Abstract
Key message
FtMYB18 plays a role in the repression of anthocyanins and proanthocyanidins accumulation by strongly down-regulating the CHS and DFR genes in Tartary buckwheat, and the C5 motif plays an important role in this process.
Abstract
Anthocyanins and proanthocyanidins (PAs) are important flavonoids in Tartary buckwheat (Fagopyrum tataricum Gaertn.), which provides various vibrant color and stronge abiotic stress resistance. Their synthesis is generally regulated by MYB transcription factors at transcription level. However, the negative regulations of MYB and their effects on flavonol metabolism are poorly understood. A SG4-like MYB subfamily TF, FtMYB18, containing C5 motif was identified from Tartary buckwheat. The expression of FtMYB18 was not only showed a negative correlation with anthocyanins and PAs content but also strongly respond to MeJA and ABA. As far as the transgenic lines with FtMYB18 overexpression, anthocyanins and PAs accumulations were decreased through down-regulating expression levels of NtCHS and NtDFR in tobacco, AtDFR and AtTT12 in Arabidopsis, FtCHS, FtDFR and FtANS in Tartary buckwheat hairy roots, respectively. However, FtMYB18 showed no effect on the FLS gene expression and the metabolites content in flavonol synthesis branch. The further molecular interaction analysis indicated FtMYB18 could mediate the inhibition of anthocyanins and PAs synthesis by forming MBW transcriptional complex with FtTT8 and FtTTG1, or MYB-JAZ complex with FtJAZ1/-3/-4/-7. Importantly, in FtMYB18 mutant lines with C5 motif deletion (FtMYB18-C), both of anthocyanins and PAs accumulations had recovered to the similar level as that in wild type, which was attributed to the weakened MBW complex activity or the deficient molecular interaction between FtMYB18ΔC5 with FtJAZ3/-4. The results showed that FtMYB18 could suppress anthocyanins and PAs synthesis at transcription level through the specific interaction of C5 motif with other proteins in Tartary buckwheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins and proanthocyanidins (PAs) are two important components of flavonoids, a class of plant secondary metabolites with various physiological functions that also help plants to resist adverse environmental damage (Vogt 2010; WinkelShirley 2002). Plants with high levels of anthocyanins and PAs are considered to have the better cultivation traits and economic value (Jia et al. 2020). Actually, anthocyanins and PAs can provide diverse benefits for the quality of the fruit and seed. Anthocyanins often give fruit or seed a vivid color, whereas PAs have an inhibitory effect on their decay and bitterness (Cipollini and Stiles 1993; Isabelle and Noble 2005).
In the plant flavonoid synthesis pathway, anthocyanins and PAs biosynthesis share part of common pathways, but belong to distinct branches (Kang et al. 2014). Anthocyanins is synthesized in the endoplasmic reticulum through the joint action of chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT) (Aron and Kennedy 2008). PAs is a condensation product of the flavan-3-ol subunits catechin and epicatechin, which are synthesized from direct precursors of leucoanthocyanins and anthocyanins by leucoanthocyanin reductase (LAR) and anthocyanin reductase (ANR), respectively (Jia et al. 2020).
Some transcription factors (TFs), such as myeloblastosis (MYB), basic Helix Loop Helix (bHLH) and WD repeats (WD40), are able to regulate the expression of the biosynthesis genes of anthocyanins and PAs individually or in the form of protein complex at transcriptional level (Hichri et al. 2011). Being the largest TF family, MYB TFs have been extensively studied in various plants, they are classified based on the conserved MYB domain repeats and fundamental motifs (Dubos et al. 2010; Jiang et al. 2017; Xiang et al. 2019). According to the specific C-terminal regions, the R2R3-MYB TFs, which contain two imperfect repeat sequences, are classified into 25 subgroups. Subgroup 6 R2R3-MYBs are anthocyanins-related accelerators: AtMYB75 (PAP1), AtMYB90 (PAP2), AtMYB113 and AtMYB114 in Arabidopsis (Borevitz et al. 2000; Gonzalez et al. 2010; Stracke et al. 2010, 2001; Zimmermann et al. 2010), VvMYBA1 and VvMYBA2 in grapevine (Walker et al. 2010), MdMYB10 and MdMYB110a in apple (Espley et al. 2010; Chagne et al. 2013), and PpMYB10.1 in peach (Rahim et al. 2014). The PAs-related activating R2R3-MYBs belong to subgroup 5, such as AtMYB123 (TT2) and AtMYB5 in Arabidopsis (Feng et al. 2009; Nesi et al. 2001), and VvMYBPA1, VvMYBPA2, VvMYB5a and VvMYBPAR in grapevine (Jochen et al. 2007; Koyama et al. 2014; Laurent et al. 2006; Deluc et al. 2008; Nancy et al. 2009). The subgroup 4 (SG4) and SG4-like TFs are anthocyanins and PAs repressors, such as AtMYB3, AtMYB4, AtMYB7 and AtMYB32 in Arabidopsis, and PpMYB18 in peach (Dubos et al. 2010; Huang et al. 2019; Jin et al. 2014; Preston et al. 2010; Zhou et al. 2019). For anthocyanins and PAs related R2R3-MYBs, the differential functional motifs can affect their specific biological functions and activities (Stracke et al. 2001).
Actually, it has been widely and well studied that MYBs are involved in the regulation of anthocyanins/PAs synthesis and the development of trichomes in plants through MYB-bHLH-WD40 ternary complex (MBW) (Kim et al. 2015; Lepiniec et al. 2010; Matsui et al. 2008; Xu et al. 2015). In Arabidopsis, the MYB (PAP1/PAP2/TT2/MYB113/MYB114)-bHLH (GL3/EGL3/TT8)-WD40(TTG1) complex could promote anthocyanins accumulation (Gonzalez et al. 2010), where TT2-TT8-TTG1 complex was able to control the accumulation of PAs in the seed coat by up-regulating the expression of DFR, LDOX, TT19, TT12, AHA10 and BAN (BANYULS) (Lepiniec et al. 2010). Moreover, the MYB proteins could also interact with jasmonate ZIM-domain (JAZ) protein, a signaling molecule induced by jasmonate acid (JA), and regulate the synthesis of anthocyanins. Through competing with the E3 ubiquitin ligase SCFCOI1 complex, Arabidopsis MYBs of AtMYB21, AtMYB24, AtMYB90, AtMYB113 and AtMYB114 were able to bind with JAZ proteins and regulate the anthocyanins synthesis related genes DFR and ANS. (Fernandez et al. 2011; Qi et al. 2011; Song et al. 2011). Thus it can be seen that the regulation of anthocyanins and PAs presents a considerable complexity and diversity in plant.
Tartary buckwheat (Fagopyrum tataricum Gaertn.) is a part of the Fagopyrum genus (Polygonaceae). It is an excellent medicinal cereal and famous for its balanced nutrient and rich flavonoid content (Gao et al. 2016; Li et al. 2012). As the content of flavonoids is an important index of the economic value and stress resistance for a crop, the regulation of flavonoid metabolism has become the focal point of Tartary buckwheat research. In previous studies, data showed that MYB TFs have the diversified biological effects on the synthesis of Tartary buckwheat flavonoids. For example, the activator FtMYB1 and FtMYB2 could improve anthocyanins and PAs accumulation by up-regulating the expression of DFR and TT12 genes in tobacco and Arabidopsis (Bai et al. 2014; Luo et al. 2017); the suppressor FtMYB11, FtMYB13 and FtMYB14 could inhibit the accumulation of anthocyanins and flavonols by inhibiting the expression of DFR and FLS in Tartary buckwheat (Zhang et al. 2018; Zhou et al. 2017a); the suppressor FtMYB8 and FtMYB15 could inhibit the accumulation of anthocyanins and promote the accumulation of flavonols by inhibiting the expression of DFR and TT12 in tobacco and Arabidopsis (Huang et al. 2019; Luo et al. 2017). In addition, it has been reported that anthocyanins synthesis related TFs FtMYB11, FtMYB13 and FtMYB14 were able to regulate plant metabolism by responding to JA (Zhang et al. 2018). However, the reported Tartary buckwheat MYB inhibitors usually down-regulate anthocyanins biosynthesis and enhance the accumulation of flavonols, but anthocyanins/PAs-specific inhibitor has not been reported yet.
In this study, we report a R2R3-MYB TF from Tartary buckwheat, designated FtMYB18 (NCBI GenBank Accession No.: MK990568). The molecular features of FtMYB18 were characterized by bioinformatics analysis, qRT-PCR, yeast-monohybrid technology and stimulation factors. Using transgenic technology, the biological function and target genes of FtMYB18 were identified. The functional motif C5 and interaction proteins of FtMYB18 were evaluated in both yeast and plant. This work found a specific TF inhibitor to anthocyanidins and PAs from Tartary buckwheat and provided an interesting insight to the biosynthesis of flavonoids in this crop.
Materials and methods
Plant materials and growth conditions
The wild-type (WT) Tartary buckwheat seeds (Xiqiao No. 2) were obtained from Professor Anhu Wang of Xichang University. The WT tobacco seeds (NC89) were provided by Professor Jinwen Zhang of Gansu Agricultural University. The WT Arabidopsis thaliana ecotype Columbia-0 (Col-0) seeds were kept in our laboratory. The Tartary buckwheat seeds were germinated in the greenhouse with a light period of 16 h. The hormone concentrations and stress conditions used on eight-day-old Tartary buckwheat seedlings were as follows: 2 mM MeJA, 100 µM ABA, UV-B illumination (302 nm, 0.1 mW/cm2) and 4 ℃. In this study, there are three biological repeats in each experiment, and each biological repetition has three technical repeats.
Analysis of the FtMYB18 promoter
The total RNA of Tartary buckwheat was obtained by EASYspin Plant RNAiso reagent (Aidlab, China). Obtain Tartary buckwheat cDNA through used PrimeScript™ RT Reagent Kit (TaKaRa, Japan). The DNAquick Plant System (Tiangen, China) was used to obtain genomic DNA. The cDNA of FtMYB18 (FtPinG0009308100.01.T01) and the 5′ upstream sequence of PFtMYB18 were obtained from the Tartary buckwheat genomic data (Zhang et al. 2017), then cloned using specific primers (PYFP-FtMYB18F/R and PFtMYB18F/R, Supplementary Table S3). In addition, the C5 (TLLLFR) motif in the FtMYB18 C-terminus was cut off to obtain the cDNA of FtMYB18-C. The cDNA sequence alignment and phylogenetic tree construction was achieved with the DNAMAN software and Mega5 software, respectively. The PFtMYB18 sequence (Supplementary Table S3) was analysised in the PlantCARE database.
GUS analysis FtMYB18 promoter under environment effect
The plant expression vector pBI101-PFtMYB18-GUS was constructed using primer pairs PBI-PFtMYB18-F/R (Supplementary Table S3), and transiently transfected into 45-day-old Tartary buckwheat leaves using Agrobacterium tumefaciens strain GV3101 (Gandhi et al. 1999). After a 2-day co-culture, the Tartary buckwheat leaves were kept on ½ MS medium at 4 ℃ and with UV-B illumination (302 nm, 0.1mW/cm2), or the leaves were transferred to ½ MS plates containing 0.8 μM ABA or 2 mM MeJA. The leaf disks (diameter 1 cm) were obtained by punching machine for GUS staining.
FtMYB18 transcriptional activity and subcellular localization assays
The CDS (coding sequence) of FtMYB18 and FtMYB18-C was inserted into the ADH1 promoter region of the pBridge vector to investigate the transcriptional activity by β-galactosidase staining (Dai et al. 2012). The pBridge-FtMYB1 (Bai et al. 2014) (positive control), pBridge (empty vector as negative control), the pBridge-FtMYB18 and pBridge-FtMYB18-C plasmids were introduced into yeast AH109 cells according to the method of Gietz et al. (1992), and positive yeast colony will grow on SD/-Trp-His medium.
The CDS of FtMYB18 or FtMYB18-C was inserted into the pCHF3-YFP vector kept in our laboratory to obtain the pCHF3-FtMYB18-YFP or pCHF3-FtMYB18-C-YFP recombinant vectors. Then, Agrobacterium cells containing these plasmids were transiently transformed into onion epidermal cells and YFP fluorescence was observed by fluorescence microscope as previously described in proven report (Liu et al. 2009).
Generation of transgenic plants overexpressing FtMYB18 and FtMYB18-C
The cells of Agrobacterium tumefaciens strain GV3101 containing pCHF3-FtMYB18-YFP and pCHF3-FtMYB18-C-YFP vectors were used to transform Tartary buckwheat, tobacco and Arabidopsis (Huang et al. 2019; Xuan et al. 2016). Transgenic T3 homozygous Arabidopsis plants and tobacco plants were selected on MS medium containing 50 μg/mL kanamycin (Huang et al. 2019); the FtMYB18-overexpressing Tartary buckwheat hairy root lines were selected in ½ MS liquid medium with 50 μg/mL kanamycin (Xuan et al. 2016).
Determination of anthocyanins and PAs content in transgenic plants
The anthocyanins and PAs were extracted from transgenic tobacco flowers, Arabidopsis seedling and Tartary buckwheat hairy root using method as described in the literature (Luo et al. 2017; Yao et al. 2017). The anthocyanins and PAs contents were measured as previously reported (Luo et al. 2017), and three repeats were set for each experimental group.
Yeast two-hybrid and three-hybrid experiments
Yeast two-hybrid was achieved as reported earlier (James et al. 1996) to screen for interaction of FtMYB18 with FtTT8, FtGL3, FtEGL3 and FtTTG1 (Huang et al. 2019). Briefly, the FtMYB18 or FtMYB18-C CDS were recombined into the pGADT7 prey vector, and the FtTT8, FtGL3, FtEGL3 (bHLH) and FtTTG1 (WD40) CDS into the pGBKT7 bait vector. The co-transformation experiment of pGADT7 and pGBKT7 was carried out using yeast strain AH109 (Clontech). The control groups were that NC1, wild-type AH109 cells; NC2, empty pBridge plasmid.
To investigate the formation of MBW complex, a yeast three-hybrid method was performed as described by Tao et al. (2017). FtTT8 and FtTTG1 CDS were cloned into the pBridge vector (Clontech) to producefusions with the GAL4 DNA-binding domain (BD) and Met promoter, respectively; and transformed into yeast strain Y187. The pGADT7-FtMYB18 and pGADT7-FtMYB18-C vectors were co-transformed or double transformants were selected on SD/-Met/-Leu/-Trp medium; the positive protein interaction yeast was selected on selective SD/-Met/-Leu/-Trp/-His/-Ade medium, for which a serial dilution analysis was performed. The primers used are shown in Supplementary file1 Table S2.
Transient determination of anthocyanins and PAs in tobacco leaves
Dual-LUC assays in tobacco were performed according to the method of Huang et al. (2019). The CDS of FtMYB18/FtMYB18-C were inserted into the pGreenII 62-SK vector, the promoter of FtCHS and FtDFR into the pGreenII0800LUC vector. The pGreenII 62-SK vector was used as an effector to act on the pGreenII0800LUC vector. The transformed leaves were sprayed with 0.1 m fluorescein and soaked, then placed in the dark for 6 min for luminescence detection. The LUC image was detected and the luminous intensity was quantized under the imaging device (Nightowl II LB983 combined with Indigo software). Three repetitions were measured in each experimental group.
The instantaneous color determination experiment was performed on 3-week-old tobacco leaves (Zhou et al. 2019). It has been reported that the complex of TT8 and TTG1 can promote anthocyanins/PAs accumulation (Baudry et al. 2006). So, the Agrobacterium cultures containing pGreenII0800LUC-FtMYB18, pGreenII0800LUC-FtTT8 and pGreenII0800LUC-FtTTG1 were mixed with a ratio of 1:1:1 to test the repression of FtMYB18. The mixed culture of A. tumefaciens was injected into the back of tobacco leaves (Jiang et al. 2017; Xiang et al. 2019). The seedlings were placed in the dark for 12 h, and then moved to the greenhouse (25 ℃; 16 h/8 h light/dark). The photos were obtained 7 days after infiltration. 0.2 g of infiltrated tobacco leaves were used to determine the anthocyanins and PAs contents (Wei et al. 2009; Zhou et al. 2019).
Quantitative real-time PCR analysis
Total RNA was extracted from fresh plant materials. Quantitative real-time PCR (qRT-PCR) was performed using the TB Green PreMix Ex Taq II kit (Tli RNAseH Plus) and the CFX96 RT-PCR machine (Bio-Rad, USA), following the manufacturer's instructions. The PCR amplification procedure was as follows: 40 cycles at 95 ℃ for 45 s, 95 ℃ for 16 s and 58 ℃ for 45 s. The analysis of each sample was repeated three times. Relative expression level was computed using 2−ΔΔCt method (Luigi and Faggioli 2011). The primers used in qRT-PCR analysis are shown in Supplementary file1 Table S2.
Statistical analysis
The obtained data, such as anthocyanins and PAs content and qRT-PCR result data were statistically analyzed by SPSS software (SPSS13.0). Statistical significance was evaluated by Duncan's test.
Results
FtMYB18 is highly expressed in Tartary Buckwheat seeds
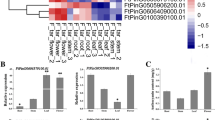
The expression levels of SG4 and SG4-like R2R3-MYB subfamily genes (Zhang et al. 2017), which were widely demonstrated to regulate anthocyanins and PAs biosynthesis, were visualized as a heat map during the whole growth period of Tartary buckwheat (Fig. 1a). The results showed that a SG4-like R2R3-MYB, FtMYB18 (FtPinG0009308100), expressed higher in young seeds, but lower in the pre-filling stage. To avoid differences in gene expression among different varieties, the FtMYB18 gene expression was analysed in “Xiqiao No. 2”, a main cultivar of Tartary buckwheat in Sichuan Province, P. R. China. At the full-bloom stage (s4 stage), FtMYB18 expression was significantly higher in the seeds (Fig. 1b), which is consistent with the transcript group data of the Tartary buckwheat genome (Zhang et al. 2017). Furthermore, the anthocyanins and PAs content were very low at the s4 stage (Fig. 1c , d). Thus, we speculate that FtMYB18 expression was opposite trend with plant anthocyanins and PAs accumulation.
Analysis of FtMYB18 expression and anthocyanins/PAs content in Tartary buckwheat. a Heat map of SG4 and SG4-like R2R3-MYBs. FtPinG0000941300, FtPinG0002366200, FtPinG0004494100, FtPinG0007642100 and FtPinG0008794300 belong to the subgroup 4 (SG4); FtMYB8, FtPinG0004848100, FtPinG0007101500, FtMYB15, FtMYB18 belong to SG4-like. The mRNA accumulation with high Z-score is represented in red color while the mRNA with low Z-score is represented in green color. PSS, seeds in prefilling stage; FSS, seeds in filling stage; MSS, seeds in mature stage; BR, basal root; RT, root tip. b Expression analysis of FtMYB18 in Tartary buckwheat whole growth period: s1, cotyledon stage; s2, 5–7 leaves period; s3, initial bloom stage; s4, full-bloom stage; s5, filling stage; s6, maturity stage. c Anthocyanins content at full-bloom stage; d PAs content at full-bloom stage. **P < 0.01, and *P < 0.05
Molecular and subcellular characterization of FtMYB18
FtMYB18 encodes a MYB protein with 243 amino acids. Two introns (145–228 and 359–525 bp) and three exons (1–144, 229–358 and 359–983 bp) constitute the 983 bp FtMYB18 gene (Fig. 2a). Multiple sequence alignment based on other SG4 MYB TFs and FtMYB18 protein had two classical MYB repeats and a bHLH motif at its N-terminal (Grotewold et al. 2001), while two conserved SG4-MYB motifs (C1 and C2) (Stracke et al. 2001) and a repression motif (C5) were close to its C-terminal (Dubos et al. 2010; Matsui et al. 2008) (Fig. 2b). In addition, according to the results of phylogenetic analysis, FtMYB18 was clustered in the SG4 group and shared high homology with AtMYB6, AtMYB8 and AtMYB32 (Cavallini et al. 2015), which worked as suppressors in anthocyanins biosynthesis. In multiple species relationship comparison, it can be seen that SG4-Like MYB has a unique C5 motif, while FtMYB18 has closely related to VvMYBC2-L1 and VvMYBC2-L2 (Fig. 2c).
Molecular characteristic Analysis of FtMYB18. a A schematic diagram of the structure of FtMYB18. b Multiple sequence alignment. The R2 and R3 SANT repeat domains are indicated with gray boxes, and the conserved motifs are marker by black rectangles. c The R2R2-MYB TFs Phylogenetic relationships of Tartary buckwheat and Arabidopsis thaliana (upper panel). The detailed phylogenetic tree for Fagopyrum tataricum SG4s and SG4-likes MYBs with the known SG4s MYBs from other species (lower panel). GenBank accession numbers are provided in Supplementary1 Table S5. d Transactivational activity detection of FtMYB18 and FtMYB18-C in AH109 yeast cells (FtMYB1, positive control; NC1, wild-type AH109 cells; NC2, empty pBridge plasmid). e Localization of FtMYB18 and FtMYB18-C in onion epidermal cells
According to the research of Matsui et al. (2008), the C5 motif plays an important role in the secondary metabolism of Arabidopsis thaliana, so we studied the C5 motif by creating FtMYB18-C gene (lost the C5 motif of FtMYB18). The transcriptional activity of FtMYB18 and FtMYB18-C was confirmed by a yeast two-hybrid assay (Fig. 2d). The results showed that FtMYB18/FtMYB18-C had individual transcriptional activity. In order to show the localization of FtMYB18/FtMYB18-C protein at the subcellular level, fusions with YFP were created and the cells of FtMYB18-YFP and FtMYB18-C-YFP transgenic onion were examined by confocal microscopy (Fig. 2e). Both proteins showed accumulation in the nucleus, suggesting that FtMYB8/FtMYB18-C could be nuclear proteins.
Molecular characterization of the FtMYB18 promoter
The FtMYB18 N-terminal upstream sequence was determined from Tartary buckwheat genome data and analyzed by PlantCare, a online software. The 2457-bp FtMYB18 promoter (PFtMYB18) sequence was obtained by PCR (supplementary file1 Table S1). Several cis-regulatory elements were identified in PFtMYB18 that may be involved in plant environmental and hormonal responses (Supplementary file1 Fig S1A, Supplementary file1 Table S3). The results showed that PFtMYB18 contains 27 cis-acting elements and classified into 15 groups. Most of them are involved in the response to the environment and hormones, such as some light-responsive cis-acting elements (GT1 motifs) and low-temperature responsiveness cis-acting elements (LTR motif). Besides, the cis-acting elements in response to phytohormone were also found in PFtMYB18 including ABRE (involved in ABA responsiveness) and CGTCA motif (involved in MeJA responsiveness). These results suggest that the activity of the PFtMYB18 promoter may be affected by UV-B, cold, ABA and MeJA.
FtMYB18 is induced by hormones and stress
To study the in vivo activities of FtMYB18 promoter under UV-B, cold, MeJA and ABA conditions, the recombinant plasmid pBI101-PFtMYB18-GUS was constructed and transformed into Tartary buckwheat leaves. By transiently expressing GUS gene, the empty vector Gus activity did not change under normal circumstances (P > 0.05) (Supplementary file1 Table S4). Under MeJA treatment, there was a significant inhibition in GUS activity already within 5 h (P < 0.05), whereas both ABA and UV-B treatment significantly induced GUS activity and GUS expression. Cold treatment did not lead to any significant changes, even after 15 h of treatment (P > 0.05) (Supplementary file1 Figure S1).
In Tartary buckwheat, the transcript-level of FtMYB18 showed a significant raise and reached the maximum at 6 h (P < 0.05) after cold and UV-B conditions, and then decreased to near pre-stress levels (Fig. 3). After ABA treatment, the expression of FtMYB18 increased rapidly to a maximum within the first two hours (P < 0.01), then dropped and maintained a higher abundance than that before treatment (P < 0.05). In addition, FtMYB18 expression increased sharply within 0.5 h after MeJA treatment (P < 0.05), and then gently decreased to the pre-treatment level.
Response of FtMYB18 expression to environmental factors in Tartary buckwheat. FtH3 was used as internal reference gene. The level of FtMYB18 expression at 0 h is set to "1". The average value is calculated repeatedly by three times of technology, and ± SD represents the error. **P < 0.01, and *P < 0.05
In a word, the above experiments showed that the response of PFtMYB18 to hormone was obvious, but the significant change of PFtMYB18 response was limited to a specific time under stress treatments.
The C5 motif is necessary for FtMYB18 function
To clarify the connection between FtMYB18 and anthocyanins and PAs biosynthesis, FtMYB18-overexpressing lines of tobacco and Arabidopsis were generated and verified by qRT-PCR (Supplementary file1 Figure S2 and Figure S3). The FtMYB18-overexpressing tobacco lines showed a lighter petal pigmentation compared with the wild-type (WT) (Fig. 4a). Similarly, for FtMYB18-overexpressing Arabidopsis seedlings grown on MS medium supplemented with 3% sucrose, the bottom of leaf buds seemed to be lighter than those from WT lines (Fig. 4b), which may reveal that there were less anthocyanins in FtMYB18-overexpressing plants compared with WT lines.
Overexpression of FtMYB18 reduces the anthocyanins and PAs content in transgenic tobacco and Arabidopsis plants. a Flower petal pigmentation of transgenic tobacco. b Color of transgenic Arabidopsis seedlings. c Seed coat pigmentation of transgenic Arabidopsis before (upper panel) and after vanillin-HCl staining (lower panel). d Anthocyanins, PAs and rutin contents in transgenic tobacco flowers. e Anthocyanins, PAs and rutin contents in transgenic Arabidopsis plants. f Detection of transcriptional levels of flavonoid biosynthetic genes in transgenic tobacco flowers (left panel) and Arabidopsis seedlings (right panel). The mRNA accumulation of WT tobacco flower/Arabidopsis seedlings is expressed as "1". Ntβ-actin gene and Atactin2 gene are used as reference gene in tobacco and Arabidopsis, respectively. The average value is calculated repeatedly by three times of technology, and ± SD represents the error. *P < 0.05, and **P < 0.01
To confirm these visual observations, total anthocyanins and PAs levels were determined. The anthocyanins content of the FtMYB18-overexpressing lines was significantly lower than that of the WT tobacco/Arabidopsis (P < 0.05); the PAs content was suppressed in FtMYB18-overexpressing Arabidopsis (P < 0.05), but no significant difference was observed with the WT tobacco flowers (P > 0.05); besides, the rutin (Fig. 4d, e) contents were no significant difference with the WT tobacco/Arabidopsis (P > 0.05) as well as the other flavonols (Supplementary file1 Figure S2C and Figure S3C). There was also no significant difference between the FtMYB18-C-overexpressing and WT plants, suggesting that the C5 motif is necessary to regulate anthocyanins and PAs accumulation. The seed coat colour of FtMYB18-overexpression transgenic lines was lighter than that of WT Arabidopsis neither direct observation nor vanillin-HCl staining (Fig. 4c). However, the color phenotypes of FtMYB18-C-overexpressing lines were similar to those of WT, which indicated the important role of C5 motif in the FtMYB18 protein again.
In order to explore the effects of FtMYB18/FtMYB18-C overexpression on anthocyanins and PAs biosynthesis, some flavonoid biosynthetic genes were monitored at the transcriptional levels (Fig. 4f). The expression levels of the early biosynthetic genes (EBGs, including CHS, CHI and F3′H) and the late biosynthetic genes (LBGs, including DFR and ANS) in the flavonoids synthesis branch (Dubos et al. 2008) were significantly decreased in FtMYB18-overexpressing tobacco plants (P < 0.05). Surprisingly, only AtDFR and AtTT12 expression decreased significantly in transgenic Arabidopsis with FtMYB18 (P < 0.05) when the expression level of AtBAN, a PAs inhibition related gene, increased remarkably (P < 0.05). Seen from the above, FtMYB18 had different effects on the expression level of flavonoid metabolism-related genes in tobacco and Arabidopsis due to different target genes in different species.
In contrast, FtMYB18-C-overexpressing tobacco and Arabidopsis plants only showed a significant difference in expression levels of DFR. We can conclude that the loss of the C5 motif will weaken FtMYB18′s inhibitory function.
The anthocyanins and PAs levels in FtMYB18-overexpressing Tartary buckwheat plants are decreased
To accurately elucidate the effect of FtMYB18 on Tartary buckwheat plants, FtMYB18- and FtMYB18-C-overexpressing Tartary buckwheat hairy root lines were generated and verified by qRT-PCR (Supplementary file1 Figure S4). The pigment deposition in WT hairy roots was greater than that in FtMYB18 transgenic hairy roots, but similar to FtMYB18-C transgenic hairy roots (Fig. 5a). The hairy root proliferation rate of FtMYB18 plants was lower than that of the WT after 30 days (Fig. 5b). These results indicate reduced anthocyanins accumulation and inhibited root development in the FtMYB18 transgenic Tartary buckwheat hairy roots compared with the WT roots. The flavonoids content analysis indicated that the anthocyanins and PAs accumulation was significantly lower in FtMYB18-overexpressing hairy roots than in WT (P < 0.05); while the rutin (Fig. 5c) content showed no significant difference with the WT (P > 0.05) as well as the other flavonols (Supplementary file1 Figure S4C). However, there was only a slight reduction in flavonoids content in hairy root lines with FtMYB18-C-overexpression (Fig. 5c) (P > 0.05).
Overexpression of FtMYB18 and FtMYB18-C in Tartary buckwheat hairy root. a Phenotypic observation. b Total weight of hairy root. Multiplication coefficient = the weight after proliferation/the weight before proliferation. c Anthocyanins, PAs and rutin contents detection. The average value is calculated repeatedly by three times of technology, and ± SD represents the error. *P < 0.05. d, e Expression detection of flavonoid pathway genes. The mRNA accumulation of WT Tartary buckwheat is expressed as "1". We used FtH3 gene as experiment reference genes. The average value is calculated repeatedly by three times of technology, and ± SD represents the error. *P < 0.05
At the transcription level, the expression of FtCHI, FtF3′H and FtFLS in transgenic lines showed no significant difference compared with those in WT Tartary buckwheat hairy roots (P > 0.05), but FtCHS and FtDFR expression was significantly decreased (Fig. 5d) (P < 0.05). As same as the Arabidopsis and tobacco, the expression levels of these genes in FtMYB18-C-overexpressing hairy roots were close to those in WT lines.
FtMYB18 suppresses anthocyanins and PAs biosynthesis through transcriptional repression of anthocyanins and PAs biosynthetic genes
In order to confirm the suppressive effect of FtMYB18 on anthocyanins and PAs pathway gene transcription, the promoter region of two candidate anthocyanins and PAs biosynthetic genes, FtCHS and FtDFR, was used for the dual luciferase assay system in transiently transformed tobacco leaves. As shown in Fig. 6, FtMYB18 significantly suppressed the luminescence intensity of PFtCHS/PFtDFR:LUC compared with the empty control (P < 0.05), whereas FtMYB18-C only slightly suppressed the luminescence intensity (P > 0.05). The results show that FtCHS and FtDFR were the target genes of FtMYB18/FtMYB18-C.
FtMYB18 inhibits FtCHS and FtDFR expression in transient expression assays. a Representative images of N. benthamiana leaves are shown at 48 h after infiltration. 62SK means the empty pGreenII 62SK vector. b Quantitative analysis of luminescence intensity. The average value is calculated repeatedly by three times of technology, and ± SD represents the error. *P < 0.05
The C5 motif is not necessary for FtMYB18 to form the MBW complex
Sequence analysis showed that FtMYB18 contains a conserved bHLH binding motif (Fig. 2b). It has been established that the regulation of flavonoid biosynthesis by MYBs dependents on forming MBW complexes (Lepiniec et al. 2010; Nesi et al. 2001). To identify with which bHLH and WD40 proteins MYB18 interacts, yeast two-hybrid and three-hybrid assays were performed with three bHLH genes (FtGL3, FtEGL3 and FtTT8) and one WD40 gene (FtTTG1) (Huang et al. 2019). We showed that FtMYB18 can interact with FtTT8 and FtTTG1, and FtMYB18 can form an MBW complex with FtTT8-FtTTG1 (Supplementary file1 Figure S6). The absence of C5 does not affect the formation of the MBW complex in yeast, but according to the results of the gradient dilution experiment, the C5 motif will weaken the activity of MBW after 10–3 dilution (Fig. 7a, b).
Molecular interaction identification of FtMY18 and FtMY18-C. a FtMYB18 and FtMYB18-C interact with FtTT8, FtGL3, FtEGL3, and FtTTG1 in yeast. FtMYB18 or FtMYB18-C was fused to the pGADT7 vector, and FtTT8, FtGL3, FtEGL3, or FtTTG1 was fused to the pGBKT7 vector. SD/-Trp-Leu medium was used to screen the transformation for plasmids, and SD/-Leu-Leu-His-Ade medium was used to screen the transcriptional activation of HIS3 gene. Growth was monitored after 7 days. Yeast cells transformed with the empty plasmids pGADT7 and pGBKT7 were used as controls. b FtMYB18 and FtMY18-C interact with FtTT8-FtTTG1 to form MBW complex. FtMYB18 or FtMY18-C was fused to the PGADT7 vector. -FtTT8/-FtTTG1/-FtTT8 + FtTTG1 was fused to the pBridge vector, respectively. SD/-Met-Trp-Leu medium was used to screen the transformation for plasmids, and SD/-Met-Ade-His-Trp-Leu medium was used to screen the transcriptional activation of the HIS3 gene. Positive hybrid yeast was screened by Met-free medium, and its activity was detected by dilution gradient of 1, 10–1, 10–2 and 10–3. Yeast cells transformed with the empty plasmids pGADT7 and pGBKT7 were used as controls. c Detection of anthocyanins/PAs content in transiently transformed tobacco leaves. SK represents the empty vector. The average value is calculated repeatedly by three times of technology, and ± SD represents the error. **P < 0.01
We next verified if C5 is necessary for the function of the MBW complex. Tobacco leaves co-infiltrated with TT8, TTG1 and FtMYB18 expression vectors produced a weak pigmentation (Fig. 7c), whereas that with TT8, TTG1 and FtMYB18-C resulted in a moderate pigment accumulation. Meanwhile, both of their pigmentations were slightly weaker than that of the controls co-infiltrated with SK-TT8-TTG1 or TT8-TTG1. The further analysis showed that anthocyanins/PAs was highly accumulated in the control groups mentioned above, followed by FtMYB18-C-TT8-TTG1 in amount (P > 0.05), while lower in FtMYB18-TT8-TTG1 group (P < 0.05) (Fig. 7c). These findings suggest the C5 motif is necessary for FtMYB18 involved in the suppression of the anthocyanins synthesis, although there is no effect on binding activity of FtMYB18-C with the other MBW complex members.
C5 could affect the formation and function of the FtMYB18–FTJAZ complex
Previous studies demonstrated that JAZ proteins are involved in plant anthocyanins accumulation (An et al. 2015; Liu et al. 2017; Wen et al. 2018). In order to verify the role of FtMYB18 in the JA signal transduction pathway, seventeen JAZ genes were screened from Tartary buckwheat flowering transcriptome (data not shown). Seven JAZ genes with high expression (Supplementary file1 Table S1) were obtained and used to verify their interaction with FtMYB18 (Fig. 8a). The results indicated that FtMYB18 could interact with FtJZA1, FtJZA3, FtJZA4 and FtJZA7 but FtMYB18-C could only interact with FtJZA1 and FtJZA7 (Fig. 8b).
Molecular interaction identification of FtMYB18/FtMYB18-C with FtJAZ proteins. a Heat map of FtJAZs. The color intensity of the blue and red rectangle reflects low and high Z-scores for mRNA accumulation, respectively. b FtMYB18 and FtMY18-C interact with FtJAZ1, FtJAZ3, FtJAZ4 and FtJAZ7 in yeast. Images showing the growth of transformed AH109 yeasts on SD/-Trp-Leu medium and SD/-Leu-Trp-His-Ade medium. c. Detection of anthocyanins content in transiently transformed tobacco leaves. The average value is calculated repeatedly by three times of technology, and ± SD represents the error. *P < 0.05
Investigation of anthocyanins in tobacco leaf disc revealed that FtMYB18 + FtJAZ1 and FtMYB18 + FtJAZ7 did not cause a significant decrease in anthocyanins accumulation compared with FtMYB18-C + FtJAZ1 and FtMYB18-C + FtJAZ7 (P > 0.05). FtMYB18-C + FtJAZ3 and FtMYB18-C + FtJAZ4 caused a significant increase in anthocyanins accumulation compared with FtMYB18 + FtJAZ3 and FtMYB18 + FtJAZ4 (P < 0.05) (Fig. 8c). This suggests that deletion of the C5 motif disable FtMYB18 to interact with some JAZ proteins, which could promote anthocyanins accumulation.
Discussion
FtMYB18 as a special suppressor and its violent response to hormones
Clarifying the negative regulatory mechanism of anthocyanins/PAs synthesis will help reveal the fundamental reason of this phenomenon in Tartary buckwheat (Cui and Wang 2012; Yuwei et al. 2017). Previous studies have shown that MYB SG4 subfamily plays an important role in the anthocyanins/PAs synthesis and is widely involved in stress response (Stracke et al. 2001). Based on the transcriptome analysis of Tartary buckwheat, five SG4 MYBs and five SG4-like MYBs (Huang et al. 2019; Zhang et al. 2017) were obtained. Among them, FtMYB18 has typical molecular structure characteristics of SG4 subfamily TFs, such as C1, C2 and C5 conservative motifs, but there are few reports about the C5 motif specific mechanism at present. The correlation between tissue-specific expression of FtMYB18 and anthocyanins/PAs accumulation at flowering stage suggest that FtMYB18 might be involved in anthocyanins and PAs biosynthesis. Besides, The FtMYB18 expression could be affected by abiotic factors, which is similar to other SG4 MYB proteins, such as more anthocyanins will be accumulated when plant was treated by ABA/cold/UV-B/MeJA conditions, while the repressor AtMYB7 (Kim et al. 2015), VvMYBC2-L1 (Cavallini et al. 2015), and FtMYB11 (Zhou et al. 2019) was activated in these conditions. Meanwhile, the FtMYB18 expression was reduced after 0.5 h of ABA and MeJA treatments, which may imply that FtMYB18 has an inhibition function in anthocyanins synthesis.
Functional identification of FtMYB18 through overexpression in plants
Overexpression FtMYB18 inhibited the accumulation of anthocyanins and PAs in transgenic plants, which was similar to previously reported MYB repressors, such as strawberry FaMYB1 (Aharoni et al. 2001), poplar PtMYB182 (Yoshida et al. 2015), Medicago MtMYB2 (Jun et al. 2015), apple MdMYB16 (Linwang et al. 2011), and grapevine VvMYBC2-L2 (Zhu et al. 2018). The further phenotype analysis indicated that similar biologic effects of FtMYB18 on flavonoid metabolites were observed in different host species, although there were different target genes among them. In general, FtMYB18 could inhibit anthocyanins and PAs accumulation by down-regulation of DFR and CHS expression while there were no significant effects on flavonol content and FLS expression. Actually, it has been proven that anthocyanins content was decreased due to the down-regulation of CHS, DFR and ANS expression in petunia with PhMYB27 overexpression, but there were no significant change in rutin content and FLS expression (Albert et al. 2014). Moreover, PtrMYB182 could down-regulate the expression of CHS, F3H and DFR to inhibit the synthesis of anthocyanins and PAs in poplar hairy roots, while there were no significant effects on rutin accumulation and FLS expression (Yoshida et al. 2015). Thus it can be seen that FtMYB18, PhMYB27 and PtrMYB182 have similar regulation patterns of flavonoids biosynthesis. To sum up, we speculate that FtMYB18 could specifically inhibit the anthocyanins and PAs synthesis by down-regulating DFR expression. In addition, FtMYB18 could hinder the early products synthesis of flavonoid pathway by down-regulating CHS expression, which may be a reason why there is no significant change in flavonol content.
Possible mechanism behind the negative regulation of anthocyanins and PAs biosynthesis by FtMYB18
Generally, many MYB transcription factors are involved in the biosynthesis of anthocyanins and PAs alone or through MBW ternary complexes in plants (Wenjia et al. 2014). For example, strawberry suppressor FaMYB1 could not only independently exercise its inhibitory function, but also form complexes with GL3/EGL3 to inhibit anthocyanins accumulation by down-regulation of DFR and ANS expression (Aharoni et al. 2001). AtCPC, an anthocyanin-related suppressor in Arabidopsis, can inhibit the CHS, CHI, DFR and ANS expression, it also can form a complex with EGL3/GL3(bHLH) and TTG1(WD40) to hinder the anthocyanins accumulation (Tominagawada et al. 2014). In this study, protein interaction experiments reveal that FtMYB18 could decrease the anthocyanins and PAs accumulation by forming a complex with FtTT8 and FtTTG1. Since FtTT8 is an important member of anthocyanin-activated MBW complex (Huang et al. 2019), we speculate that the FtMYB18 may inhibit anthocyanins synthesis by competitive binding FtTT8 to interfere with the formation of the MBW activation complex or form the MBW complex with an active repressive action ( Schwinn et al. 2016).
Meanwhile, the metabolism of flavonoids could be regulated by JA signaling pathway in plants (Yao et al. 2018). GhMYB25-like could form a complex with GhJAZ2 to inhibit lignin synthesis and to reduce flavonoids accumulation in cotton (Hu et al. 2016). Actually, through competitive binding with JAZ protein in JAZ-bHLH complex, MYB-JAZ complex could be formed to suppress anthocyanins synthesis (Qi et al. 2011). Similar to AtMYB21/AtMYB24 reported previously (Song et al. 2011), FtMYB18 could interact with FtJAZ1/-3/-4/-7 and obviously suppress anthocyanins/PAs synthesis in this study. Furthermore, FtTT8 would be released from JAZ-bHLH complex because of JAZ degradation mediated by SCFCOI1 in JA signaling pathway. After that, FtMYB18 could competitively bind with FtTT8 to form a suppressive MBW complex for anthocyanins synthesis. The detailed mechanism of FtMYB18 in JA acid signal pathway still needs more studies to be identified in the future.
The C5 motif play an important role in FtMYB18 regulation anthocyanins and PAs biosynthesis
It has been reported that C5 motif (TLLLFR) was found in some MYB transcription factors, such as AtMYBL2 (Matsui et al. 2008), VvMYBC2-L1/VvMYBC2-L3 (Cavallini et al. 2015), PtrMYB182 (Yoshida et al. 2015) and PpMYB18 (Zhou et al. 2019). However, the detailed role of C5 motif in biological effects of MYB has not been well investigated. So far, the only PtrMYB182 mutant with TLLLFR motif deletion has been confirmed to diminish the inhibitory activity of MYB134-bHLH131-MYB182 complex on anthocyanins synthesis in poplar (Yoshida et al. 2015). In this study, C5 motif deletion could weaken the inhibiting effects of FtMYB18 on anthocyanins and PAs synthesis mediated by MBW complex and FtMYB18-JAZ complex. Therefore, we believed that the C5 motif plays a crucial role in the biological function of FtMYB18.
Conclusions
In this study, FtMYB18, a transcription factor gene isolated from Tartary buckwheat, belongs to the SG4 subfamily. The expression level of FtMYB18 is negatively correlated with the accumulation of anthocyanins and PAs. FtMYB18 could decrease the accumulation of anthocyanins/PAs in transgenic plants due to decreases in the expression of CHS and DFR genes. Furthermore, FtMYB18 achieves its biological functions by forming MYB-bHLH-WD40 ternary complex (MBW) or interacting with JAZ proteins. Particularly, the deletion of C5 motif could weaken the inhibitory effects of FtMYB18 on anthocyanins/PAs synthesis by reducing the MBW complex activity and removing its interaction with JAZs. We have proposed a potential working model for FtMYB18 (Fig. 9).
Schematic diagram of FtMYB18 function. The expression of FtMYB18 gene is slightly enhanced by cold and UV-B, and suppressed by ABA and MeJA. FtMYB18 could compete with other MYB proteins to form MYB-TT8-TTG1 complexes, which leads to a lower anthocyanins/PAs accumulation through down-regulating CHS and DFR expression. Besides, FtMYB18 may inhibit the synthesis of anthocyanins/PAs by interacting with JAZ protein. Particularly, the C5 motif deletion could weaken the biological function of FtMYB18
Abbreviations
- ABA:
-
Abscisic acid
- ANS:
-
Anthocyanidin synthase
- CHI:
-
Chalcone isomerase
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol reductase
- EBGs:
-
Early biosynthetic genes
- F3′H:
-
Flavonoid 3′-hydroxylase
- FLS:
-
Flavonol synthase
- LBGs:
-
Late biosynthetic genes
- MBW:
-
MYB-bHLH-WD40
- MeJA:
-
Methyl jasmonate
- PAs:
-
Proanthocyanidins
- SG4:
-
Sub-group 4
- SG5:
-
Sub-group 5
- SG6:
-
Sub-group 6
- SG7:
-
Sub-group 7
- SPSS:
-
Statistical product and service solutions
- TFs:
-
Transcription factors
- TT12:
-
Transparent testa 12
- UFGT:
-
UDP-flavonoid glucosyltransferase
- X-gal:
-
5-Bromo-4-chloro-3indolyl β-d-galactopyranoside
- X-α-Gal:
-
5-Bromo-4-chloro-3-indoxyl a-Dgalactoside
- JAZ:
-
Jasmonate ZIM-domain
- HCl:
-
Hydrochloric acid
- WT:
-
Wild type
References
Aharoni A, Vos CHRD, Wein M, Sun Z, Greco R, Kroon A, Mol JN, OConnell AP (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28(3):319–332
Albert NW, Davies KM, Lewis DH, Zhang H, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26(3):962–980
An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ (2015) MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol 56(4):650–662
Aron PM, Kennedy JA (2008) Flavan-3-ols: Nature, occurrence and biological activity. Mol Nutr Food Res 52(1):79–104
Bai Y, Li CL, Zhang JW, Li SJ, Luo XP, Yao HP, Zhao HX, Park S, Wu Q (2014) Characterization of two Tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis. Physiol Plant 152(3):431–440
Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46(5):768–779
Borevitz JO, Xia YJ, Blount JW, Dixon RA, Lamb CJ (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12(12):2383–2393
Cavallini E, MatusJT FL, Zenoni S, Loyola R, Guzzo F, Schlechter RO, Ageorges A, Arcejohnson P, Tornielli GB (2015) The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol 167(4):1448–1470
Chagne D, Linwang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle C, Kumar S, Silva ND, Micheletti D, Mcghie TK, Crowhurst RN, Storey R, Velasco R, Hellens RP, Gardiner SE, Allan AC (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161(1):225–239
Cipollini ML, Stiles EW (1993) Fruit rot, antifungal defense, and palatability of fleshy fruits for frugivorous birds. Ecology 74(3):751–762
Cui XD, Wang ZH (2012) Optimum reaction conditions for rutin-hydrolysis enzyme (RHE) from Tartary buckwheat seeds and inhibitory effect of cu~(2+) on its activity. Food Sci 26(4):271–276
Dai X, Wang Y, Yang A, Zhang WH (2012) OsMYB2P1, an R2R3 MYB transcription factor, is involved in the regulation of phosphate-starvation responses and root architecture in rice. Plant Physiol 159(1):169–183
Deluc LG, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon J, Robinson SP, Barrieu F (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol 147(4):2041–2053
Dubos C, Gourrierec JL, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul J, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55(6):940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lc L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Espley R, Hellens R, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2010) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49(3):414–427
Feng LS, Milliken ON, Pham H, Seyit R, Napoli RS, Preston J, Koltunow AM, Parish RW (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21(1):72–89
Fernandez P, Chini A, Fernandezbarbero G, Chico J, Gimenezibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Francozorrilla JM, Pauwels L, Witters E, Puga MI, Pazares J, Goossens A, Reymond P, Jaeger GD, Solano R (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23(2):701–715
Gandhi R, Maheshwari SC, Khurana P (1999) Transient gene expression and influence of promoters on foreign gene expression in Arabidopsis thaliana. Vitro Cell Dev Biol Plant 35(3):232–237
Gao F, Zhao HX, Yao HP, Li CL, Chen H, Wang AH, Park SU, Wu Q (2016) Identification, isolation and expression analysis of eight stress-related R2R3-MYB genes in Tartary buckwheat ( Fagopyrum tataricum ). Plant Cell Rep 35(6):1385–1396
Gietz D, Jean AS, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20(6):1425–1425
Gonzalez A, Zhao M, Leavitt J, Alan M (2010) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J 53(5):814–827
Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Chandler VL (2001) Identification of the residues in the MYB domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci USA 97(25):13579–13584
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62(8):2465–2483
Hu H, He X, Tu L, Zhu L, Zhu S, Ge Z, Zhang X (2016) GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J 88(6):921–935
Huang Y, Wu Q, Wang S, Shi JQ, Dong QX, Yao PF, Shi GN, Xu SX, Deng RY, Li CL, Chen H, Zhao HX (2019) FtMYB8 from Tartary buckwheat inhibits both anthocyanin/proanthocyanidin accumulation and marginal trichome initiation. BMC Plant Biol 19(1):1–16
Isabelle L, Noble AC (2005) Polyphenols: factors influencing their sensory properties and their effects on food and beverage preferences. Am J Clin Nutr 81(1):330–335
James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient Two-Hybrid selection in yeast. Genetics 144(4):1425–1436
Jia Y, Selva C, Zhang YJ, Li B, Mcfawn LA, Broughton S, Zhang XQ, Westcott S, Wang PH, Tan C, Angessa TT, Xu YH, Whitford R, Li CD (2020) Uncovering the evolutionary origin of blue anthocyanins in cereal grains. Plant J 101(5):1057–1074
Jiang Y, Liu C, Yan D, Wen X, Liu Y, Wang H, Dai J, Zhang Y, Zhou B, Ren XL (2017) MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J Exp Bot 68(5):1055–1069
Jin H, Cominelli E, Bailey P, Parr AJ, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2014) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19(22):6150–6161
Jochen B, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143(3):1347–1361
Jun JH, Liu C, Xiao X, Dixon RA (2015) The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation inmedicago truncatula. Plant Cell 27(10):2860–2897
Kang YL, Pei J, Cai WL, Liu W, Wu QH (2014) Research progress on flavonoid metabolic synthesis pathway and related function genes in medicinal plants. Chin Trad Herb Drugs 45(9):1336–1341
Kim JH, Hyun WY, Nguyen HN, Jeong CY, Xiong L, Hong SW, Lee H (2015) AtMYB7, a subgroup 4 R2R3 MYB, negatively regulates ABA-induced inhibition of seed germination by blocking the expression of the bZIP transcription factor ABI5. Plant Cell Environ 38(1):559–571
Koyama K, Numata M, Nakajima I, Gotoyamamoto N, Matsumura H, Tanaka N (2014) Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J Exp Bot 65(15):4433–4449
Laurent D, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde J, Merillon J, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140(2):499–511
Lepiniec L, Baudry A, Weisshaar B, Heim RA, Dubreucq B, Caboche M (2010) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39(3):366–380
Li CL, Bai YC, Li SJ, Chen H, Han XY, Zhao HX, Shao JR, Park SU, Wu Q (2012) Cloning, characterization, and activity analysis of a flavonol synthase gene FtFLS1 and its association with flavonoid content in Tartary buckwheat. J Agric Food Chem 60(20):5161–5168
Linwang K, Micheletti D, Palmer JW, Volz RK, Lozano L, Espley RV, Hellens RP, Chagne D, Rowan DD, Troggio M, Iglesias I, Allan AC (2011) High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ 34(7):1176–1190
Liu HY, Feng DR, Liu B, Yan-Min HE, Wang HB, Wang JF (2009) Studies on subcellular localization of mpasr in onion epidermal cells mediated by Agrobacterium. J Trop Subtrop Bot 17(3):218–222
Liu XJ, An XH, Liu X, Hu DG, Wang XF, You CX, Hao YJ (2017) MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J Exp Bot 68(11):2977–2990
Luigi M, Faggioli F (2011) Development of quantitative real-time RT-PCR for the detection and quantification of Peach latent mosaic viroid. Eur J Plant Pathol 130(1):109–116
Luo XP, Zhao HX, Yao PF, Li QQ, Huang YJ, Li CL, Chen H, Wu Q (2017) An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and proanthocyanidins from Tartary buckwheat. J Plant Growth Regul 37(1):76–84
Matsui K, Umemura Y, Ohme TM (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55(6):954–967
Nancy T, Laurent T, Agnès A, Sandrine V, Clotilde V, Véronique C, Charles R (2009) Ectopic expression of VvMYBPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149(2):1028–1041
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13(9):2099–2114
Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2010) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40(6):979–995
Qi T, Song SH, Ren QG, Wu DW, Huang H, Chen Y, Fan M, Peng W, Ren CM, Xie DX (2011) The Jasmonate-ZIM-Domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23(5):1795–1814
Rahim MA, Busatto N, Trainotti L (2014) Regulation of anthocyanin biosynthesis in peach fruits. Planta 240(5):913–929
Schwinn KE, Ngo H, Kenel F, Brummell DA, Albert NW, Mccallum J, Pitherjoyce M, Crowhurst RN, Eady C, Davies KM (2016) The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front Plant Sci 9(7):1865–1875
Song S, Qi TC, Huang H, Ren QC, Wu DW, Chang CQ, Peng W, Liu YL, Peng JR, Xie DX (2011) The Jasmonate-ZIM Domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23(3):1000–1013
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456
Stracke R, Ishihara HG, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2010) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50(4):660–677
Tao T, Zhou CJ, Wang Q, Chen XR, Han CG (2017) Rice black streaked dwarf virus P7–2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS ONE 12(5):177–185
Tominagawada R, Wada T (2014) Regulation of root hair cell differentiation by R3 MYB transcription factors in tomato and Arabidopsis. Front Plant Sci 5(1):91–102
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3(1):2–20
Walker AR, Elizabeth L, Jochen B, Mcdavid DAJ, Thomas MR, Robinson SP (2010) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J 49(5):772–785
Wei Z, Ning G, Lv H, Liao L, Bao M (2009) Single MYB-type transcription factor AtCAPRICE: a new efficient tool to engineer the production of anthocyanin in tobacco. Biochem Biophys Res Commun 388(4):742–747
Wen JF, Li Y, Qi TC, Gao H, Liu B, Zhang M, Huang H, Song SH (2018) The C-terminal domains of Arabidopsis GL3/EGL3/TT8 interact with JAZ proteins and mediate dimeric interactions. Plant Signal Behav 13(1):23–32
Wenjia X, Grain D, Bobet S, Gourrierec JG, Thevenin J, Kelemen Z, Lepiniec L, Dubos C (2014) Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol 202(1):132–144
WinkelShirley B (2002) Biosynthesis of flavonoids and effects of stress current. Opin Plant Biol 5(3):218–223
Xiang L, Liu X, Li H, Yin X, Grierson D, Li F, Chen K (2019) CmMYB#7, an R3 MYB transcription factor, acts as a negative regulator of anthocyanin biosynthesis in chrysanthemum. J Exp Bot 70(12):3111–3123
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20(3):176–185
Xuan H, Jingwen Y, Yangyang Z, Dengfeng X, Xue J, Ziqin X (2016) Efficient rutin and quercetin biosynthesis through flavonoids-related gene expression in Fagopyrum tataricum Gaertn. hairy root cultures with UV-B irradiation. Front Plant Sci 7(4):63–76
Yao PF, Zhao HX, Luo XP, Gao F, Li CL, Yao HP, Chen H, Park SU, Wu Q (2017) Fagopyrum tataricum FtWD40 functions as a positive regulator of anthocyanin biosynthesis in transgenic tobacco. J Plant Growth Regul 36(3):755–765
Yao N, Jing L, Zheng H, Chen M, Shen Y (2018) Research progress of jasmonate-responsive transcription factors in regulating plant secondary metabolism. China J Chin Mater Med 43(5):897–903
Yoshida K, Ma D, Constabel CP (2015) The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol 167(3):693–710
Yuwei Z, Li J, Yuan Y, Gu J, Chen P (2017) Purification, characterization and partial primary structure analysis of rutin-degrading enzyme in Tartary buckwheat seeds. Chin J Biotechnol 33(5):796–802
Zhang LL, Ma B, Gao Q, Du HL, Han YH, Li Y, Cao YH, Qi M, Zhu YX, Lu HW, Ma MC, Liu LL, Zhou JP, Nan CH, Qin YJ, Wang J, Cui L, Liu HM, Liang CZ, Qiao ZJ (2017) The Tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol Plant 10(9):1224–1237
Zhang K, Logacheva MD, Meng Y, Hu JP, Wan DP, Li L, Janovska D, Wang ZY, Georgiev MI, Yu Z, Yang FY, Yan ML, Zhou ML (2018) Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum. J Exp Bot 69(8):1955–1966
Zhou M, Sun ZM, Ding MQ, Logacheva MD, Kreft I, Wang D, Yan ML, Shao JR, Tang YX, Wu YM, Zhu XM (2017a) FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytol 216(3):814–828
Zhou H, Linwang K, Wang FW, Espley RV, Ren F, Zhao JB, Ogutu C, He HP, Jiang Q, Allan AC, Han YP (2019) Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol 221(4):1919–1934
Zhu Z, Li G, Liu L, Zhang Q, Han Z, Chen X, Li B (2018) A R2R3-MYB transcription factor, VvMYBC2L2, functions as a transcriptional repressor of anthocyanin biosynthesis in grapevine (Vitis vinifera L.). Molecules 24(1):92–99
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2010) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins. Plant J 40(1):22–34
Acknowledgements
We thank the Annick Bleys for critical reading and editing of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Project No. 31871698).
Author information
Authors and Affiliations
Contributions
QXD and HXZ conceived the original screening and research plans; YJH, YC, MW and ZXZ carried out part of material collection, RNA extraction; CLL carried out flavonoid quantification analysis; XLW and HC performed most of the experiments; PFY analyzed the data, QXD and QW design most of the experiments and wrote the article; All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, Q., Zhao, H., Huang, Y. et al. FtMYB18 acts as a negative regulator of anthocyanin/proanthocyanidin biosynthesis in Tartary buckwheat. Plant Mol Biol 104, 309–325 (2020). https://doi.org/10.1007/s11103-020-01044-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01044-5