Abstract
MYB transcription factors play important roles in the abiotic stress response in plants, but their characteristics and functions in buckwheat (Fagopyrum tataricum) have not been fully investigated. Here, a novel R2R3-type MYB gene, designated FtMYB12, was isolated from the cultivated tartary buckwheat F. tataricum. Using quantitative real-time PCR, we found that the FtMYB12 was greatly induced by low temperature. Sub-localization and yeast transactivity assay demonstrated that the FtMYB12 gene encodes a nuclear transcription activator. Overexpression of FtMYB12 in transgenic Arabidopsis plants resulted in enhanced cold tolerance. The FtMYB12 overexpressing Arabidopsis lines showed higher root length and had elevated levels of proline content and lower levels of malondialdehyde under cold stress conditions compared to the wild-type plants. The results revealed that FtMYB12 may play an essential role in regulation of cold stress-responsive signaling in F. tataricum.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Buckwheat is an ancient dicotyledonous crop belonging to the Polygonaceae family. The most widely cultivated buckwheat species include Fagopyrum esculentum (common buckwheat) and Fagopyrum tataricum (tartary buckwheat), constituting raw material for the production of buckwheat tea, groats, flour, and noodles (Zhang and others 2012a, b). Fagopyrum tataricum is an important crop in the high mountain areas of western China and in the Himalayan hills (up to 4,500 m high-altitude areas) because of its cold resistant nature. In addition, it has a higher nutritional value, especially in respect to a high antioxidant activity in terms of its flavonoid content compared to common buckwheat (Wang and Campbell 2007; Zhang and others 2012a, b; Zhou and others 2013). It has been reported that rutin content in the seeds of tartary buckwheat is approximately 100 times higher than that found in common buckwheat (Fabjan and others 2003). Thus, more researchers focus on tartary buckwheat as a beneficial health crop for the production of nutraceutical products and functional foods (Zhou and others 2013).
Low temperature presents a major challenge in our quest for sustainable food production as it reduces the potential yields in crop plants. Surviving low temperatures leads plants to acquire mechanisms by which they can sensitively perceive freezing tolerance and regulate their physiology accordingly over a long evolutionary scale. Deciphering the mechanism by which plants perceive low temperature is of critical importance for the development of rational breeding and transgenic strategies (Zhou and others 2010a, b). Transcription factors (TFs), including the MYB family, which have been widely studied in plants, play essential roles in regulating gene expression in response to low temperature (Zhou and others 2010a, b). MYB proteins contain the conserved SANT (for SWI3, ADA2, N-CoR, and TFIIIB) DNA-binding domain (Rosinski and Atchley 1998; Jin and Martin 1999). According to the number of tandem repeats of the SANT domains, MYB proteins can be classified into three subfamilies: MYB-like proteins (MYB1R), R2R3-type MYB factors, and R1R2R3 MYB (MYB3R) factors, with one, two, and three repeats, respectively (Rosinski and Atchley 1998).
Several plant MYB factors involved in the responses to abiotic stresses such as drought, salt, and cold have been studied. In Arabidopsis thaliana, AtMYB2, AtMYB44, AtMYB102, AtMYB60, AtMYB96, and AtMYB14 function in drought or salt-stress response (Abe and others 2003; Denekamp and Smeekens 2003; Cominelli and others 2005; Agarwal and others 2006; Jung and others 2008; Seo and others 2009; Ding and others 2009), while AtMYBC1 and AtMYB14 play important roles in the plant response to cold stress (Zhai and others 2010; Chen and others 2013). Over-expression of OsMYB4, OsMYB3R-2, and OsMYB2 have been shown to increase the freezing and multiple abiotic stress tolerance of Arabidopsis (Vannini and others 2004; Dai and others 2007; Ma and others 2009; Yang and others 2012). In wheat, several MYB TFs have also been found to play important roles in response to abiotic stresses. TaMYB1 is involved in the response to hypoxia (Lee and others 2007). Over-expression of TaMYB2A and TaMYB56-B have been shown to increase the salt, drought, and cold stress tolerance of Arabidopsis (Zhang and others 2012a, b). TaMYBsdu1, TaMYB73, TaMYB3R1 and TaMYB4 function in drought and salt tolerance (Rahaie and others 2010; Cai and others 2011; He and others 2012; Al-Attala and others 2014). Recently, the identification of MYB factor genes from other plants and their roles in abiotic stress tolerance have been attempted. Over-expression of chrysanthemum CmMYB2 in Arabidopsis significantly increased tolerance to drought (Shan and others 2012). Over-expression of sheepgrass LcMYB1 in Arabidopsis improved salt tolerance (Cheng and others 2013). In addition, over-expression of apple MdoMYB121 and MdSIMYB1 remarkably enhanced the tolerance to high salinity, drought, and cold stress in transgenic plants (Cao and others 2013; Wang and others 2014). However, not much is known about MYB factors involved in abiotic stress from buckwheat, and more attempts at isolating MYB genes and characterizing their functions are needed.
In the present study, we isolated and investigated the function of a new R2R3-MYB TF FtMYB12 from F. tataricum. We characterized its gene expression patterns, sub-cellular localization, and trans-activity. Transgenic Arabidopsis plants over-expressing FtMYB12 were evaluated for cold stress tolerance. Our data indicate that FtMYB12 may play an important role in the cold stress signaling pathway in buckwheat.
Materials and Methods
Transcriptomic Data Analyses and Isolation of the FtMYB12 Gene
The publicly available, annotated set of F. tataricum genes expressed in the flower and inflorescence was collected (Logacheva and others 2011). The conserved MYB domain DNA sequence (At5g35550 and FtMYB123L) search was employed to confirm the candidate sequences as MYB genes (Zhou and others 2013). To confirm the full length of putative cDNA sequence, FtMYB12 cDNA was amplified using the primers FtMYB12F 5′-ATG AGG AAT CCG GCG GTA-3′ and FtMYB12R 5′-TTA GAT TTC TGA TGG GAT CAA AG-3′. The amplification conditions were as follows: 94 °C for 5 min, followed by 30 cycles of 94 °C for 45 s, 57 °C for 45 s, 72 °C for 2 min, followed by a final extension step of 72 °C for 10 min. The PCR products were cloned into the pMD19-T vector and sequenced. The nucleotide and amino acid sequences were analyzed by the DNAMAN software and compared with those released in GeneBank databases by using the BLAST program.
Phylogenetic Tree
Protein sequences were aligned by clustalW and a tree was constructed with the MEGA version 5 program with the neighbor-joining (NJ) method (Saitou and Nei 1987; Tamura and others 2011).
Plant Materials and Stress Treatments
Two-week-old F. tataricum seedlings were treated under different stress conditions. Cold treatment was conducted by transferring the young trees to a growth chamber set at 4 °C under a 16 h light/8 h dark cycle (2,500 lux). For salt, drought, and UV stress treatments, seedlings were grown in soil irrigated with 100-mM NaCl, 30 % PEG6000 and exposed to UV-B (302 nm, 0.1 mW/cm2), respectively. The length of time for the above treatments was 0, 12, and 24 h. The control plants were mock-treated with water. Tissue-specific expression of the FtMYB12 genes was analyzed in 3-week-old seedlings that were separated into root, stem, and leaf tissues. All samples were quickly frozen in liquid nitrogen and stored at −80 °C until use.
Subcellular Localization and Transcriptional Activity Assay
To determine its exact sub-cellular location, FtMYB12 was combined with the N-terminal of green fluorescent protein (GFP) to yield a fused FtMYB12-GFP protein. The entire coding region of the target gene amplified by PCR was inserted into the SalI and BamHI sites of vector p163-GFP. Protoplasts were isolated from Arabidopsis cell suspension ecotype Col-0, and 10-µg plasmid DNA was introduced by polyethylene glycol (PEG)-mediated transfection as previously described (Schirawski and others 2000). The transformed cells were cultured on Murashige-Skoog (MS) medium at 28 °C for 24 h in the dark and observed under a laser confocal scanning microscope (Leica DM IRBE).
To investigate the transcriptional activity of FtMYB12, the full length coding region was inserted into the EcoRI and SalI sites of the yeast expression vector pBridge containing the binding domain (BD) of GAL4. The plasmids were introduced into the yeast strain AH109 with the reporter gene His3 and LacZ following the manufacturer’s instructions (Clontech, Palo Alto, CA, USA). Yeast cells containing pBridge empty vector were used as negative control. The colony-lift filter β-galactosidase assay was carried out according to the Yeast Protocols Handbook (Clontech Laboratories, Inc.).
Generation of Transgenic Arabidopsis
The FtMYB12 open reading frame (ORF) was PCR-amplified from F. tataricum cDNA using the primer set 5′-CGG GATC CAT GAG GAA TCC GGC GGT ATC-3′ and 5′-CGG AAT TCG ATT TCT GAT GGG ATC AAA G-3′, digested with BamHI and EcoRI and cloned into pRT101 (Töpfer and others 1987). For the construction of transgenic lines constitutively overexpressing FtMYB12, the cauliflower mosaic virus (CaMV) 35S cassette containing the FtMYB12 ORF was digested from pRT101 with SphI and cloned in pCAMBIA1300 digested with SphI. The binary vector pCAMBIA1300-FtMYB12 was introduced into the Agrobacterium tumefaciens strain LBA4404. Arabidopsis ecotype Col-0 plants were transformed using the floral dip method. Seeds were surface-sterilized by incubation for 1 min in 70 % ethanol, 15 min in 50 % bleach, and five rinses with sterile water. Surface-sterilized seeds were grown on plates containing MA medium supplemented with 0.6 % w/v agar. Transgenic plants were selected on solid MA medium containing 100 mg/L timentin and 50 mg/L hygromycin. Transgenic plants from T2 generations were selected on MA medium (Masson and Paszkowski 1992) containing only 50-mg/L hygromycin. Following stratification for 3 days at 4 °C, seeds were incubated at 25 °C in a growth chamber under the 16 h light/8 h dark cycle (2,500 lux). Positive homozygous lines were test for expression of FtMYB12 by RT-PCR.
Determination of Physiological and Biochemical Properties
The malondialdehyde (MDA) content was determined as previously described (Zhou and others 2012). The proline content was calculated as described by Bates and others (1973).
Quantitative Real-Time PCR
Total RNA was extracted from pulverized frozen tissue by phenol/chloroform extraction followed by overnight precipitation with 2 M lithium chloride, washed with 70 % ethanol, and resuspended in water. Reverse transcription was carried out using the M-MuLV Reverse Transcriptase (Revert AidTM First Strand cDNA Synthesis Kit, Fermentas) according to the manufacturer’s protocol. The quantitative PCR (Q-PCR) amplification was carried out as previously described (Zhou and others 2010a, b). The following primer sets for Q-PCR were used: 5′-CGT CTC CGA TGG ATG AAC TA-3′ and 5′-TGC CAG CTA TGA GAG ACC AC-3′ for FtMYB12; 5′-CGC AAG TAC CAG AAG AGC AC-3′ and 5′-GAG AGC AGA CAC AGC AGA GC-3′ for FtH3 (histone H3); 5′-CAA GGA AGG TAT CCC ACC G-3′ and 5′-TTA GAA TCC ACC ACG AAG ACG-3′ for Arabidopsis polyubiquitin gene. The primers of some genes (COR15a, DREB2A, RD29A, CBF1, CBF2, CBF3, and COR47) in Arabidopsis were previously described (Dai and others 2007). The genes encoding FtH3 and polyubiquitin were used as reference genes in buckwheat and Arabidopsis, respectively. Relative gene expression was quantified using the comparative threshold cycle (CT) method (Zhou and others 2010a, b).
Statistical Analysis
Data were analyzed using the Student’s t test. A P value of <0.05 was considered to be significant.
Results
Identification and Phylogenetic Analysis of FtMYB12
To identify putative MYB genes that may be closely involved in the cold response in buckwheat, two MYB genes At5g35550 and FtMYB123L as probes were screened in publicly available transcriptomic data (Logacheva and others 2011). According to a set of unigenes with annotation as MYB TFs, more than 5 full length MYB genes were cloned (data not shown). Of them, the gene (FtMYB12) encoded R2R3-type MYB factor, which clusters with the OsMYB4 factor involved in freezing tolerance in rice (Fig. 1a) (Vannini and others 2004). The FtMYB12 gene encodes a protein of 171 amino acid residues with a predicted molecular mass of 19.46 kDa and a calculated pI of 10.32. FtMYB12 has an open reading frame of 516 bp (GenBank Accession No. KJ586579, Supplemental Fig. 1). Genomic PCR products amplified by primers designed from the 5′ and 3′ untranslated region revealed that FtMYB12 had 1 intron of 88 bp. Analyses of the primary structure revealed that FtMYB12 contains two imperfect MYB repeats within its N-terminal region that correspond approximately to the R2 and R3 MYB repeats (Fig. 1b). The highly conserved W residues, which are thought to be involved in the folding of the DNA-binding domain, are present in FtMYB12. In addition, the short linker sequence between the two MYB repeats also displays amino acid conservation (Zhou and others 2013).
Phylogeny and alignment of FtMYB12. a Phylogenetic tree shows the relationships of FtMYB12 to MYB factors from other plants, the tree is based on alignment of complete protein sequences (AtMYB2 (AT2G47190), AtMYB44 (AT5G67300), AtMYB96 (AT5G62470), AtMYB15 (AT3G23250), AtMYB60 (AT1G08810), AtMYB102 (AT4G21440), OsMYB4 (D88620), TaMYB73 (JN969051), TaMYB1 (DQ353858), and TaMYB2 (AY615199)). b Alignment of R2 and R3 SANT domains. Identical amino acid residues are shown by stars and conserved amino acid residues of SANT domain motifs are indicated by arrows
FtMYB12 Localizes in Nuclei and has Trans-Activation Activity
The transient expression assay using Arabidopsis protoplasts showed that FtMYB12 fused to the GFP was localized in the nucleus (Fig. 2a). The control GFP protein was distributed throughout the cell, as expected (Fig. 2a). These results confirm that FtMYB12 is a nuclear protein. The transactivation assay indicated that all transformants grew well on the transformant selective SD/Trp plates (Fig. 2b), whereas only transformants of pBridge that were fused with FtMYB12 grew on the transactivation selective SD/Trp-/Ade-/His plates (Fig. 2b), showing that FtMYB12 activated the transcription of reporter genes Ade and His in the genome of the yeast AH109 strain. Consistently, the β-galactosidase activity assay indicated that the β-galactosidase units of the transformant with pBridge-FtMYB12 were higher by about fivefold than that of pBridge (Fig. 2c), demonstrating that FtMYB12 also activated the reporter gene LacZ in the genome of AH109. These results demonstrate that FtMYB12 is a nuclear transcription activator.
Subcellular localization and transactivation assay of FtMYB12. a Transiently expressed FtMYB12 in Arabidopsis protoplasts shows its subcellular localization. Bars 10 µm. b Transactivation assay of FtMYB12. The full length of FtMYB12 was fused with pBridge, and transformed yeasts were selected on both SD-Trp and SD-Trp-His-Ade media, respectively. c β-galactosidase activity assay (Miller units). The mean value is from three independent measurements, and error bars indicate the standard variation. pBridge was used as a negative control
Expression of FtMYB12 is Activated by Cold Stress
The regulation of FtMYB12 expression by various abiotic stresses was investigated by subjecting two-week-old buckwheat seedlings to drought (30 % PEG6000), cold (4 °C), salt (100 mM NaCl), and UV-B treatments. The FtMYB12 mRNA accumulated quickly in response to cold and reached its maximal level at 12 h (Fig. 3a). In addition, expression of FtMYB12 was also induced by PEG and salt treatment, but the highest levels of transcription occurred at 12 h (Fig. 3a). To further study FtMYB12 gene expression patterns, we used QRT-PCR analysis with RNA from roots, stems, and leaves. The result indicated that FtMYB12 was found to be ubiquitous in all tissues but the highest level was in young stems (Fig. 3b). Taken together, these results suggest that FtMYB12 is induced under cold, salt, and drought stimulation, which suggests that it functions during these stresses.
Expression of FtMYB12 as assessed by QRT-PCR. a The expression of FtMYB12 in response to various stresses. b The expression pattern of FtMYB12 in buckwheat root, stem, and leaf. FtH3 was used as an internal control. The data were normalized to wild-type or root (set at 1) using the comparative threshold cycle method. Error bars represent the standard deviation of triplicate runs for QRT-PCR
Cold Tolerance of Transgenic Arabidopsis
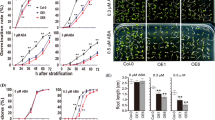
Since FtMYB12 was mainly induced by cold, its role in cold tolerance in Arabidopsis was explored by the gain-of-function approach. Transgenic Arabidopsis plants were developed with the FtMYB12 gene under the control of the CaMV35S promoter. Overall, nine lines of transgenic Arabidopsis were positively confirmed by PCR (data not shown). From the nine transgenic lines, we selected two higher FtMYB12 expression lines (OX#3 and OX#9) for further analysis (Supplemental Fig. 2). The transgenic plants (T2) showed no morphological difference in the vegetative and floral tissues, as compared to wild-type (WT) plants (data not shown). To examine the freezing tolerance of transgenic Arabidopsis, we tested the effect of low temperature on seedling root growth. The primary root lengths of the WT and transgenic lines were relatively consistent at room temperature (25 °C) (Fig. 4a, b). However, when seedlings were grown at low temperature (4 °C), the root length of the WT lines was significantly retarded compared to transgenic lines (Fig. 4a, b). The cold treatment caused root growth retardation in both WT and transgenic plants but the effect was more pronounced in WT seedlings. These results suggested that overexpression of FtMYB12 can improve cold tolerance in transgenic plants.
Stress response of Arabidopsis transgenic plants overexpressing FtMYB12. a and b Phenotypic response to low temperature, three-day-old seedlings grown at room temperature (25 °C) and at low temperature (4 °C), 14 days before the images shown were taken and primary root growth was detected. c and d MDA and proline contents were at room temperature and low temperature. Each experiment was repeated three times. Error bars represent the standard error of triplicate analyses. b, c, d The significance of difference between indices of WT and each OX was calculated using Student’s t test (*P < 0.05)
To cope with low temperature, the primary effects of stress are a reduction in cellular membrane damage and accumulation of proline content. We discovered that cold stress decreased MDA content and increased proline content. The levels of MDA content and proline content in lines FtMYB12 overexpressing Arabidopsis plants did not show much difference as compared with those in control plants under normal growth conditions (Fig. 4c, d). However, when seedlings were grown at low temperature (4 °C), after 14 days of cold stress, the MDA content in WT plants was higher than that in transgenic plants (Fig. 4c), whereas the proline content of the WT lines was significantly retarded compared to transgenic lines (Fig. 4d). The result suggested that the cold tolerance of the transgenic line was enhanced effectively because of the expeditious accumulation of the substance content compared to control plants (Zhou and others 2012).
FtMYB12 Promotes Transcription of COR15a in Arabidopsis
To elucidate the molecular mechanism of FtMYB12 in the cold response, we monitored the expression of cold-responsive genes identified in the regulated pathways by QRT-PCR analysis. Under cold treatment for 14 days, the tested marker genes, including COR15a, DREB2A, RD29A, CBF1, CBF2, CBF3, and COR47 (Fig. 5), showed significant induction in both WT and transgenic plants which is consistent with previous studies (Stockinger and others 1997; Dai and others 2007). However, under room temperature (25 °C), the expression of COR15a in FtMYB12 overexpressed transgenic plants was substantially higher than that in wild-type plants (Supplemental Fig. 3), whereas there was no significant induction in expression of DREB2A, RD29A, CBF1, CBF2, CBF3, and COR47 in transgenic or wild-type plants (Fig. 5). These results indicate that the induction strength of the COR15a gene expression can be correlated to the transcription abundance of transgenic FtMYB12 (Supplemental Fig. 3).
Expression patterns of stress-responsive genes in WT and transgenic Arabidopsis using QRT-PCR. Total RNA was extracted from 17-d-old plants grown at room temperature (25 °C) and low temperature (4 °C) for 14 days, respectively. Transcript levels measured by QRT-PCR of COR15a (a), DREB2A (b), RD29A (c), CBF1 (d), CBF2 (e), CBF3 (f), and COR47 (g). The polyubiquitin gene was used as an internal control. Data represent means and standard error of three replicates
Discussion
In plants, TF including the MYB family function in various pathways to confer stress tolerance. Several plant MYB factors involved in the responses to cold stress have been studied, such as AtMYBC1 (Zhai and others 2010), AtMYB14 (Chen and others 2013), OsMYB4 (Vannini and others 2004), and OsMYB3R-2 (Dai and others 2007). In this study, we identified a cold-inducible R2R3 MYB TF, FtMYB12, from cold-tolerance buckwheat. Sub-localization, β-galactosidase, and transactivation assays indicate that FtMYB12 is a transcription activator (Fig. 2). FtMYB12 clusters with R2R3 type MYB proteins of other plants (Fig. 1a), indicating that FtMYB12 is a R2R3 type MYB transcription factor. In addition, FtMYB12 can be identified on the basis of conserved SANT domains, which are typically located in the N-terminal region and critical for DNA-binding activity (Fig. 1b) (He and others 2012). This domain generally consists of up to four imperfect amino acid sequence repeats (R) of about 52 amino acids, each forming three α-helices, and form a helix–helix–turn–helix motif, and are involved in transcriptional regulation mechanisms (Dubos and others 2010).
Usually, TFs are induced rapidly during the early phase of the response to abiotic stresses, reach maximal induction at several hours, and then decrease in expression level (Dai and others 2007). For example, CBF1 and CBF3 showed peak induction at 6 h in wild-type plants and CBF2 showed peak induction at 3 h (Gong and others 2002), whereas TaMYB73 showed peak induction at 0.5 h during stress treatment (He and others 2012). However, the transcript level of FtMYB12 increases after cold treatment for 12 h and gradually accumulates within 24 h. Furthermore, FtMYB12 is induced by drought and salt stress (Fig. 3a), which is dissimilar to OsMYB4 involved in cold stress in rice that was induced only by cold stress (Vannini and others 2004). This is the first report, to our knowledge, showing that an R2R3- type MYB from buckwheat is involved in cold, drought, and salt stress.
TFs are involved in the response to stress in plants which, in turn, facilitate the ability to cope with these stresses. For example, cold-, salt-, and drought-induced OsMYB3R-2 promote tolerance to these stresses in Arabidopsis (Dai and others 2007), salt- induced TaMYB73 promotes tolerance to this stress in Arabidopsis (He and others 2012). Here, the over-expression of FtMYB12 increased the tolerance of Arabidopsis to cold stress (Fig. 4). Previous reports showed that COR15a and DREB2A are involved in cold stress signaling by CBF/DREB1 pathways (Artus and others 1996; Liu and others 1998). However, our results reveal that FtMYB12 only activates the expression of COR15a but not DREB2A and CBFs (Fig. 5). Thus, overexpression of FtMYB12 increases expression of COR15a which is involved in plant tolerance by the CBF/DREB1-independent pathways. In addition, we found that MDA content decreased under cold stress (Fig. 4c). Leaf oxidative damage to lipids was expressed as equivalents of MDA. The increases in MDA content suggested that cold-stressed plants encountered cellular damage and lipid peroxidation. After 14 days of cold stress, the MDA content in transgenic plants was lower than that in WT plants (Fig. 4c). It has been reported that increased proline content is positively correlated with freezing tolerance (Ma and others 2009). Under normal growth conditions (25 °C), the levels of cellular free proline did not differ between WT and transgenic plants (Fig. 4d). In contrast, after cold treatment the levels of free proline in FtMYB12 overexpressing transgenic lines increased substantially compared with WT plants. These results were similar to the alterations observed in the transgenic rice overexpressing the freezing resistant OsMYB3R-2 (Ma and others 2009). Taken together, these results indicate that the FtMYB12 protein may play an important role under cold stress through changing a variety of physiological and biochemical processes.
Low temperature lowers plant production worldwide. The responses of plants to cold stress have evolved a variety of alterations in physiological, biochemical, and gene expression processes. Here, we cloned and characterized a novel R2R3-type MYB protein FtMYB12 localized at the nucleus and induced by cold, drought, and salt stress. The enhanced freezing tolerance of CaMV35S-FtMYB12 Arabidopsis plants reveals that FtMYB12 could mediate low-temperature signal transduction. Taken together, this report provides beneficial information for understanding of the mechanism of cold regulation in buckwheat.
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Al-Attala MN, Wang X, Abou-Attia MA, Duan X, Kang Z (2014) A novel TaMYB4 transcription factor involved in the defence response against Puccinia striiformis f. sp. tritici and abiotic stresses. Plant Mol Biol 84:589–603
Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93:13404–13409
Bates LS, Waldren R, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207
Cai H, Tian S, Liu C, Dong H (2011) Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene 485:146–152
Cao ZH, Zhang SZ, Wang RK, Zhang RF, Hao YJ (2013) Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 8(7):e69955
Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G (2013) AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biol Rep 31:87–97
Cheng L, Li X, Huang X, Ma T, Liang Y, Ma X, Peng X, Jia J, Chen S, Chen Y, Deng B, Liu G (2013) Overexpression of sheepgrass R1-MYB transcription factor LcMYB1 confers salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem 70:252–260
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132:1415–1423
Ding Z, Li S, An X, Liu X, Qin H, Wang D (2009) Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics 36:17–29
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Fabjan N, Rode J, Košir IJ, Wang ZH, Zhang Z, Kreft I (2003) Tartary buckwheat (Fagopyrum tataricum Gaertn.) as a source of dietary rutin and quercitrin. J Agric Food Chem 51:6452–6455
Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99:11507–11512
He Y, Li W, Lv J, Jia Y, Wang M, Xia G (2012) Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J Exp Bot 63:1511–1522
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146:623–635
Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, Seo YW (2007) A MYB transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol Plant 129:375–385
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Logacheva MD, Kasianov AS, Vinogradov DV, Samigullin TH, Gelfand MS, Makeev VJ, Penin AA (2011) De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genom 12:30
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150:244–256
Masson J, Paszkowski J (1992) The culture response of Arabidopsis thaliana protoplasts is determined by the growth conditions of donor plants. Plant J 2:829–833
Rahaie M, Xue GP, Naghavi MR, Alizadeh H, Schenk PM (2010) A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt tolerant and sensitive genotypes. Plant Cell Rep 29:835–844
Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46:74–83
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schirawski J, Planchais S, Haenni AL (2000) An improved protocol for the preparation of protoplasts from an established Arabidopsis thaliana cell suspension culture and infection with RNA of turnip yellow mosaic tymovirus: a simple and reliable method. J Virol Methods 86:85–94
Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151:275–289
Shan H, Chen S, Jiang J, Chen F, Chen Y, Gu C, Li P, Song A, Zhu X, Gao H, Zhou G, Li T, Yang X (2012) Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol Biotechnol 51:160–173
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Töpfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res 15:5890
Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37:115–127
Wang Y, Campbell CG (2007) Tartary buckwheat breeding (Fagopyrum tataricum L. Gaertn.) through hybridization with its Rice-Tartary type. Euphytica 156:399–405
Wang RK, Cao ZH, Hao YJ (2014) Overexpression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol Plant 150:76–87
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Zhai H, Bai X, Zhu Y, Li Y, Cai H, Ji W, Ji Z, Liu X, Liu X, Li J (2010) A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem Biophys Res Commun 394:1018–1023
Zhang L, Zhao G, Xia C, Jia J, Liu X, Kong X (2012a) Overexpression of a wheat MYB transcription factor gene, TaMYB56-B, enhances tolerances to freezing and salt stresses in transgenic Arabidopsis. Gene 505:100–107
Zhang ZL, Zhou ML, Tang Y, Li FL, Tang YX, Shao JR, Xue WT, Wu YM (2012b) Bioactive compounds in functional buckwheat food. Food Res Int 49:389–395
Zhou ML, Ma JT, Pang JF, Zhang ZL, Tang YX, Wu YM (2010a) Regulation of plant stress response by dehydration responsive element binding (DREB) transcription factors. Afr J Biotechnol 9:9255–9279
Zhou ML, Zhu XM, Shao JR, Wu YM, Tang YX (2010b) Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with MeJA/nitric oxide elicitation in Catharanthus roseus hairy root culture. Appl Microbiol Biotechnol 88:737–750
Zhou ML, Ma JT, Zhao YM, Wei YH, Tang YX, Wu YM (2012) Improvement of drought and salt tolerance in Arabidopsis and Lotus corniculatus by overexpression of a novel DREB transcription factor from Populus euphratica. Gene 506:10–17
Zhou ML, Tang Y, Zhang KX, Li FL, Yang PY, Tang YX, Wu YM, Shao JR (2013) Identification of TT2 gene from floral transcriptome in Fagopyrum tataricum. Food Res Int 54:1331–1333
Acknowledgments
This research was supported by the Key Project of Science and Technology of Sichuan, China (Grant No. 04NG001-015, ‘‘Protection and exploitation of wild-type buckwheat germplasm resource’’), the grant from the National Transgenic Program (2013ZX08005-004) and National Natural Science Foundation of China (Grant No. 31372361).
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.

344_2014_9472_MOESM1_ESM.jpg
Fagopyrum tataricum FtMYB12 sequence (red represents exon sequence, dark represents intron sequence). Supplementary material 1 (JPEG 157 kb)


344_2014_9472_MOESM3_ESM.jpg
COR15a gene expression in 9 trangenic FtMYB12 overexpressing Arabidopsis lines under room temperature (25 °C). Supplementary material 3 (JPEG 33 kb)
Rights and permissions
About this article
Cite this article
Zhou, M., Wang, C., Qi, L. et al. Ectopic Expression of Fagopyrum tataricum FtMYB12 Improves Cold Tolerance in Arabidopsis thaliana . J Plant Growth Regul 34, 362–371 (2015). https://doi.org/10.1007/s00344-014-9472-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-014-9472-7