) are enzymes that de-methylesterify plant cell wall pectins. WAK 1 is a cytoplasmic serine/threonine kinase that spans the plasma membrane and extends into the extracellular region to bind tightly to the cell wall. WAK 1 is induced by pathogen infection and salicylic acid or its analogue INA (He et al. 1998). These results suggested that an EAA application improved mechanical strengthening and re-modeling of the cell wall to protect the plants from salinity stress. Among the transcription factors, a few GR-RBP were down-regulated. In Arabidopsis thaliana, there are eight GR-RBP family members and these have a role in stress responses including cold, high salinity, and dehydration (Kyung et al. 2005). It has been reported that some members are negative regulators of salt stress (Kyung et al. 2005). We carried out phenotypic assays using T-DNA mutants down-regulated in these genes in medium containing 125 mM NaCl to study the relative importance of each of these. The results presented by Jithesh et al. (2012) showed that loss of functional mutation of GR-RBP and invertase/pectin methylesterase inhibitor family protein showed tolerances to the concentrations of NaCl tested, and further support our findings that an application of EAA mitigated the salinity stress in Arabidopsis by decreasing expression of genes involved in the negative regulation of salinity tolerance.
Interestingly, several genes in the phenylpropanoid pathway, especially flavonoid synthesis was up-regulated by EAA, both on days 1 and 5. This included phenylalanine ammonia lyase 1 (PAL1) and PAL2 (that catalyzes the conversion of phenylalanine to cinnamate); chalcone synthase (CHS), (required for the condensation of 4-coumaroyl-CoA and malonyl-CoA to yield naringenin chalcone); chalcone isomerase (CHI); flavonoid 3′-hydroxylase (F3′H); and dihydroflavonol 4-reductase (DFR) (Fig. 5). Flavonoids are a diverse group of secondary metabolites with a wide array of biological functions such as pigmentation, facilitators of plant–microbe interactions, and reproduction. These flavonoids inhibited the generation of reactive oxygen species, and have also been linked to defense responses against biotic and abiotic stresses, such as pathogens, wounding, and UV light damage (Fini et al. 2011). ANE improved the phenolic and flavonoid content of spinach and cabbage [Fan et al. (2011); Lola-Luz et al. (2013)]. ANE application also induced the expression of genes involved in secondary metabolism in spinach, thereby improving nutritional quality, and yield (Fan et al. 2013). Glutathione S-transferase (GST) plays an important role in oxidative stress (Chen et al. 2012). A previously published report showed that the over-expression of GST enhanced the growth of transgenic tobacco (Roxas et al. 2000). The application of EAA induced the expression of GST in Arabidopsis under salinity stress, at both the time points. In addition to their role in oxidative stress, GST also helped in transportation of flavonoids to vacuoles by multi-drug resistance-associated protein (MRP)-type ABC transporters (Agati et al. 2012). EAA also induced the expression of the ABC transporter protein in plants treated with salt. Thus, these studies support our findings that bioactive components of the EAA extract enhanced the expression of the genes involved in biosynthesis and transportation of flavonoids which in turn alleviated oxidative stresses imposed by salinity stress.
Flow diagram of representative metabolic pathways regulated by the EAA treatments. Each pathway is shown consisting of nodes and lines, which represent the metabolites and reactions, respectively. Expression-level change of each reaction is shown in a color relative to the expression level, as indicated in the color scale bar (also shown). Triangles denote amino acids; squares denote carbohydrates; diamonds denote proteins; open circle, others; closed circle, phosphorylated
EAA-treated Arabidopsis plants showed increased plant growth under salinity stress conditions. It is possible that the EAA treatment increased the potential of active extrusion of Na+ from the cytosol back through the roots to the external medium. SOS1, a plasma membrane Na+/H+ antiporter performs this essential function in Arabidopsis and other plants (Shi et al. 2000). Indeed, EAA treatment of the sos1 mutant did not reverse the lethal phenotype, confirming the critical role of SOS1 in sodium extrusion. Alternatively, reducing net K+ loss and maintaining active K+ transport from roots could aid in retaining cytosolic K+ levels. Isoforms belonging to the HAK/KUP/KT family are involved in both low- and high-affinity K+ transport (Bañuelos et al. 2002). The transcript abundance of KUP6 was found to increase in EAA-treated plants. KUP6 may be possibly involved in maintaining cytoplasmic K+ levels during conditions where external Na+ inhibited K+ uptake.
In conclusion, climate change, urbanization, and global warming and associated reduction in global arable land pose an imminent threat to meet the food requirements of an ever-increasing human population. Since abiotic stress tolerance is an oligogenic trait, genetically engineering salt and drought-tolerant crops is challenging (Agarwal et al. 2013). This study demonstrated the efficacy of the EAA extract in improving salinity stress tolerance in Arabidopsis, and provides a holistic molecular and fundamental explanation of EAA-mediated regulation of different stress-responsive genes.
References
Abercrombie JM, Matthew DH, Ranjan P, Rao MR, Saxton AM, Yuan JS, Stewart CN (2008) Transcriptional responses of Arabidopsis thaliana plants to As (V) stress. BMC Plant Biol 8(1):87
Agarwal PK, Jha B (2010) Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol Plant 54:201–212. https://doi.org/10.1007/s10535-010-0038-7
Agarwal PK, Shukla PS, Gupta K, Jha B (2013) Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol 54:102–123. https://doi.org/10.1007/s12033-012-9538-3
Agarwal PK, Gupta K, Lopato S, Agarwal P (2017) Dehydration responsive element binding transcription factors and their applications for the engineering of stress tolerance. J Exp Bot 68:2135–2148
Agati G, Azzarello E, Pollastri S, Tattini M (2012) Flavonoids as antioxidants in plants: location and functional significance. Plant Sci 196:67–76. https://doi.org/10.1016/j.plantsci.2012.07.014
Al-Maskri A, Al-Kharusi L, Al-Miqbali H (2010) Effects of salinity stress on growth of lettuce (Lactuca sativa) under closed-recycle nutrient film technique. Int J Agric Biol 12:377–380
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256–1258. https://doi.org/10.1126/science.285.5431.1256
Babu RC, Zhang J, Blum A, Ho THD, Wu R, Nguyen HT (2004) HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 166:855–862. https://doi.org/10.1016/j.plantsci.2003.11.023
Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130:784–795. https://doi.org/10.1104/pp.007781
Battacharyya D, Babgohari MZ, Rathor P, Prithiviraj B (2015) Seaweed extracts as biostimulants in horticulture. Sci Hortic 196:39–48. https://doi.org/10.1016/j.scienta.2015.09.012
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. https://doi.org/10.2307/2346101
Blunden G, Jenkins T, Liu YW (1997) Enhanced leaf chlorophyll levels in plants treated with seaweed extract. J Appl Phycol 8:535–543. https://doi.org/10.1007/BF02186333
Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol 63(4):491–503
Boyer JS (1982) Plant productivity and environment. Science 218:443–448. https://doi.org/10.1126/science.218.4571.443
Candat A, Paszkiewicz G, Neveu M, Gautier R, Logan DC, Avelange-Macherel MH, Macherel D (2014) The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell 26:3148–3166
Chandran D (2015) Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 67(7):461–471
Chen CN, Chu CC, Zentella R, Pan SM, Ho THD (2002) AtHVA22 gene family in Arabidopsis: phylogenetic relationship, ABA and stress regulation, and tissue-specific expression. Plant Mol Biol 49:633–644
Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP (2012) Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol 158(1):340–351
Chinnusamy V, Jagendorf A, Zhu JK (2005) Understanding and improving salt tolerance in plants. Crop Sci 45:437. https://doi.org/10.2135/cropsci2005.0437
Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8:314–325. https://doi.org/10.1055/s-2006-924120
Coello P, Hey SJ, Halford NG (2011) The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J Exp Bot 62:883–893
Craigie JS (2011) Seaweed extract stimuli in plant science and agriculture. J Appl Phycol 23:371–393. https://doi.org/10.1007/s10811-010-9560-4
Delauney AJ, Verma DPS (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4:215–223
Dong MA, Farré EM, Thomashow MF (2011) Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA 108:7241–7246. https://doi.org/10.1073/pnas.1103741108
dos Reis SP, Lima AM, de Souza CRB (2012) Recent molecular advances on downstream plant responses to abiotic stress. Int J Mol Sci 13:8628–8647. https://doi.org/10.3390/ijms13078628
Elansary HO, Yessoufou K, Abdel-Hamid AM, El-Esawi MA, Ali HM, Elshikh MS (2017) Seaweed extracts enhance Salam turfgrass performance during prolonged irrigation intervals and saline shock. Front Plant Sci 8:830. https://doi.org/10.3389/fpls.2017.00830
Fahad S, Hussain S, Matloob A, Khan FA, Khaliq A, Saud S, Hassan S, Shan D, Khan F, Ullah N, Faiq M (2015) Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul 75:391–404. https://doi.org/10.1007/s10725-014-0013-y
Fan D, Hodges DM, Zhang J, Kirby CW, Ji X, Locke SJ, Critchley AT, Prithiviraj B (2011) Commercial extract of the brown seaweed Ascophyllum nodosum enhances phenolic antioxidant content of spinach (Spinacia oleracea L.) which protects Caenorhabditis elegans against oxidative and thermal stress. Food Chem 124(1):195–202
Fan D, Hodges DM, Critchley AT, Prithiviraj B (2013) A commercial extract of brown macroalga (Ascophyllum nodosum) affects yield and the nutritional quality of spinach in vitro. Commun Soil Sci Plant Anal 44(12):1873–1884
Fini A, Brunetti C, Ferdinando MD, Ferrini F, Tattini M (2011) Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal Behav 6:709–711
Garg R, Shankar R, Thakkar B, Kudapa H, Krishnamurthy L, Mantri N, Varshney RK, Bhatia S, Jain M (2016) Transcriptome analyses reveal genotype-and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci Rep 6:9228
Goñi O, Fort A, Quille P, McKeown PC, Spillane C, O’Connell S (2016) Comparative transcriptome analysis of two Ascophyllum nodosum extract biostimulants: same seaweed but different. J Agric Food Chem 64(14):2980–2989. https://doi.org/10.1021/acs.jafc.6b00621
Goñi O, Quille P, O’Connell S (2018) Ascophyllum nodosum extract biostimulants and their role in enhancing tolerance to drought stress in tomato plants. Plant Physiol Biochem. https://doi.org/10.1016/j.plaphy.2018.02.024
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157. https://doi.org/10.1042/BJ20041931
Gupta AK, Kaur N (2005) Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J Biosci 30:761–776
He ZH, He D, Kohorn BD (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14:55–63. https://doi.org/10.1046/j.1365-313X.1998.00092.x
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12(8):343–351
Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AA (2007) Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30:333–349
Hundertmark M, Hincha DK (2008) LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom 9:118–139
Hwang SM, Kim DW, Woo MS, Jeong HS, Son YS, Akhter S, Choi GJ, Bahk JD (2014) Functional characterization of Arabidopsis HsfA6a as a heat-shock transcription factor under high salinity and dehydration conditions. Plant Cell Environ 37:1202–1222
Ishitani M, Majumder AL, Bornhouser A, Michalowski CB, Jensen RG, Bohnert HJ (1996) Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J 9:537–548. https://doi.org/10.1046/j.1365-313X.1996.09040537.x
Jithesh MN, Wally OSD, Manfield I, Critchley AT, Hiltz D, Prithiviraj B (2012) Analysis of seaweed extract-induced transcriptome leads to identification of a negative regulator of salt tolerance in Arabidopsis. Hortic Sci 47:704–709
Khan W, Rayirath UP, Subramanian S, Jithesh MN, Rayorath P, Hodges DM et al (2009) Seaweed extracts as biostimulants of plant growth and development. J Plant Growth Regul 28:386–399. https://doi.org/10.1007/s00344-009-9103-x
Kumari M, Taylor GJ, Deyholos MK (2008) Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol Genet Genomics 279(4):339. https://doi.org/10.1007/s00438-007-0316-z
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5:325–331. https://doi.org/10.1016/S1369-5266(02)00275-3
Kyung JK, Yeon OK, Kang H (2005) Characterization of transgenic Arabidopsis plants overexpressing GR-RBP4 under high salinity, dehydration, or cold stress. J Exp Bot 56:3007–3016. https://doi.org/10.1093/jxb/eri298
Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress response. Proc Natl Acad Sci USA 109:17129–17134
Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20:951–962. https://doi.org/10.1007/BF00027165
Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants 4:112–166. https://doi.org/10.3390/plants4010112
Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L (2012) Functional and transcriptome analysis 696 reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lola-Luz T, Hennequart F, Gaffney M (2013) Enhancement of phenolic and flavonoid compounds in cabbage (Brassica oleraceae) following application of commercial seaweed extracts of the brown seaweed (Ascophyllum nodosum). Agric Food Sci 22(2):288–295
Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P (2008) Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol 179:1004–1016
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498. https://doi.org/10.1016/j.tplants.2004.08.009
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Nair P, Kandasamy S, Zhang J, Ji X, Kirby C, Benkel B et al (2012) Transcriptional and metabolomic analysis of Ascophyllum nodosum mediated freezing tolerance in Arabidopsis thaliana. BMC Genom 13:643. https://doi.org/10.1186/1471-2164-13-643
Negrão S, Schmöckel SM, Tester M (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119(1):1–11. https://doi.org/10.1093/aob/mcw191
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M et al (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351. https://doi.org/10.1104/pp.104.059147
Peng Z, Lu Q, Verma DP (1996) Reciprocal regulation of ∆ 1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Genet Genomics 253:334–341
Rayirath P, Benkel B, Hodges DM, Allan-Wojtas P, MacKinnon S, Critchley AT et al (2009) Lipophilic components of the brown seaweed, Ascophyllum nodosum, enhance freezing tolerance in Arabidopsis thaliana. Planta 230:135–147. https://doi.org/10.1007/s00425-009-0920-8
Rayorath P, Jithesh MN, Farid A, Khan W, Palanisamy R, Hankins SD et al (2008) Rapid bioassays to evaluate the plant growth promoting activity of Ascophyllum nodosum (L.) Le Jol. using a model plant, Arabidopsis thaliana (L.) Heynh. J Appl Phycol 20:423–429. https://doi.org/10.1007/s10811-007-9280-6
Rengasamy KR, Kulkarni MG, Stirk WA, Van Staden J (2015) Eckol-a new plant growth stimulant from the brown seaweed Ecklonia maxima. J Appl Phycol 27:581–587
Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12:707–720
Rosa M, Prado C, Podazza G, Interdonato R, González JA, Hilal M, Prado FE (2009) Soluble sugars: metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal Behav 4(5):388–393
Roxas VP, Lodhi SA, Garrett DK, Mahan JR, Allen RD (2000) Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol 41:1229–1234. https://doi.org/10.1093/pcp/pcd051
Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97:11655–11660. https://doi.org/10.1073/pnas.97.21.11655
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31(3):279–292. https://doi.org/10.1046/j.1365-313X.2002.01359.x
Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I et al (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98:4788–4793. https://doi.org/10.1073/pnas.081557298
Seo PJ, Park JM, Kang SK, Kim SG, Park CM (2011) An Arabidopsis senescence-associated protein SAG29 regulates cell viability under high salinity. Planta 233:189–200
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97:6896–6901. https://doi.org/10.1073/pnas.120170197
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Shukla PS, Shotton K, Norman E, Neily W, Critchley AT, Prithiviraj B (2018) Seaweed extract improve drought tolerance of soybean by regulating stress-response genes. AoB Plants. https://doi.org/10.1093/aobpla/plx051
Subramanian S, Sangha JS, Gray BA, Singh RP, Hiltz D, Critchley AT et al (2011) Extracts of the marine brown macroalga, Ascophyllum nodosum, induce jasmonic acid dependent systemic resistance in Arabidopsis thaliana. against Pseudomonas syringae pv. tomato DC3000 and Sclerotinia sclerotiorum. Eur J Plant Pathol 131:237–248. https://doi.org/10.1007/s10658-011-9802-6
Swidzinski JA, Sweetlove LJ, Leaver CJ (2002) A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J 30(4):431–446. https://doi.org/10.1046/j.1365-313X.2002.01301.x
Szabolcs I (1994) Soils and salinisation. In: Pessarakali M (ed) Handbook of plant and crop stress. Marcel Dekker, New York, pp 3–11
Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K et al (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135:1697–1709. https://doi.org/10.1104/pp.104.039909
Thalmann M, Santelia D (2017) Starch as a determinant of plant fitness under abiotic stress. New Phytol 214:943–951
Urano K, Maruyama K, Ogata Y, Morishita Y, Takeda M, Sakurai N, Suzuki H, Saito K, Shibata D, Kobayashi M, Yamaguchi-Shinozaki K (2009) Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J 57(6):1065–1078
Valliyodan B, Nguyen HT (2006) Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9(2):189–195
Van Oosten MJ, Pepe O, De Pascale S, Silletti S, Maggio A (2017) The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem Biol Technol Agric 4:5. https://doi.org/10.1186/s40538-017-0089-5
Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45:523–539. https://doi.org/10.1111/j.1365-313X.2005.02593.x
Wally OSD, Critchley AT, Hiltz D, Craigie JS, Han X, Zaharia LI et al (2013) Regulation of phytohormone biosynthesis and accumulation in Arabidopsis following treatment with commercial extract from the marine macroalga Ascophyllum nodosum. J Plant Growth Regul 32:324–339. https://doi.org/10.1007/s00344-012-9301-9
Wang X (2005) Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol 139(2):566–573
Wise MJ, Tunnacliffe A (2004) POPP the question: what do LEA proteins do? Trends Plant Sci 9:13–17. https://doi.org/10.1016/j.tplants.2003.10.012
Wright GW, Simon RM (2003) A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 19:2448–2455. https://doi.org/10.1093/bioinformatics/btg345
Yadav NS, Shukla PS, Jha A, Agarwal PK, Jha B (2012) The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol 12:188. https://doi.org/10.1186/1471-2229-12-188
Yamaguchi-shinozakiaib K, Shinozaki K (1994) A nove1 cis-acting element in an Arabidopsis genes involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6:251–264. https://doi.org/10.1105/tpc.6.2.251
Zhang X, Ervin EH (2004) Cytokinin-containing seaweed and humic acid extracts associated with creeping bentgrass leaf cytokinins and drought resistance. Crop Sci 44:1737–1745. https://doi.org/10.2135/cropsci2004.1737
Zhang X, Schmidt RE (1999) Antioxidant response to hormone-containing product in Kentucky bluegrass subjected to drought. Crop Sci 39:545–551
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445. https://doi.org/10.1016/S1369-5266(03)00085-2
Acknowledgements
Authors are grateful for the valuable suggestions of Dr. Dhirti Battacharya and Emily Mantin for reading the manuscript. The work reported in this paper was partly funded by Atlantic Innovation Fund program of Atlantic Canada Opportunities Agency and Acadian Seaplants Limited.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered as a potential conflict of interest. Authors declare no conflict of interest, financial or non-financial.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Methanolic extract of A. nodosum (MEA) impart protection against salinity stress. Two-week-old Arabidopsis were subjected to two different concentrations 100 and 150 mM NaCl. (A) Leaf Area (sq cm), (B) Plant Height (cm), (C) Leaf Number and (D) Fresh weight (mg). Fig. S2 Growth parameters of A. thaliana in the presence of salinity supplemented with different organic sub-fraction of A. nodosum (ANE): (A) Plant Height (cm) (B) Fresh weight (mg) (C) Leaf Number and (D) Leaf Area (sq cm) of Arabidopsis. Experiments without the NaCl treatments and quantification performed with or without the different sub-fraction treatment served as a control. Fig. S3 Relative root growth of Arabidopsis with different concentrations of EAA under salt stress. Relative root length of Arabidopsis upon treatment with different concentrations of EAA (0.1, 0.25, 0.5, 1.0 g L-1 equivalents of seaweed extract). The average root length (from 30 seedlings) was measured after 3 days of transferring to ½ MS plates supplemented with different concentrations of EAA under NaCl stress (100mM). Fig. S4 Salt stress sensitivity of wild-type (Col-0), sos mutants grown on vertical plates. Four-day-old seedlings of WT, cat2hp1, cat2hp2, sos1, sos2, sos3 were transferred from MS medium (1/2 x) to MS media containing 0 mM NaCl (A), 75 mM NaCl (B) or 75 mM NaCl supplemented with EAA (C). Seedlings were allowed to grow for 5 d. Fig. S5 Comparison of level of gene expression between EAA (A, B) and MEA (C, D) treatments. The relation between the 1.5-fold difference and statistical significance using t test are presented by volcano plots in EAA and MEA treatments at day 1 and day 5 post treatments. Fig. S6 Heat map representing the clustering of differentially expressed genes on day1 and day 5 of the application of EAA in the presence of 150 mM NaCl. The fold change expression data represented in this figure is obtained from three replicates. Color scale represents the normalized fold induction is presented in the figure: green represents high expression and red represents low expression. Fig. S7 Heat map representing the clustering of differentially expressed genes on day1 and day 5 of the application of MEA in the presence of 150 mM NaCl. The fold change expression data represented in this figure is obtained from three replicates. Color scale represents the normalized fold induction is presented in the figure: green represents high expression and red represents low expression. Fig. S8 MIPS classification of genes regulated by EAA treatments. Functional Classification (FunCat) of genes performed based on MIPS (http://bbc.botany.utoronto.ca/ntools/cgi-in/ntools_classification_superviewer.cgi) was used to analyze the GO categories of differentially regulated genes increased by EAA (A, B) and decreased by the treatment (C, D) over two days (day 1 and day 5). Figure shows the results of normalizing the frequency for the number of gene in each category to the frequency of the number of genes in each category present on the ATH1 gene chip. IDs falling into classification categories other than unclassified and classification not yet clear-cut were removed from these categories. (PDF 332 KB)
Rights and permissions
About this article
Cite this article
Jithesh, M.N., Shukla, P.S., Kant, P. et al. Physiological and Transcriptomics Analyses Reveal that Ascophyllum nodosum Extracts Induce Salinity Tolerance in Arabidopsis by Regulating the Expression of Stress Responsive Genes. J Plant Growth Regul 38, 463–478 (2019). https://doi.org/10.1007/s00344-018-9861-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-018-9861-4