Abstract
This work presents a highly sensitive silver (Ag) based Surface Plasmon Resonance (SPR) sensor with graphene-coated over Silicon nitride (Si3N4) for sensing of Deoxyribonucleic acid (DNA) hybridization. The design consists of five layers namely Ag, Si3N4, graphene, and sensing medium along with BK7 glass prism that couple light at the metal–dielectric interface. The performance of the present structure has been evaluated using the angular interrogation method. The present investigation reveals the difference in complementary DNA strands and mismatched DNA strands as well as single-nucleotide polymorphisms (SNP) event by examining the variation of resonance angle in the reflectivity spectrum. The performance of the present structure is compared with the contemporary structures and found better than existing sensors. Therefore, the proposed structure is suitable in the biochemical field for the detection of biomolecules interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In present days, SPR technology has emerged as leading sensing technology in the biochemical field in the analysis of various substances like DNA, protein binding, and different enzymes, etc. [1,2,3,4]. DNA hybridization is an exercise of combining two single-stranded DNA (ssDNA) and this results in the formation of double-stranded DNA (dsDNA).
Combining two ssDNA is performed by the hydrogen bonds formed between the nucleotide bases. DNA hybridization measures the genetic similarity between two pools of the DNA sequence which can be used to diagnose more than 400 diseases directly. The dsDNA hybridization event and the mismatched DNA hybridization event are illustrated in Fig. 1a, b, respectively. In DNA hybridization, probe ssDNA is used to identify unknown, target ssDNA that results in DNA hybridization event by generating dsDNA as shown in Fig. 1a.
The hybridization process is a bio-recognition process of identifying nucleotide-binding between the base pairs, i.e., guanine (G) and cytosine (C), adenine (A), and thymine (T) [5, 6].
Surface plasmons (SPs) are the collective free electrons oscillation at the interface between metal and dielectric and are excited using the p-polarized light beam at the interface. This results in the generation of the evanescent wave at the interface that decays exponentially. This collective phenomenon is known as SPR. SPR sensing is the label-free sensing that works on the principle of the shift of the resonance angle when there is a variation in the refractive index (RI) of the sensing medium [7, 8]. In the prism-based SPR sensing approach, the excitation of the SPs is carried out by the Kretschmann based attenuated total reflection (ATR) method. The angle of the incident light in the ATR method can be best controlled by the angular interrogation approach [9]. Recently, some transition metal dichalcogenides (TMDCs) materials including WS2, WSe2, MoSe2, and MoS2, etc. are seen in the SPR biosensing. These TMDCs act as plasmonic supporting materials that improve the performance of the sensor [10,11,12,13]. A new 2D material, black phosphorus (BP) has also been seen in research in biosensing applications, optoelectronics, and electronics field, etc. [14, 15]. For researchers, performance improvement with a less complex structure is always the primary choice in designing these sensors. Lately, SPR sensors with improved performance have been seen in the research [16,17,18]. This study aims to provide the possibility of having better performance. The selection of plasmonic active metals such as gold, silver, and copper. plays a significant role in the designing of these sensors. Gold is highly resistive to the oxidation but the poor binding capabilities of molecules and large full-width half maxima (FWHM) limit the performance of the gold-based SPR sensor. Silver, a plasmonic active material provides better performance in terms of FWHM [18,19,20,21]. To overcome the oxidation problem of this material, the anti-reflection coating of the Si3N4 layer can be deposited over the silver layer. Si3N4 has come in attention because of its excellent thermal and chemical stability with a large bandgap of ~ 5 eV [22, 23]. Graphene layer, a very popular 2D nanomaterial of a one-atomic thin planar sheet of sp2 bonded carbon atoms arranged in a hexagonal honeycomb lattice, is chosen as a top layer because of its high biomolecules absorption capabilities with the large value of the surface-to-volume ratio (Surface/Volume) that gives the assurance of better absorption of molecules compared with other carbon materials [24, 25]. The presence of void and point defects on the graphene surface strongly influences the optical properties of the graphene layer that can be characterized with the help of Raman spectroscopy [26, 27]. Further, the significant number of defects on the graphene sheet can induce some transformation of graphene phase into a rippled H6 diamond-like phase with higher dielectric properties. Further, these defects may have influence on the performance of the proposed structure. So it is essential to reduce these defects before consideration of such material [28]. The proposed structure is used to sense DNA hybridization and may be used for the detection of other analytes such as formalin, protein, etc.
This work is arranged as: Sect. 2 outlines the structure consideration and parameters analysis. The key parameters of the proposed structure are discussed in Sect. 3. The numerical analysis and discussion are given in Sect. 4. The conclusion is given in Sect. 5. Finally, references have been added at the end.
2 Structure considerations and parameters analysis
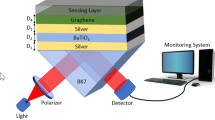
The proposed structure of the SPR biosensor is illustrated in Fig. 2a, b. Here, a He–Ne laser is used to incident a TM- polarized light of 633 nm wavelength. The first layer is a BK7 prism. The RI of the prism is 1.5151 [29]. The second layer of silver of optimized thickness (dAg = 50 nm) is grown over the BK7 prism. The RI of the silver layer is 0.056253 + 4.2760i which is taken from Johnson and Christy [30]. Further, the anti-reflection coating of the Si3N4 layer of the optimized thickness (dSi3N4 = 5 nm) is grown over the Ag layer that is the third layer. This makes the Ag layer highly resistive to the oxidation. The refractive index value of this Si3N4 layer is calculated by [31],
The fourth layer is the monolayer graphene (0.34 nm) that is deposited over the Si3N4 layer and the complex RI of this layer is 3 + 1.1487i [32]. Finally, the fifth layer is the sensing medium that is phosphate-buffered saline (PBS) solution. The shift in RI (\({{\delta n}}^{{S}})\) in the sensing medium is given as,
where, \({n}_{1}^{S}\) is the RI of the sensing medium (PBS solution, \({n}_{1}^{S}=1.334\)) in the absence of probe or target DNA in this medium [2], \({C}_{a}\) is the concentration of adsorbed molecules, \(\frac{\mathrm{dn}}{\mathrm{dc}}\) (~ 0.182 \({\mathrm{cm}}^{3}/\mathrm{g}\) for PBS solution) is the increment in the RI due to the adsorbate [33, 34] and \({n}_{2}^{S}\) is the RI of sensing medium after adsorption of DNA molecules. Suppose \({\mathrm{C}}_{\mathrm{a}}\) is taken 1000 nM. Due to this 1000 nM adsorbate, the RI of sensing medium changes to \({n}_{2}^{S}\). This is because the sensing medium contains 1000 nM adsorbate molecules. The proposed study is based on the transfer matrix method (TMM) and the Fresnel multilayer reflection theory is used for calculation of reflectivity at the multiple interfaces assuming all layers in this proposed structure are stacked along the z-axis.
The reflection intensity \(({R})\) of multi-layer structure can be given as [35],
where \({r}_{p}\) is the reflection coefficient of p-polarized light.
3 Key parameters of the proposed structure
In this study, sensitivity, detection accuracy, and quality factor have been evaluated for the performance of the proposed biosensor.
The sensitivity \((S)\) is expressed in resonance angle shift (\({{\delta \theta }}_{\mathrm{spr}}\)) and RI shift (\({\delta n}_{s}\)). Mathematically [36],
It is generally expressed in \({{degree}/\mathrm{RIU}}^{-1}\).
The detection accuracy (D.A.) is the ratio of \({{\delta \theta }}_{\mathrm{spr}}\) to FWHM. Mathematically [37],
It is a unitless parameter.
Quality factor \(\left(Q\right)\) can be given by the ratio of sensitivity \((S)\) to FWHM [38],
It is generally expressed in \({\mathrm{RIU}}^{-1}\).
4 Numerical analysis and discussion
The proposed sensor structure with Ag-Si3N4-graphene for sensing probe DNA and non-complementary (mismatched) DNA is shown in Fig. 1a. Initially, we have optimized the thicknesses of layers for minimum reflectivity in the proposed structure. For this, we have plotted Fig. 3a–d of thicknesses 40, 45, 50, and 50 nm of the Ag layer with thicknesses 1, 3, 5, 7, and 10 nm of the Si3N4 layer. It is evident from these figures that minimum reflectivity is obtained with 50 nm thickness of the Ag layer and 5 nm thickness of the Si3N4 layer.
After optimizing the thickness of the Ag layer and Si3N4 layer, we have plotted the SPR curves with and without the Si3N4 layer as shown in Fig. 4. In this figure, with the Ag-graphene-sensing medium (PBS solution, 1.334), the black solid line represents the SPR curve before the adsorption of DNA molecules while the black dot line represents the SPR curve for DNA concentration in PBS solution that provides the RI of 1.40 of the sensing medium. For the numerical analysis of our proposed sensor, the RI sensitivity is considered for the large range of RI change from 1.334 to 1.40. The value for minimum reflectivity and resonance angle before and after adsorption of the DNA molecules are 0.007908, 68.19°, and 0.00464, 78.09° respectively.
Again, in this figure, with the Ag-Si3N4-graphene-sensing medium (PBS solution, n = 1.334), the red solid line represents the SPR curve before the adsorption of DNA molecules while the red dot line represents the SPR curve for DNA concentration in PBS solution that provides the RI of 1.40 of the sensing medium. The value for minimum reflectivity and resonance angle before and after adsorption of the DNA molecules are 0.0003897, \({70.79}^{^\circ }\), and 0.1029, \({83.99}^{^\circ }\) respectively. Further, the values of FWHM calculated from Fig. 4 are \({1.88}^{^\circ }\) and \({1.33}^{^\circ }\) for the structure with and without the Si3N4 layer. The values of sensitivity and detection accuracy are obtained 200 degree/RIU and 7.02 respectively while the quality factor is obtained 106.38 \({\mathrm{RIU}}^{-1}\) for the proposed structure. Table 1 lists the performance parameters with and without the Si3N4 layer. It is noted from Table 1 that the sensitivity of the proposed structure with the Si3N4 layer is obtained as 200 degree/RIU which is ~ 33% higher than the SPR structure without the Si3N4 layer.
The reflection intensity plot for the bare sensor (before adsorption the probe DNA) and SPR curve with the probe DNA are presented in Fig. 5. The resonance angle and the minimum value of reflectivity of the bare sensor and with the probe DNA are \({70.79}^{^\circ }\), 0.0003897, and \({73.89}^{^\circ }\), 0.0008299, respectively. It is noted from Fig. 5 that the shift of 3.10° in resonance angle represents the change in the RI of the sensing medium. The presence of electron-rich molecules in DNA alters the carrier concentration on the Si3N4-graphene surface which alters the RI at the Si3N4-graphene surface and this change in the RI will turn to lead to altering the propagation constant of the surface plasmon wave (SPW) that finally shifts the SPR angle [2].
Figure 6 represents the presence of the non-complementary DNAs over the Ag-Si3N4-graphene surface of the proposed sensor. The resonance angle and the minimum reflectivity for the bare sensor, with the probe DNAs and with the non-complementary (mismatched) DNAs are\({70.79}^{^\circ }\), 0.0003897,\({73.89}^{^\circ }\), 0.0008299 and\({73.98}^{^\circ }\), 0.001964, respectively. Here, the small resonance angle shift (\({0.09}^{^\circ })\) is seen for the non-complementary DNAs that is because of mismatch hydrogen bonding between the base pairs. This indicates the mismatched DNA hybridization event. Hence, the present structure is capable of discriminating between the perfectly matched (complementary) DNAs and the mismatched (non-complementary) DNAs [14].
The proposed model for the sensing of DNA hybridization between two ssDNAs (one is probe and the other is the target) is illustrated in Fig. 2b. Here, the sensing medium is composed of probe DNA, complementary DNA, and PBS solution. The SPR curves of Fig. 7 shows the dsDNA hybridization event between the probe DNA and different concentrations of ct-Target DNA (1000, 1001, 1010, and 1100 nM). The values of the resonance angle and minimum reflectivity for the different DNA concentrations in the dsDNA event are given in Table 2.
Table 3 gives the resonance angle shift \({\Delta\uptheta }_{\mathrm{spr}}^{\mathrm{P}-\mathrm{T}} \left(\mathrm{degree}\right)\) and minimum reflectivity difference \({\Delta \mathrm{R}}_{\mathrm{min}}^{\mathrm{P}-\mathrm{T}}\) for different concentrations of ct-Target DNA. It is seen from this table, as the concentration of ct-Target DNA increases, the resonance angle shift \({\Delta\uptheta }_{\mathrm{spr}}^{\mathrm{P}-\mathrm{T}} \left(\mathrm{degree}\right)\) increases which gives the strong matching combination between the target DNA and the probe DNA [14, 37]. In Table 3, the minimum resonance angle shift \({({\Delta\uptheta }_{\mathrm{spr}}^{\mathrm{P}-\mathrm{T}} \left(\mathrm{degree}\right))}_{\mathrm{min}}={1.30}^{^\circ }\) and the minimum reflectivity difference \({({\Delta \mathrm{R}}_{\mathrm{min}}^{\mathrm{P}-\mathrm{T}})}_{\mathrm{min}}=0.000339\) are two indicating factors. Based on the conditions of these two indicating factors, Table 4 is prepared and can be utilized for deciding whether DNA hybridization occurred or not [24, 33].
Furthermore, we have shown the effect of increasing the number of graphene layers in Fig. 8. This figure reveals that the maximum sensitivity of 212.12 degree/RIU is obtained with three graphene layers while without the graphene layer, the maximum detection accuracy and quality factor are obtained 9.124 and 138.24 RIU−1, respectively. In addition, the performance of the proposed structure has been compared to other contemporary structures as shown in Table 5, which shows better performance than other existing sensors. Therefore, this proposed sensor with high performance finds its application in sensing of DNA hybridization.
5 Conclusion
In conclusion, a highly sensitive SPR sensor based on Ag-Si3N4-graphene has been designed and simulated for sensing of DNA hybridization and mismatched DNA event. The presence of the anti-reflection coating of the Si3N4 layer over the surface of the Ag layer enhances the sensitivity of the proposed sensor (~ 33%). Sensitivity is obtained as 200 degree/RIU for the proposed sensor. While quality factor and detection accuracy are found to be 106.38 RIU−1 and 7.02 respectively. Although the presented sensor is modeled for DNA sensing application. This structure can be used for sensing a wide range of biological analytes. Hence, this study opens a new path of sensing in the field of medical science.
References
S. Zeng, D. Baillargeat, H.-P. Ho, K.-T. Yong, Chem. Soc. Rev. 43, 3426 (2014)
S. Pal, Y.K. Prajapati, J.P. Saini, Opt. Rev. 27, 57 (2020)
M. Çalışır, M. Bakhshpour, H. Yavuz, A. Denizli, Sens. Actuators B: Chemical 306, 127561 (2020)
H.-S. Lee, T.-Y. Seong, W.M. Kim, I. Kim, G.-W. Hwang, W.S. Leea, K.-S. Lee, Sensors and Actuators B: Chemical 266, 311 (2018)
K. Tamersit, F. Djeffal, IEEE Sens. J. 16, 4180 (2016)
H. Karimi, R. Yusof, R. Rahmani, H. Hosseinpour, M.T. Ahmadi, Nanoscale Res Lett 9, 71 (2014)
A. Shalabney, I. Abdulhalim, Sens. Actuators, A 159, 24 (2010)
H. Fu, S. Zhang, H. Chen, J. Weng, IEEE Sens. J. 15, 5478 (2015)
E. Kretschmann, H. Raether, Zeitschrift für Naturforschung A 23, 2135 (1968)
N. Mudgal, A. Saharia, A. Agarwal, J. Ali, P. Yupapin, G. Singh, Opt. Quant. Electron. 52, 307 (2020)
J. Banerjee, M. Ray, Sens. Actuators B: Chemical 281, 520 (2019)
Q. Ouyang, S. Zeng, X.-Q. Dinh, P. Coquet, K.-T. Yong, Procedia Eng. 140, 134 (2016)
A.K. Sharma, B. Kaur, V.A. Popescu, Opt. Mater. 102, 109824 (2020)
S. Pal, A. Verma, S. Raikwar, Y.K. Prajapati, J.P. Saini, Appl. Phys. A 124, 394 (2018)
B. Meshginqalam, J. Barvestani, Opt. Mater. 86, 119 (2018)
A. Panda, P.D. Pukhrambam, G. Keiser, Appl. Phys. A 126, 153 (2020)
C.M. Das, Q. Ouyang, L. Kang, Y. Guo, X.-Q. Dinh, P. Coquet, K.-T. Yong, Photonics Nanostruct. Fundam. Appl. 38, 100760 (2020)
R. Kumar, S. Pal, A. Verma, Y.K. Prajapati, J.P. Saini, Superlattices Microstruct. 145, 106591 (2020)
A. Srivastava, A. Verma, R. Das, Y.K. Prajapati, Optik 203, 163430 (2020)
Y.K. Prajapati, A. Srivastava, Superlattices Microstruct. 129, 152 (2019)
A. Kumar, A.K. Yadav, A.S. Kushwaha, S.K. Srivastava, Sen. Actuators 2, 100015 (2020)
O. Blázquez, J. López-Vidrier, S. Hernández, J. Montserrat, B. Garrido, Energy Procedia 44, 145 (2014)
S.-L. Ku, C.-C. Lee, Opt. Mater. 32, 956 (2010)
M.B. Hossain, M.M. Rana, Sensor Letters 14, 145 (2016)
A. Keshavarz, S. Zangenehzadeh, A. Hatef, Appl. Nanosci. 10, 1465 (2020)
S. Neuville, Carbon structure Analysis with Differentiated Raman spectroscopy (Refined Raman Spectroscopy fundamentals for improved Carbon Material Engineering), (LAMBERT ACADEMIC PRESS, 2014)
S. Neuville, Surf. Coat. Technol. 206, 703 (2011)
S. Neuville, Micromachines 10(8), 539 (2019)
Z. Lin, L. Jiang, L. Wu, J. Guo, X. Dai, Y. Xiang, D. Fan, IEEE Photon J 8, 1 (2016)
P.B. Johnson, R.W. Christy, Phys Rev B 6, 4370 (1972)
K. Luke, Y. Okawachi, M.R.E. Lamont, A.L. Gaeta, M. Lipson, Opt. Lett. 40, 4823 (2015)
H. Vahed, C. Nadri, Opt. Mater. 88, 161 (2019)
M.S. Rahman, M.R. Hasan, K.A. Rikta, M.S. Anower, Opt. Mater. 75, 567 (2018)
T. Tumolo, L. Angnes, M.S. Baptista, Anal. Biochem. 333, 273 (2004)
P.K. Maharana, R. Jha, P. Padhy, Sens. Actuators B 207, 117 (2015)
N. Mudgal, P. Yupapin, J. Ali, G. Singh, Plasmonics (2020). https://doi.org/10.1007/s11468-020-01146-2
M.S. Rahman, M.S. Anower, M.R. Hasan, K.A. Rikta, Sensor Letters 15, 510 (2017)
P. Sun, M. Wang, L. Liu, L. Jiao, W. Du, F. Xia, M. Liu, W. Kong, L. Dong, M. Yun, Appl. Surf. Sci. 475, 342 (2019)
Acknowledgements
Authors would like to thank, ECE Department, Malaviya National Institute of Technology, Jaipur for infrastructure support to accomplish this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors state that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mudgal, N., Saharia, A., Choure, K.K. et al. Sensitivity enhancement with anti-reflection coating of silicon nitride (Si3N4) layer in silver-based Surface Plasmon Resonance (SPR) sensor for sensing of DNA hybridization. Appl. Phys. A 126, 946 (2020). https://doi.org/10.1007/s00339-020-04126-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00339-020-04126-9