Abstract
Different rhizobacteria may regulate plant growth using different mechanisms, including production of signal molecules that modulate plant morphogenesis and gene expression. Iron (Fe) is an essential micronutrient for plant growth and is frequently limited in plants. Plants with Strategy I Fe-uptake systems enhance root ferric reductase (FRO) activity to promote Fe absorption. Plants with Strategy II Fe-uptake systems increase Fe absorption by phytosiderophore production. However, recent reports have shown that plants with Strategy II systems also possess FRO genes that are expressed not in roots, but in shoots. Different rhizobacteria trigger plant Strategy I Fe-uptake systems via emission of volatile organic compounds (VOCs). In this work, we show that the plant growth-promoting rhizobacterium Arthrobacter agilis UMCV2 modulates the morphogenesis and FRO transcription of Sorghum bicolor, a plant with a Strategy II Fe-uptake system, via VOC emission. We found that in a system with separate compartments, VOCs emitted by A. agilis promoted plant growth, caused an increase in chlorophyll concentration, and modified the root architecture system. We tested the effect of the pure bacterial volatile compound dimethylhexadecylamine, produced by the UMCV2 strain, on plant growth and found an increase of 1.8-fold on shoot fresh weight, shoot length, chlorophyll concentration, and lateral root number at a concentration of 8 μM. This effect was dose dependent and was comparable to the effects produced by A. agilis VOCs. Simultaneously, we analyzed SbFRO1 expression using quantitative polymerase chain reaction and found that SbFRO1 expression was strongly modulated by VOCs produced by A. agilis, specifically dimethylhexadecylamine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron (Fe) is an essential element for plant growth and development. It plays a central role in the energetic metabolism of plant cells because of its participation in the respiratory and photosynthetic electron transport chains in the mitochondria and chloroplasts, respectively (Vigani and others 2013). Although Fe is not present in the chlorophyll molecule, it is involved in the synthesis of chlorophyll (Chereskin and Castelfranco 1982; Balk and Schaedler 2014). Thus, Fe deficiency is directly related to the low concentrations of chlorophyll in plants (Abadía and others 2011).
Fe is highly abundant in soil, but its low solubility restricts its availability at neutral or alkaline pH conditions in aerobic soils to levels low enough to limit plant growth. The scarcity of Fe reduces agricultural production and causes a decline in the nutritional value of crops (Zuo and Zhang 2011). In response to this deficiency, higher plants have evolved two known strategies to acquire Fe. Plants using Strategy I acidify the root environment to improve Fe solubilization by employing an ATPase. The activity of a root ferric reductase protein (FRO) is elevated to reduce Fe(III) to Fe(II), and Fe(II) is then internalized by IRT transporters (Hindt and Gerinot 2012). A subgroup of higher plants constituted by the order Poales evolved a different mechanism to obtain Fe, namely Strategy II. Roots of plants with Strategy II Fe-uptake systems secrete compounds known as phytosiderophores that chelate Fe(III) in the rhizosphere, and the phytosiderophore–Fe complex is then internalized in the roots by YS transporters (Abadía 2011; Enomoto and Goto 2013).
Some components of the Strategy I Fe-uptake system are conserved in plants using Strategy II. The Poaceae Oryza sativa (rice) possesses OsIRT1 and OsIRT2 transporter genes, which are expressed in the roots and induced by low-Fe conditions, and the OsFRO2 gene, whose transcript is detectable only in the leaves of Fe-deficient rice (Ishimaru and others 2006). The Poaceae Hordeum vulgare (barley) and Sorghum bicolor (sorghum) also possess FRO genes that are expressed in the leaves but not in the roots (Mikami and others 2011).
Some rhizospheric bacteria (rhizobacteria) emit volatile organic compounds (VOCs), promoting growth activity in plants (Ryu and others 2003) by modulating auxin homeostasis (Zhang and others 2007), and are able to induce the activity of Strategy I components (Zhang and others 2009; Orozco-Mosqueda and others 2013a, 2013b). Arthrobacter agilis UMCV2 is a rhizobacterium that promotes the growth of the legumes (plants with Strategy I Fe-uptake systems) Phaseollus vulgaris, Medicago truncatula, and Medicago sativa. Previously, it has been shown that A. agilis is a dissimilatory Fe-reducing bacterium capable of Fe reduction in non-flooded soil, causing plants to uptake Fe (Valencia-Cantero and others 2007), thereby triggering M. sativa growth via emission of the VOC dimethylhexadecilamine (DMHDA) (Velázquez-Becerra and others 2011), and elevating components of the Strategy I Fe-uptake system by the same VOC (Orozco-Mosqueda and others 2013b).
Cereals (Poaceae) are the first source of human and livestock consumption (Alexandratos and Bruinsma 2012). Therefore, determining the mechanisms by which rhizobacteria can promote cereal development and improve their nutritional uptake is of utmost importance to increase production. In the present work, we used the plant S. bicolor as a Poaceae model with a Strategy II Fe-uptake system because of its relative ease of in vitro manipulation and its completely sequenced genome (Mace and others 2013), thus facilitating the search for genes in future studies. We tested the hypothesis that A. agilis VOCs, and specifically DMHDA, induce the growth of plants with Strategy II Fe-uptake systems in a manner similar to how they trigger the growth of plants with Strategy I Fe-uptake systems by modulating morphogenesis and up-regulating FRO gene expression on leaves, with consequent improvement in the Fe nutritional status in aerial tissues.
Materials and Methods
Plant Material and Growth Conditions
Commercially available seeds of S. bicolor var. “Emerald Green” were employed. Seeds were stored at 4 °C until use, and they were then surface sterilized as previously described (Velázquez-Becerra and others 2011). S. bicolor seeds were treated with 70 % alcohol for 8 min, washed with sterile water, and immersed in 20 % sodium hypochlorite for 2 min. They were then rinsed five times with sterile water.
Seeds were germinated in Petri dishes containing 0.2 × Murashige and Skoog medium (MS base medium) at pH 6.5, plus 6 g of agar (Phytotechnology; Shawnee Mission, KS, US) per L in a Percival growth chamber. The seeds were incubated for 16 h with light and 8 h in darkness, with a light intensity of 200 μmol m2 s−1 at 22 °C. Petri plates were placed vertically at an angle of 65° to allow root growth along the agar surface and unimpeded aerial growth of the hypocotyls. Square Petri plates with dimensions of 100 × 15 mm (Becton, Dickinson and Company NJ 07417 U.S.A. catalogue 351112) were employed. Seedlings were employed 96 h after germination to obtain embryonic axes, as modified from Peña-Uribe and others (2012) Seedlings were gently separated from the rest of their endosperm using a scalpel, and then the embryonic axes were treated with 10 % commercial sodium hypochlorite for 5 min and rinsed three times with sterile water. Finally, axenic embryonic axes were placed in Petri dishes or glass flasks with MS base medium and grown in a growth chamber.
Cocultivation of A. agilis UMCV2 and S. bicolor
We used A. agilis UMCV2 isolated from a slightly acidic soil (Valencia-Cantero and others 2007). 16S ribosomal sequence data are available in GenBank under accession numbers AY553857 and AY553858. The bacteria were stored in 30 % glycerol at −20 °C until use. The bacteria were grown routinely in nutrient agar (NA) at 25 °C. Germinating seedlings of S. bicolor were placed in Petri dishes with MS base medium plus 0.1 × NA. A. agilis UMCV2 was inoculated (or uninoculated, as controls) to form a line, and after 24 h, seedlings were placed 3 cm from the bacterial line on the plate. Controls and inoculated plants were placed in a growth chamber for 96 h for further analysis.
To investigate the role of bacterial VOCs on plant growth, embryonic axes were placed, 96 h after germination, within 170-mL glass flasks containing 25 mL of MS base medium and a vial with 5 mL of NA. Vials inside the flasks were inoculated with 0.1 mL of a suspension of A. agilis UMCV2 (approximately 1 × 107 CFU/mL) to avoid physical contact between S. bicolor and the bacteria. Other vials were filled with 10 μL of an ethanolic solution of DMHDA (Sigma-Aldrich; St. Louis, MO, USA; catalogue 40460, CAS: 112-69-6) at a concentration of 1, 10, or 100 µM, or with 10 μL of ethanol (as controls). Plants were grown in a growth chamber for further analysis.
Effect of DMHDA on Plant Growth
DMHDA was added to MS base medium just before the moment of medium solidification and mixed carefully. This medium was used to test the growth and development of S. bicolor embryonic axes. The Petri plates with plants under different treatments were placed on a Percival growth chamber for 16 h with light and 8 h in darkness at 22 °C for five days.
S. bicolor Growth Analysis
Measurements of fresh weight biomass were performed using an analytical balance after carefully removing the plants from the culture medium, whereas the length of the plants was determined on a scale (in millimeters). Chlorophyll quantification was conducted as previously described (Orozco-Mosqueda and others 2013a) using a CCM-200 chlorophyll meter (Opti-Sciences, Inc.; Hudson, NH, USA) to measure chlorophyll concentration based on the rates of transmitted radiation (940 and 660 nm) through a leaf in arbitrary units.
In Silico Search and Identification of FRO Gene Copies
To search for FRO copies, we performed a homology Basic Local Alignment Search Tool (BLAST) search by employing the previously identified OsFRO1 and OsFRO2 gene sequences (Ishimaru and others 2006) as a probe, obtained from GenBank with accession numbers AB126084 and AB126085, respectively. The BLAST searches were carried out in the database of the S. bicolor Genome Sequencing Resources using the BLASTN program at the following web pages: http://www.plantgdb.org/SbGDB and http://blast.ncbi.nlm.nih.gov. Other FRO sequences from Hordeum vulgare, Arabidopsis thaliana, and Medicago truncatula were also employed. Six putative FRO sequences were obtained with different identities. Predictions of transmembrane domains were performed using the HMMTOP 2.0 transmembrane topology prediction program at http://www.enzim.hu/hmmtop (Tusnády and Simon 2001). The obtained putative gene sequences were also analyzed to search for domains of FAD, NADPH, transmembrane regions, and the oxidoreductase signature motifs as previously described (Orozco-Mosqueda and others 2012) through the BLAST; Conserved Domain Architecture Retrieval Tool (cdart) at http://www.ncbi.nlm.nih.gov/ (Geer and others 2002).
To search for IRT copies, we performed a BLAST search employing the previously identified OsIRT1 and OsIRT2 gene sequences (Ishimaru and others 2006) as a probe, obtained from GenBank with accession numbers AB070226 and AB126086, respectively.
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 6 (Tamura and others 2013) employing the “maximum parsimony” algorithm with 500 repetitions for bootstraps calculation.
Analysis of SbFRO1 Gene Expression
Embryonic axes of S. bicolor were transferred to flasks containing MS base medium with 30 µM FeIII-ethylenediaminetetraacetic acid (EDTA) (Fe-rich) or 0.3 µM FeIII-EDTA (Fe-deficient), and vials inoculated with A. agilis UMCV2 or with DMHDA as described above. After 24 h, total RNA extraction was carried out with TRI reagent (Sigma-Aldrich). The RNA was treated with RNAase-free DNase I to remove residual genomic DNA (Mukherjee and others 2006). The RNA samples were run on a 1.2 % agarose gel and stained with GelRed (Biotium; USA) to confirm the quality of the RNA. Finally, we carried out cDNA synthesis with the Super Script First-Strand Synthesis System for reverse transcription polymerase chain reaction (RT-PCR) (Life Technologies/Gibco-BRL CA U.S.A.).
The oligonucleotides SbFRO1F (5′ TGA CAG CCT GCG TGG AAG 3′), SbFRO1R (5′ GAG CAT CAA CAG CGG AC 3′), SbAct1F (5′ TCT GGC ATC ACA CCT TCT AC3′), and SbAct1R (5′ GTA CGA CCG CTG GCA TAG 3′) were designed on the basis of the putative sequence of SbFRO1 (GenBank XM_002446494) and the S. bicolor actin sequence (GenBank X79378.1). Semi-quantitative PCR amplifications were performed with a TC-412 Techne Thermal Cycler. GoTaq® Master Mixes tubes (Promega) were used (tubes were supplied with enzyme, magnesium, dNTPs, and buffer). Only 0.1 μg of template cDNA and 50 pmol of each primer were added to each tube. The following PCR conditions were used: initial denaturation at 95 °C for 3 min, denaturation at 95 °C for 30 s, annealing at 57.5 (63) °C for 30 s, and extension at 72 °C for 25 (15) s, for 30 cycles. The RT-PCR products were analyzed by agarose gel (1.2 %) electrophoresis.
The RT-PCR products were additionally purified using the Wizard® SV Gel and PCR Clean-Up System (Promega WI 53711 U.S.A), following the manufacturer’s instructions and sequenced at the National Laboratory of Genomics for Biodiversity, CINVESTAV-Irapuato, Mexico. The sequences were employed to corroborate the homology with the SbFRO1 putative gene using the BLASTN program as described above.
For real-time PCR (qPCR), the oligonucleotides SbFRO1UP (5′TGG CGA TCT TGA GCG ACA T 3′), SbFRO1LO (5′TCT TGG GCA TGC ATT GCT T 3′), SbACTUP (5′ GGC TGA CGC CGA GGA TAT C 3′), and SbACTLO (5′ TGA CCA TAC CGG TTC CAT TGT 3′) were designed with Primer Express Software Version 3.0 by Applied Biosystems (Foster City, CA, USA). The cDNA was diluted at 1:5 to conduct standard curves. The quantification of the initial SbFRO1 template concentration was performed with the ΔΔCT method (Puckette and others 2008) using the actin gene as a control. The qPCR was performed using the iQ SYBR-Green kit (Applied Biosystems) as follows: for a 20-μL reaction, 10 μL of Master mix, 0.5 μL of 10 μM forward and reverse primers, 5 μL of cDNA template (20 ng/μL in the first dilution), and 4 μL of water were added. Samples were run and analyzed using the Applied Biosystems Real-Time PCR machine with the cycling protocol at 95 °C for 4 min, followed by 95 °C for 15 s, then 63 °C for 30 s, for 40 cycles, and for a melting curve stage at 95 °C for 15 s, 63 °C for 1 min, and then ramped to 95 °C at a rate of 0.3 °C/s. Cycle threshold (Ct) values of SbActin were subtracted from Ct values of control samples (ΔCt), and the average Ct values of the different treatment samples were subtracted from the average ΔCt value (ΔΔCt).
Statistical Analysis
All experiments were independently reproduced two to three times, including at least n = 6 per treatment. The results were analyzed using the Student’s t test when comparing two means, or with analysis of variance and Duncan’s means separation test for multiple comparisons (p < 0.05). When appropriate, the results were analyzed using a factorial design, comprising two factors with two and three levels (factor 1: exposure to A. agilis VOCs, exposure to DMHDA, or unexposed; and factor 2: Fe-rich and -deficient media), followed by Duncan’s multiple range test (p ≤ 0.05).
Results
A. agilis UMCV2 Compounds Modulate the Growth of S. bicolor
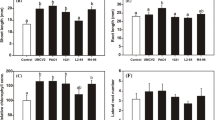
To determine the potential direct effects of A. agilis on the promotion of Poaceae plant growth, we studied the effect of A. agilis UMCV2 inoculation in S. bicolor by placing seeds over the surface of Petri plates containing solidified MS medium, and inoculated A. agilis 3 cm from the seeds. Plant fresh weight, stem length, root length, and lateral root number were determined 96 h after inoculation. Inoculation with A. agilis UMCV2 produced 60 % inhibition on the growth of primary root length, correlating to a 42 % inhibition of root biomass growth (Fig. 1c–f) when compared with uninoculated seedlings. However, no significant effects were observed on shoot biomass or chlorophyll content (data not shown), and a slightly increased shoot length (15 %) was observed when compared with controls (Fig. 1a). In this way, the ratio of shoot biomass to root biomass was increased to approximately 87 % in response to A. agilis inoculation (Fig. 1b).
A. agilis UMCV2 compounds modulate the growth of S. bicolor. A. agilis UMCV2 was inoculated 24 h before S. bicolor seedlings were placed on plates. After 96 h, shoot length (a), shoot-to-root biomass ratio (b), primary root length (c), and root fresh weight (d) were plotted. Bars represent the mean ± standard error values (n = 24). Asterisks indicate statistically significant differences (p < 0.05; Student’s t test). The images show a representative general view of the control, uninoculated system (e), and the plant–bacteria interaction system (f)
A. agilis UMCV2 VOCs Promote S. bicolor Growth
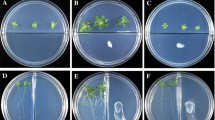
We aimed to determine whether VOCs produced by A. agilis UMCV2, including DMHDA, modulate the growth of S. bicolor (plant with Strategy II Fe-uptake system) as it does in plants with Strategy I Fe-uptake systems. To do this, we designed a system with S. bicolor embryonic axes and A. agilis UMCV2 or DMHDA in separate compartments to avoid direct plant–bacteria contact while allowing plant–bacteria interaction via the emission of VOCs. Plants exposed to A. agilis UMCV2 VOCs for 48 h showed significantly higher shoot and root biomass production compared with control plants (Fig. 2a, b). Plants exposed to volatilized DMHDA from a vial with doses of 10 µL at 1, 10, or 100 µM (Fig. 2c) also showed higher biomass production, although only at the shoots. Treatments with DMHDA induced no significant differences in the biomass production among them (Fig. 2a, b).
A. agilis UMCV2 VOCs promote S. bicolor growth. S. bicolor embryonic axes were placed in glass flasks containing 0.2 × MS medium. A. agilis UMCV2 was inoculated in vials containing NA medium. Other vials were filled with 10 μL of dimethylhexadecylamine at 1, 10, or 100 μM, and the systems were incubated. After 48 h, shoot fresh weight (a) and root fresh weight (b) were plotted. Bars represent the mean ± standard error values (n ≥ 12). Letters above bars are used to indicate means that differ significantly by a Duncan’s multiple range test (p < 0.05). The images (c) show a representative general view of S. bicolor plants exposed to the control, uninoculated system
Modulation of S. bicolor Morphogenesis by DMHDA
To observe the effect of direct application of DMHDA in the plant culture media, the embryonic axes of S. bicolor were placed in culture media mixed with the increasing concentrations of DMHDA. After 5 days of incubation, DMHDA produced clear effects on plant growth. S. bicolor plants grown with 8 µM DMHDA produced shoots that were 1.7-fold taller with 2.1-fold higher shoot biomass compared with the controls without DMHDA (Fig. 3a, b). The chlorophyll concentration also was 1.6-fold higher in plants incubated with 8 µM DMHDA compared with controls (Fig. 3c). DMHDA concentrations of 2 and 4 µM also produced plants with taller shoots, higher biomass, and higher chlorophyll concentrations compared with the control plants, but these effects were less pronounced than those observed in plants incubated with 8 µM DMHDA. An increase in the DMHDA concentration to 16, 32, and 64 µM each produced plants that were progressively smaller than those incubated with 8 µM DMHDA. Plants incubated with 64 µM DMHDA were even smaller and showed lower chlorophyll concentrations than the controls without DMHDA.
DMHDA modulates S. bicolor morphogenesis. S. bicolor embryonic axes were placed in Petri plates supplied with 0.2 × MS medium and increasing concentrations of DMHDA. After 5 d, shoot fresh weight (a), shoot length (b), chlorophyll concentration (c), root fresh weight (d), root length (e), and lateral root number (f) were plotted. Bars represent the mean ± standard error values (n ≥ 16). Letters above bars are used to indicate means that differ significantly by a Duncan’s multiple range test (p < 0.05)
In general terms, results on the radicular systems were comparable with those observed in the shoots. Only roots of plants incubated with 8 µM DMHDA showed higher biomass production and were longer than roots of control plants. The rest of the treatments produced plants that were statistically similar to controls in these parameters (Fig. 3d, e). An effect on root morphogenesis was observed on lateral roots, which were also significantly increased in plants incubated with 8 µM DMHDA. However, plants incubated with 16 µM DMHDA produced only half as many lateral roots as the control plants did, and production of lateral roots was completely inhibited in plants incubated with DMHDA at 32 or 64 µM (Fig. 3d).
Identification of SbFRO1 and Putative SbIRT Genes
Previously, a similar effect on plant growth has been shown by the emission of VOCs from A. agilis UMCV2, specifically DMHDA, where ferric chelate reductase activity and Fe uptake were induced in M. truncatula plants (Orozco-Mosqueda and others 2013b). To determine the effect of A. agilis VOCs on ferric reductase gene expression in S. bicolor, we performed an in silico search to identify the FRO gene copies of this Strategy II plant. We localized six homologous and candidate sequences to the SbFRO1 gene and proceeded to analyze the open reading frames to localize the FAD union motif, NADH union motive, oxide reductase motif, and transmembrane regions to compare with the FRO genes from other Poaceae plants (Table 1). All six sequences showed protein motifs that characterized the FRO genes, but five of them showed a reduced and unusual number of predicted transmembrane regions. Complementarily, we performed phylogenetic analysis to observe the clustering of the gene sequences of proteins with the ferric reductase domain from different protein families, such as FRE, RBOH, and FRO. We found that five out of six sequences from S. bicolor belonged to a cluster with a respiratory burst oxidase-A from Arabidopsis thaliana. Only one sequence (GenBank accession number XM_002441375.1) was grouped in a cluster with all of the FRO sequences that we included in the phylogenetic tree and outside the cluster with the FRE1 of Saccharomyces cerevisiae and RBOH of A. thaliana. Inside the FRO cluster, we observed two sub-clusters. The sequences of the FRO1 genes from Poaceae plants (including our candidate sequence) as well as the AtFRO7 and MtFRO2 genes belonged to the first sub-cluster, while the sequences of the FRO2 genes from the Poaceae plants as well as the AtFRO2 and MtFRO1 genes belonged to the second sub-cluster (Fig. 4a).
Molecular characterization of SbFRO1. A phylogenetic tree of the FRO genes and some related sequences were constructed (a) employing the “maximum parsimony” algorithm. Numbers next to branches represent bootstraps in bases to 500 repetitions. Genes that were previously characterized are highlighted in bold letters. GenBank Accession Numbers are indicated in parentheses. An RT-PCR analysis of SbFRO1 (b) was performed with total RNA isolated from the roots and shoots of control plants or plants grown under Fe deficiency. The same number of actin transcripts was detected in each of the conditions
It was established that FRO genes from Poaceae plants were expressed on shoots and may be induced by Fe deficiency (Ishimaru and others 2006). With this in mind, S. bicolor plants were grown in Fe-rich and Fe-deficient conditions. Semi-quantitative RT-PCR analysis was performed using oligonucleotides designed on our candidate sequence, and cDNA from shoots and roots was obtained. We found unique amplification only in samples from the shoots, and stronger amplification was found in samples from the plants grown in Fe-deficient compared with those grown in Fe-rich conditions (Fig. 4b). Taken together, all of these data show that the sequence XM_002441375.1 corresponded to the gene SbFRO1.
We also performed an in silico search to identify putative SbIRT genes and localized three candidate sequences. Phylogenetic analysis exhibited the SbIRT sequences clustering with gene sequences closely related to the protein family that contains the cationic transporters ZIP and IRT. Two sequences (GenBank accession numbers XM_002464064.1 and XM_002466718.1) grouped together and showed the closest relationship to the OsIRT1 and OsIRT2 genes (Fig. S1). The third sequence (XM_002464063.1) grouped closer to the ZmIRT1 gene from Zea mays. The third candidate sequence formed a cluster along with the IRT genes from Poaceae plants, and a different cluster was formed with the IRT genes from plants with Strategy I Fe-uptake systems. All the IRT gene sequences (including our candidate SbIRT genes sequences) formed a bigger cluster, whereas ZIP gene sequences formed external groups. These data are in concordance with the hypothesis that S. bicolor possesses IRT genes like those present in O. sativa (Ishimaru and others 2006) and Z. mays (Li and others 2013).
A. agilis UMCV2 VOCs Modulate Morphogenesis of S. bicolor and SbFRO1 Transcription
Embryonic axes of S. bicolor were grown in Fe-rich and Fe-deficient media and exposed for 48 h to VOCs of A. agilis UMCV2, DMHDA, or solvents (controls) in our system with separate compartments. We did not observe statistical differences at the morphological level between control plants grown in Fe-rich and in Fe-deficient media. However, plants grown in Fe-rich media and exposed to DMHDA or to A. agilis UMCV2 VOCs showed significantly higher shoot biomass production and chlorophyll concentration compared with the unexposed controls. Plants grown in Fe-deficient media and exposed to DMHDA or A. agilis VOCs showed higher values of chlorophyll concentration but not of shoot biomass compared with unexposed controls (Fig. 5a, b).
A. agilis UMCV2 VOCs modulate morphogenesis of S. bicolor. Embryonic axes were placed in glass flasks containing 0.2 × MS medium. A. agilis UMCV2 was inoculated in vials containing NA medium; other vials were filled with dimethylhexadecilamine. After 48 h of exposure to bacterial VOCs, shoot fresh weight (a), chlorophyll concentration (b), root fresh weight (c), and lateral root number (d) were plotted. Bars represent the mean ± standard error values (n = 6). Letters above bars are used to indicate means that differ significantly according to a factorial analysis of variance followed by Duncan’s multiple range test (p < 0.05)
With respect to the roots, we did not observe significant differences in the root biomass between the treatments, but the lateral root number was higher on plants grown in Fe-rich conditions and exposed to DMHDA or A. agilis UMCV2 VOCs than on the unexposed plants. Again, plants grown in Fe-deficient conditions and exposed to DMHDA or A. agilis UMCV2 VOCs showed a higher number of lateral roots than the unexposed plants, but this difference was not significant (Fig. 5c, d). Thus, iron deprivation diminished the effect of A. agilis UMCV2 VOCs on plant promotion.
SbFRO1 transcription was quantified by qPCR. A factorial analysis showed that SbFRO1 transcription was modified by Fe deficiency (p < 2.6 × 10−4), but DMHDA and VOCs produced by UMCV2 induced the clearest enhancements in the relative transcription level of the SbFRO1 gene (p < 1.0 × 10−6). Plants grown in Fe-rich conditions and exposed to VOCs from A. agilis UMCV2 showed a transcription level that was almost 11-fold higher than that of the unexposed plants (plants with basal expression level). However, plants exposed to DMHDA showed a transcription level that was 35-fold higher than that of plants with basal expression. Unexposed plants grown in Fe-deficient medium showed a transcription level that was 2.6-fold higher than that of plants with basal expression, but plants grown under conditions of Fe deficiency and exposed to VOCs from A. agilis UMCV2 or DMHDA showed SbFRO1 expression levels that were 29- and 36-fold higher, respectively, than those of plants with basal expression (Fig. 6). These data showed that Fe deprivation in the media increased the expression of SbFRO1 on the shoots, but also that the bacterial VOCs (including DMHDA) more strongly increased this expression even in Fe-rich conditions. The Fe deprivation effect seemed to be additive to the effect of the bacterial VOCs in inducing SbFRO1 transcription, as clearly observed by comparing the plants exposed to VOCs from A. agilis UMCV2 that were grown in Fe-rich conditions and those grown in Fe-deficient conditions.
A. agilis UMCV2 VOCs modulate SbFRO1 transcription. A qPCR analysis of SbFRO1 was performed with total RNA isolated from the shoots of plants grown under Fe-rich or Fe-deficient conditions and exposed or unexposed to A. agilis UMCV2 VOCs or DMHDA. After 48 h of exposure to bacterial VOCs, SbFRO1 expression levels relative to controls (Fe-rich and unexposed to VOCs) were plotted. Bars represent the mean ± standard error values (n = 6). Letters above bars are used to indicate means that differ significantly according to a factorial analysis of variance followed by Duncan’s multiple range test (p < 0.05)
Discussion
Growth promotion of plants with Strategy I Fe-uptake systems due to the emission of A. agilis UMCV2 VOCs, specifically DMHDA, has been previously established (Velázquez-Becerra and others 2011). It has also been shown that A. agilis UMCV2 VOCs triggered the acidification of the rhizospheric environment and FRO activity on roots, both of which are responses of the Strategy I Fe-uptake system (Orozco-Mosqueda and others 2013b). It is not completely clear whether a better Fe nutritional status was the only reason for the growth promotion of plants with Strategy I Fe-uptake systems by A. agilis VOCs. However, evidence strongly suggested that the induction of responses to Fe deprivation played a role in this promotion. Rhizosphere acidification and FRO activity on roots are not responses of Poaceae to Fe deprivation, but A. agilis UMCV2 was originally isolated as a plant growth-promoting rhizobacteria (PGPR) from the rhizosphere of the graminaceous plant Zea mays (Valencia-Cantero and others 2007). With this in mind, we tested the hypothesis that A. agilis UMCV2 modified the growth of the model Poaceae S. bicolor to increase the SbFRO expression on shoots via the emission of VOCs as signal molecules.
A. agilis UMCV2 Acts as a PGPR on S. bicolor via VOC Emission
First, we determined the plant growth-promoting effect of A. agilis UMCV2 by direct interaction with S. bicolor in an in vitro system. When seedlings were grown on the same plate as A. agilis UMCV2 (allowing a free interchange of diffusible molecules) at an early development time (4 days), they did not produce more shoot biomass than the control plants did, and only a slight enhancement on the shoot size was observed. Simultaneously, A. agilis UMCV2 produced very strong inhibition on the root growth. In this way, strict plant growth promotion was not observed, but in turn, inhibition of the roots was seen, although these data produced a very significant enhancement on the shoot-to-root biomass ratio. An enhancement in the plant shoot-to-root biomass ratio has been correlated with better nutritional status (Kang and van Iersel 2004; Müller and others 2000). Plant inoculation with PGPR could also lead to an enhanced shoot-to root biomass ratio with the production of diffusible plant hormones such as cytokinins (Arkhipova and others 2007; Liu and others Liu et al. 2013). We employed a separate compartment system to test the effect of A. agilis UMCV2 VOCs on the growth of S. bicolor embryonic axes, avoiding the effect of direct plant–bacteria contact and the effect of the diffusible compounds. A. agilis UMCV2 VOCs promoted the production of shoot biomass as well as root biomass, confirming that A. agilis UMCV2 acted as a PGPR on S. bicolor via VOC emission. This result also strongly suggested that A. agilis UMCV2 produced a diffusible compound capable of root inhibition in the system allowing direct interaction with S. bicolor. A comparable result was obtained in a previous work performed with the M. sativa–A. agilis UMCV2 interaction system (Velezaquez-Becerra and others 2011). The effect of the commercial VOC DMHDA was also tested, as its production by A. agilis UMCV2 has been well established (Velazquez-Becerra and others 2011; Orozco-Mosqueda and others 2013b; Velazquez-Becerra and others 2013).
DMHDA Modulates S. bicolor Morphogenesis
We placed vials of DMHDA at different concentrations in the separate compartment system, expecting plants to be exposed to the volatilized DMHDA. Although all treatments produced similar growth promotion effects on shoot biomass and no significant changes in root biomass compared with controls, we inferred that volatilized DMHDA could saturate the headspace of our system and that plants could actually have been exposed to very similar DMHDA doses. To perform a better control of the DMHDA doses to which plants were exposed, DMHDA was directly added to the plant culture media. We noticed a very different effect as the addition of 8 µM DMHDA in the media produced an enhancement of over 60 % in shoot and root biomass production, shoot length, chlorophyll concentration, and number of lateral roots. Lower DMHDA concentrations produced lower or no effects, and higher DMHDA concentrations also produced lower or no effects. Therefore, it is clear that DMHDA also modulated plant growth at low micromolar concentrations and in a dose-dependent manner, suggesting the effect of a signal molecule, as observed in M. sativa, a plant with a Strategy I Fe-uptake system (Velazquez-Becerra and others 2011).
The effects of DMHDA on the growth of lateral roots and on chlorophyll concentration are of particular interest because an enhancement in lateral root number is also a response to Fe deprivation (Long and others 2010). In addition, chlorophyll concentration is considered to be an indicator of the Fe nutritional status of plants (Abadia and others 2011). These facts suggest that DMHDA may trigger responses of Fe deficiency on a plant with a Strategy II Fe-uptake system, that is, S. bicolor, as it does on plants with Strategy I Fe-uptake systems (Orozco-Mosqueda and others 2013b).
A. agilis UMCV2 VOCs Modulate S. bicolor Morphogenesis and Induce SbFRO1 Transcription
In this work, we localized SbFRO1 on the GenBank database, showing that SbFRO1 formed a cluster with the other FRO genes. Inside the FRO cluster, two sub-clusters were formed. SbFRO1 was grouped in a sub-cluster with FRO genes that are expressed in the shoots, namely HvFRO1, OsFRO1 (from Poaceae plants), and AtFRO7 (Mikami and others 2011; Ishimaru and others 2006; Jeong and others 2008). FRO genes that are expressed in the roots, namely AtFRO2 and MtFRO1 (Wu and others 2005; Andaluz and others 2009), were grouped in the second sub-cluster with the FRO2 genes from Poaceae plants. A similar clustering pattern was found by Victoria and others (2012), who named the sub-clusters as FRO1 and FRO2, respectively. It has been well established that plants with Strategy I Fe-uptake systems, like Arabidopsis and M. truncatula, have multiple FRO genes with tissue-specific expression (Mukerje and others 2006; Wu and others 2005; Orozco-Mosqueda and others 2012); however, in Poaceae plants, expression of FRO genes has only been found in the leaves. We also found SbFRO1 expression in the shoots but not in the roots, and unlike the work of Mikami and others (2011), SbFRO1 in our study showed a discrete response to Fe deprivation. The result was not too surprising because of the extraordinary intraspecific diversity of the S. bicolor genomes (Mace and others 2013). The clustering of SbFRO1 with AtFRO7 was interesting because it was demonstrated that the expression of AtFRO7 was localized in the chloroplast. Fe acquisition is essential in the chloroplast and is required for efficient photosynthesis in young seedlings under Fe-limiting conditions (Jeong and others 2008). With respect to the SbFRO1 sequence, we performed an in silico analysis (with ProtComp v.9.0), which produced a prediction of its expression on chloroplasts (data not shown).
Even when the nutritional plant reserves were eliminated, the endosperm was retired, and embryonic axes were used, plants grown in Fe deprivation conditions did not show chlorosis symptoms, probably because of the short incubation time. A discrete induction of SbFRO1 transcription of 2.6-fold compared with the basal expression was observed in plants grown in Fe deprivation. When plants were exposed to A. agilis UMCV2 or DMHDA, enhancements in chlorophyll concentration, biomass production, and lateral root number were observed. A clear induction of SbFRO1 transcription of at least ten times the basal expression was found, and it increased until it reached 30 times on plants grown in Fe deprivation. Mikami and others (2011) reported an increased sensitivity of sorghum to Fe deprivation compared with that of barley, and that the increase was not correlated with the total Fe concentration in the leaves. This difference could be because barley might allocate Fe more efficiently from the leaves to the thylakoids compared to sorghum. This observation is consistent with our results since plant energy production depends on the efficient functioning of chloroplasts (Vigani and others 2013). An increase in plant growth may be a consequence of improved functionality of the chloroplasts, which may be caused by a better supply of Fe to the chloroplasts via SbFRO1 over-transcription.
In Arabidopsis, transcription of the AtFRO2 and AtIRT1 genes is positively regulated by the basic helix–loop–helix (bHLH) protein FIT. This transcriptional regulator interacts with AtbHLH38 and AtbHLH39 in response to low-Fe conditions (Yuan and others 2008). IRT Ferrous transporters are typical of plants with Strategy I Fe-uptake systems, but have been found in at least some Poaceae plants, where they are also positively regulated under low-iron concentrations (Kobayashi and others 2014). OsIRT1 genes are expressed in vascular tissues of rice plants, suggesting that the role of these transporters is not only to uptake Fe from soil but also to translocate it from plant roots to shoots (Kobayashi and Nishizawa 2012). Although important differences exist on the regulation of Fe-deficiency responses in Poaceae and non-Poaceae plants, there are similarities at the level of bHLH transcriptional regulators (Kobayashi and Nishizawa 2012). In this work, we identified three putative SbIRT genes (Fig. S1) in S. bicolor. It is possible that these SbIRT genes are being induced by Arthrobacter agilis UMCV2 VOCs and DMHDA just as they induce SbFRO1. According to this hypothesis, the VOCs of A. agilis UMCV2 would produce a more efficient distribution of iron in leaves towards the thylakoids via SbFRO1 overexpression, while the induction of the SbIRT transporters would help supply iron to the leaves from the roots, as it happens in plants with Strategy I Fe-uptake systems (Orozco-Mosqueda and others 2013b).
The connection between plant morphogenesis and response to Fe deprivation has been documented in Arabidopsis. It was reported that interactions between bHLH transcriptional regulators were required to induce gene expression (Yuan and others 2008; Sivitz and others 2012; Enomoto and Goto 2013). bHLH transcriptional promoters such as POPEYE were involved in not only Fe deprivation responses but also in the regulation of lateral root morphogenesis (Long and others 2010). Based on the above, we speculate that DMHDA could act at a point on the gene transduction pathway common to both phenomena.
In conclusion, this work showed that A. agilis UMCV2 VOCs, specifically DMHDA, modulated S. bicolor morphogenesis, promoted plant growth, and induced SbFRO1 transcription. To the best of our knowledge, this is the first study to elucidate these effects in a graminaceous plant by bacterial volatile compounds. We propose that A. agilis UMCV2 and DMHDA facilitate the allocation of Fe to photosynthetic apparatuses, and in this way, produce a plant growth-promoting effect on plants with Strategy II Fe-uptake systems. Current efforts in our lab are aimed at characterizing the relationship between morphogenesis modulation and regulation of responses to Fe deprivation, and detailed study at the molecular level is a perspective of this work.
References
Abadía J, Vázquez S, Rellán-Álvarez R, El-Jendoubi H, Abadía A, Álvarez-Fernández A, López-Millán AF (2011) Towards a knowledge-based correction of Fe chlorosis. Plant Physiol Biochem 49:471–482. doi:10.1016/j.plaphy.2011.01.02
Alexandratos N, Bruinsma J (2012) World agriculture towards 2030/2050: the 2012 revision. Rome FAO ESA. Working paper No. 12-03
Andaluz S, Rodríguez-Celma J, Abadía A, Abadía J, López-Millán AF (2009) Time course induction of several key enzymes in Medicago truncatula roots in response to Fe deficiency. Plant Physiol Biochem 47:1082–1088. doi:10.1016/j.plaphy.2009.07.009
Arkhipova TN, Prinsen E, Veselov SU, Martinenko EV, Melentiev AI, Kudoyarova GR (2007) Cytokinin producing bacteria enhance plant growth in drying soil. Plant Soil 292:305–315. doi:10.1007/s11104-007-9233-5
Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65:125–153. doi:10.1146/annurev-arplant-050213-035759
Chereskin BM, Castelfranco PA (1982) Effects of Fe and oxygen on chlorophyll biosynthesis II. Observations on the biosynthetic pathway in isolated etiochloroplasts. Plant Physiol 69:112–116. doi:10.1104/pp.69.1.112
Enomoto Y, Goto F (2013) Long-distance signaling of iron deficiency in plants. In: Baluška F (ed) Long-distance systemic signaling and communication in plants. Springer, Berlin, pp 167–188. doi:10.1007/978-3-642-36470-9_8
Geer LY, Domrachev M, Lipman DJ, Bryant SH (2002) CDART: protein homology by domain architecture. Genome Res 12:1619–1623. doi:10.1101/gr.278202
Hindt N, Gerinot ML (2012) Getting a sense for signals: regulation of the plant Fe deficiency response. Biochim Biophys Acta 1823:1521–1530. doi:10.1016/j.bbamcr.2012.03.010
Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK (2006) Rice plants take up Fe as an Fe3+-phytosiderophore and as Fe2+. Plant J 45:335–346. doi:10.1111/j.1365-313X.2005.02624.x
Jeong J, Cohu C, Kerkeb L, Pilon M, Connolly EL, Guerinot ML (2008) Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc Natl Acad Sci USA 105:10619–10624. doi:10.1073/pnas.0708367105
Kang JG, van Iersel MW (2004) Nutrient solution concentration affects shoot: root ratio, leaf area ratio, and growth of subirrigated salvia (Salvia splendens). HortScience 39:49–54
Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63:131–152. doi:10.1146/annurev-arplant-042811-105522
Kobayashi T, Itai RN, Nishizawa NK (2014) Iron deficiency responses in rice roots. Rice 7:27
Li S, Zhou X, Huang Y, Zhu L, Zhang S, Zhao Y, Guo J, Chen J, Chen R (2013) Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol 13:10–1186
Liu F, Xing S, Ma H, Du Z, Ma B (2013) Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl Microbiol Biotechnol 97:9155–9164. doi:10.1007/s00253-013-5193-2
Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to Fe deficiency in Arabidopsis roots. Plant Cell 22:2219–2236. doi:10.1105/tpc.110.074096
Mace ES, Tai S, Gilding EK, Li Y, Prentis PJ, Bian L et al (2013) Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun 4:2320. doi:10.1038/ncomms3320
Mikami Y, Saito A, Miwa E, Higuchi K (2011) Allocation of Fe and ferric chelate reductase activities in mesophyll cells of barley and sorghum under Fe-deficient conditions. Plant Physiol Biochem 49:513–519. doi:10.1016/j.plaphy.2011.01.009
Mukherjee I, Campbell NH, Ash JS, Connolly EL (2006) Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 223:1178–1190. doi:10.1007/s00425-005-0165-0
Müller I, Schmid B, Weiner J (2000) The effect of nutrient availability on biomass allocation patterns in 27 species of herbaceous plants. Perspect Plant Ecol 3:115–127. doi:10.1078/1433-8319-00007
Orozco-Mosqueda MC, Santoyo G, Farías-Rodríguez R, Macías-Rodríguez L, Valencia-Cantero E (2012) Identification and expression analysis of multiple FRO gene copies in Medicago truncatula. Genet Mol Res 11:4402–4410. doi:10.4238/2012.October.9.7
Orozco-Mosqueda MC, Macías-Rodríguez LI, Santoyo G, Flores-Cortez I, Farías-Rodríguez R, Valencia-Cantero E (2013a) Medicago truncatula increases its Fe-uptake mechanisms in response to volatile organic compounds produced by Sinorhizobium meliloti. Folia Microbiol 58:579–585. doi:10.1007/s12223-013-0243-9
Orozco-Mosqueda MC, Velázquez-Becerra C, Macías-Rodríguez LI, Santoyo G, Flores-Cortez I, Alfaro-Cuevas R, Valencia-Cantero E (2013b) Arthrobacter agilis UMCV2 induces Fe acquisition in Medicago truncatula (strategy I plant) in vitro via dimethylhexadecylamine emission. Plant Soil 362:51–66. doi:10.1007/s11104-012-1263-y
Peña-Uribe CA, García-Pineda E, Beltrán-Peña E, de la Cruz HR (2012) Oligogalacturonides inhibit growth and induce changes in S6 K phosphorylation in maize (Zea mays L. var. Chalqueño). Plant Growth Regul 67:151–159. doi:10.1007/s10725-012-9672-8
Puckette MC, Tang Y, Mahalingam R (2008) Transcriptomic changes induced by acute ozone in resistant and sensitive Medicago truncatula accessions. BMC Plant Biol 8(1):46. doi:10.1186/1471-2229-8-46
Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA 100:4927–4932. doi:10.1073/pnas.0730845100
Sivitz AB, Hermand V, Curie C, Vert G (2012) Arabidopsis bHLH100 and bHLH101 control Fe homeostasis via a FIT-independent pathway. PLoS ONE 7(9):e44843. doi:10.1371/journal.pone.0044843
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tusnády GE, Simon I (2001) The HMMTOP transmembrane topology prediction Server. Bioinformatics 17:849–850
Valencia-Cantero E, Hernández-Calderón E, Velázquez-Becerra C, López-Meza JE, Alfaro-Cuevas R, López-Bucio J (2007) Role of dissimilatory fermentative Fe-reducing bacteria in Fe uptake by common vean (Phaseolus vulgaris L.) plants grown in alkaline soil. Plant Soil 291:263–273. doi:10.1007/s11104-007-9191-y
Velázquez-Becerra C, Macías-Rodríguez LI, López-Bucio J, Altamirano-Hernández J, Flores-Cortez I, Valencia-Cantero E (2011) A volatile organic compound analysis from Arthrobacter agilis identifies dimethylhexadecylamine, an amino-containing lipid modulating bacterial growth and Medicago sativa morphogenesis in vitro. Plant Soil 339:329–340. doi:10.1007/s11104-010-0583-z
Velázquez-Becerra C, Macías-Rodríguez LI, López-Bucio J, Flores-Cortez I, Santoyo G, Hernández-Soberano C, Valencia-Cantero E (2013) The rhizobacterium Arthrobacter agilis produces dimethylhexadecylamine, a compound that inhibits growth of phytopathogenic fungi in vitro. Protoplasma 250:1251–1262. doi:10.1007/s00709-013-0506-y
Victoria FC, Bervald CMP, da Maia LC, de Sousa RO, Panaud O, de Oliveira AC (2012) Phylogenetic relationships and selective pressure on gene families related to Fe homeostasis in land plants. Genome 55:883–900. doi:10.1139/gen-2012-0064
Vigani G, Zocchi G, Bashir K, Philippar K, Briat JF (2013) Signals from chloroplasts and mitochondria for Fe homeostasis regulation. Trends Plant Sci 18:305–311. doi:10.1016/j.tplants.2013.01.006
Wu H, Li L, Du J, Yuan Y, Cheng X, Ling HQ (2005) Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol 46:1505–1514. doi:10.1093/pcp/pci163
Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating Fe uptake gene expression for Fe homeostasis in Arabidopsis. Cell Res 18:385–397. doi:10.1038/cr.2008.26
Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS, Paré PW (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–851. doi:10.1007/s00425-007-0530-2
Zhang H, Sun Y, Xie X, Kim MS, Dowd SE, Paré PW (2009) A soil bacterium regulates plant acquisition of Fe via deficiency-inducible mechanisms. Plant J 58:568–577. doi:10.1111/j.1365-313X.2009.03803.x
Zuo Y, Zhang F (2011) Soil and crop management strategies to prevent Fe deficiency in crops. Plant Soil 339:83–95. doi:10.1007/s11104-010-0566-0
Acknowledgments
We thank the Consejo Nacional de Ciencia y Tecnología (Mexico) for funding this work via projects 165738 (LIMR) and 128341 (EVC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Diana Yazmin Castulo-Rubio and Nancy Araceli Alejandre-Ramírez have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.

344_2015_9495_MOESM1_ESM.jpg
Phylogenetic analysis of putative SbIRT genes. A phylogenetic tree of the IRT genes and some related sequences were constructed employing the “maximum parsimony” algorithm. Numbers next to branches represent bootstrap values by performing 500 repetitions. Genes that were previously characterized are highlighted in bold letters. GenBank accession numbers are indicated in parentheses (JPEG 669 kb)
Rights and permissions
About this article
Cite this article
Castulo-Rubio, D.Y., Alejandre-Ramírez, N.A., Orozco-Mosqueda, M. et al. Volatile Organic Compounds Produced by the Rhizobacterium Arthrobacter agilis UMCV2 Modulate Sorghum bicolor (Strategy II Plant) Morphogenesis and SbFRO1 Transcription In Vitro. J Plant Growth Regul 34, 611–623 (2015). https://doi.org/10.1007/s00344-015-9495-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-015-9495-8