Abstract
Objectives
To identify an 18F-fluorodeoxyglucose (18F-FDG) positron emission tomography (PET) radiomics-based model for predicting progression-free survival (PFS) and overall survival (OS) of nasal-type extranodal natural killer/T cell lymphoma (ENKTL).
Methods
In this retrospective study, a total of 110 ENKTL patients were divided into a training cohort (n = 82) and a validation cohort (n = 28). Forty-one features were extracted from pretreatment PET images of the patients. Least absolute shrinkage and selection operator (LASSO) regression was used to develop the radiomic signatures (R-signatures). A radiomics-based model was built and validated in the two cohorts and compared with a metabolism-based model.
Results
The R-signatures were constructed with moderate predictive ability in the training and validation cohorts (R-signaturePFS: AUC = 0.788 and 0.473; R-signatureOS: AUC = 0.637 and 0.730). For PFS, the radiomics-based model showed better discrimination than the metabolism-based model in the training cohort (C-index = 0.811 vs. 0.751) but poorer discrimination in the validation cohort (C-index = 0.588 vs. 0.693). The calibration of the radiomics-based model was poorer than that of the metabolism-based model (training cohort: p = 0.415 vs. 0.428, validation cohort: p = 0.228 vs. 0.652). For OS, the performance of the radiomics-based model was poorer (training cohort: C-index = 0.818 vs. 0.828, p = 0.853 vs. 0.885; validation cohort: C-index = 0.628 vs. 0.753, p < 0.05 vs. 0.913).
Conclusions
Radiomic features derived from PET images can predict the outcomes of patients with ENKTL, but the performance of the radiomics-based model was inferior to that of the metabolism-based model.
Key Points
• The R-signatures calculated by using 18 F-FDG PET radiomic features can predict the survival of patients with ENKTL.
• The radiomics-based models integrating the R-signatures and clinical factors achieved good predictive values.
• The performance of the radiomics-based model was inferior to that of the metabolism-based model in the two cohorts.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Extranodal natural killer/T cell lymphoma (ENKTL) is an aggressive malignancy of putative NK cell origin, with a minority deriving from the T cell lineage [1, 2]. ENKTL is much more common in Asia and Latin America, comprising approximately 3 to 10% of all lymphomas in East Asia, but less than 1% in Western countries. Establishing an optimal treatment strategy for ENKTL has yet to be determined, as accurate assessments are crucial for prognosis prediction and individualised treatment strategy decisions [3, 4].
18F-Fluorodeoxyglucose (18F-FDG) positron emission tomography-computed tomography (PET/CT) is routinely used for lesion detection, response monitoring, and prognostication assessments in ENKTL patients. The maximum standardised uptake value (SUVmax) is the most widely used PET imaging parameter for prediction; moreover, metabolic tumour volume (MTV) and total lesion glycolysis (TLG), which are based on both SUV and tumour volume, have also been reported to be significant prognostic biomarkers for ENKTL [5, 6]. However, these metabolic parameters do not fully reflect the spatial distribution of a tracer, which has been suggested to correlate with intra- and intertumoural heterogeneity, and even worse prognosis and survival [7]. Over the past decade, an emerging and promising field, radiomics, has been implemented to extract and analyse a large amount of advanced quantitative imaging features with high throughput from medical images to provide abundant tumour heterogeneity information [8, 9]. The most widely used medical imaging modalities in radiomic research are computed tomography (CT) and magnetic resonance imaging (MRI). Currently, radiomic features extracted from PET are considered to contain large amounts of underlying information distinct from that provided by CT and MRI [10, 11]. A continuously increasing number of studies have reported promising results regarding the value of PET-based radiomic features for various types of solid tumours [12,13,14,15,16]. Nevertheless, few published studies have reported the diagnostic, staging, or prognostic value of PET-based radiomic features in lymphoma [17,18,19,20,21]. In addition, combining multiple imaging biomarkers as a predictive signature using radiomic methods, rather than individual analyses, is a promising and useful approach for prognosis prediction and clinical management [22].
Thus, in this retrospective study, we aimed to develop the radiomic signatures (R-signatures) using 18F-FDG PET radiomic features for the prediction of progression-free survival (PFS) and overall survival (OS) in patients with ENKTL. Subsequently, the radiomics-based model integrating the R-signatures and clinical factors was established and then validated and compared with the metabolism-based model integrating metabolic parameters and clinical factors. The models are visualised as nomograms.
Materials and methods
Patients and follow-up
Ethical approval was obtained, and the requirement for informed consent from patients was waived. The inclusion criteria included as follows: (a) pathologically diagnosed ENKTL between January 2011 and January 2017, (b) pretreatment 18F-FDG PET/CT, and (c) stage I to II patients who received 2 cycles of chemotherapy with LVP (l-asparaginase, vincristine, prednisone) and concurrent radiotherapy with 2 cycles of cisplatin chemotherapy, followed by 2 cycles of LVP; stage III to IV patients who received chemotherapy with LVP with or without radiotherapy if a response was observed, and otherwise received second-line therapy. In total, 110 consecutive patients were enrolled and randomly allocated to two cohorts (82 and 28 patients in the training and validation cohorts, respectively). Follow-up was performed every 3 months after the completion of treatment. The last follow-up was conducted in April 2018. The end-points of this study were PFS and OS. PFS was defined as the interval between the date of diagnosis and the date of the first relapse, progression, or death. OS was defined as the interval between the date of diagnosis and the time of death.
PET scanner and acquisition parameters
18F-FDG PET/CT examinations were performed on a Gemini GXL PET/CT scanner equipped with a 16-slice CT (Philips Medical System). After at least 6 h of fasting, 190–375 MBq of 18F-FDG was administered intravenously. The blood glucose level was controlled to be lower than 8.0 mmol/L. Whole-body PET/CT scans were started 60 min after radiopharmaceutical injection. Emission data were acquired for 2 min per bed position. PET emission acquisition was performed in 3D mode (3D-RAMLA): the dimensions of the in-plane matrix were 4 mm × 4 mm, and the slice thickness was 4 mm. All examinations were reconstructed using an OSEM algorithm, and the CT acquisition data were used for attenuation correction.
Segmentation and feature extraction
Two nuclear medicine physicians independently performed segmentation using the Local Image Features Extraction (LifeX) package (version 4.00, http://www.lifexsoft.org) [23]. First, the lymphoma lesions were delineated manually. Then, the regions of interest (ROIs) were defined based on a threshold of 40% of the SUVmax of the defined lesions [24], and spatial resampling (2 × 2 × 2 mm), absolute intensity resampling (0–20), and intensity discretisation (number of grey levels = 64, size of bins = 0.3125) were performed [25]. A total of 41 features (Supplementary Materials) of the ROIs were extracted as follows: first-order metrics extracted from the histogram and shape; features derived from the grey-level co-occurrence matrix (GLCM), the neighbourhood grey-level different matrix (NGLDM), the grey-level run length matrix (GLRLM), and the grey-level zone length matrix (GLZLM); and conventional metabolic parameters, including the SUVmax, MTV, and TLG.
Radiomic feature selection and model building
First, the interobserver repeatability of the segmentation was evaluated using the intraclass correlation coefficient (ICC) method, and the features with an ICC greater than 0.70 were selected [26]. Then, the least absolute shrinkage and selection operator (LASSO) Cox regression algorithm was applied to the selected features [27]. Cross-validation was applied to optimise the value of λ, the coefficients of indistinctive covariates were reduced to zero, and the remaining nonzero coefficients were selected. The nonzero coefficients of the selected features were defined as radiomic scores (R-scores). We calculated the combination of R-scores for all selected radiomic features, defined as the R-signature. We determined the optimal threshold value of the R-signature by the receiver operating characteristic (ROC) curve and divided patients into high- and low-risk groups. The potential association of the R-signature with PFS and OS was evaluated using the Kaplan-Meier analysis and log-rank test.
We used the univariate Cox regression to select the significant prognostic factors of PFS and OS, respectively. R-signature and clinical variables were entered into a multivariate Cox regression to build the radiomics-based model for PFS and OS prediction. Likewise, metabolic parameters and clinical variables were entered into a multivariate Cox regression to build the metabolism-based model. Models were then visualised as nomograms. The flowchart of model building is presented in Fig. 1.
Model validation
The discrimination of the models was assessed using the Harrell concordance index (C-index) [28, 29]. Bootstrap analyses with 1000 resamples were used to obtain a corrected C-index. The calibration of the models was assessed by Hosmer-Lemeshow tests and calibration curves, and p > 0.05 accounted for a nonsignificant deviance from the theoretical perfect calibration [30].
Statistical analysis
Statistical analyses were performed using IBM SPSS Statistics (version 19.0, IBM Corp) and R software (version 3.4.2; http://www.R-project.org). All tests were two-sided, and p values of < 0.05 were considered significant.
Results
The clinical characteristics of the patients are summarised in Table 1. No differences were found between the training and validation cohorts (p = 0.149–0.945). The median follow-up time was 33 months (2–90 months). As of the final follow-up, the numbers of survivals and deaths were 63 (57.3%) and 25 (22.7%), respectively.
R-signature construction and assessment
Six radiomic features had low ICCs (ICC < 0.70), and twenty-seven radiomic features had good repeatability (ICC ≥ 0.85). The thirty-two features with ICCs ≥ 0.70 extracted in the second round were selected for further analysis. According to the LASSO results (Fig. 2), we obtained 4 and 3 radiomic features with nonzero coefficients for PFS and OS, respectively, in order to calculate the R-signaturePFS and the signatureOS. The results of the ICC analysis and the formulas for the R-signatures can be found in the Supplementary Materials.
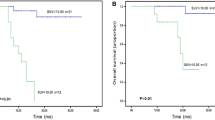
The ROC-AUCs of the R-signaturePFS were 0.788 (95% CI = 0.682–0.895) and 0.473 (p = 0.803) in the two cohorts. When analysing the association of the R-signaturePFS with PFS, the results of the log-rank test indicated significant discrimination between the high- and low-risk groups in the training cohort (Fig. 3a), but no discrimination in the validation cohort (Fig. 3c). The ROC-AUCs of the R-signatureOS were 0.637 (95% CI = 0.488–0.786) and 0.730 (95% CI = 0.548–0.912) in the two cohorts. We also observed that the R-signatureOS was significant for classifying the patients into high- and low-risk groups in the two cohorts (Fig. 3b, d).
Building of the radiomics-based model
The results of the univariate and multivariate analyses are listed in Tables 2 and 3. In the univariate analysis, the Eastern Cooperative Oncology Group performance status (ECOG PS), Ann Arbor stage, lactate dehydrogenase (LDH), International Prognostic Index (IPI), SUVmax, MTV, TLG, and the R-signaturePFS were associated with PFS; bone marrow (BM), ECOG PS, Ann Arbor stage, LDH, IPI, SUVmax, MTV, TLG, and the R-signatureOS were associated with OS.
For PFS, the R-signaturePFS and significant clinical variables in the univariate analysis were selected for inclusion in the multivariate Cox regression. The R-signaturePFS and IPI remained as prognostic factors in the multivariate analysis and were used to build the radiomics-based model (Fig. 4a). For OS, the radiomics-based model was built using the R-signatureOS and ECOG PS (Fig. 4b), which were independent prognostic factors of OS identified by the multivariate analysis.
Building of the metabolism-based model
For PFS, the significant clinical variables and metabolic parameters in the univariate analysis were entered into the multivariate Cox regression, and MTV and IPI were identified as independent prognostic factors and were used to build the metabolism-based model. Likewise, the metabolism-based model for OS prediction was built using the SUVmax, MTV, and ECOG PS.
Validation and comparison of the model
The results of the C-index and the Hosmer-Lemeshow test are shown in Table 4. The calibration curves of the models are shown in the Supplementary Materials. For PFS prediction, the radiomics-based model showed better discrimination than the metabolism-based model in the training cohort (C-index = 0.811 vs. 0.751) but poorer discrimination in the validation cohort (C-index = 0.588 vs. 0.693). The Hosmer-Lemeshow test showed that the calibration of the radiomics-based model was poorer than that of the metabolism-based model (training cohort: p = 0.415 vs. 0.428; validation cohort: p = 0.228 vs. 0.652). The calibration curves showed that the calibration of the two models was better in the training cohort than that in the validation cohort (Supplementary Figs. S1 and S2).
For OS prediction, the discrimination of the radiomics-based model was poorer than that of the metabolism-based model in the training cohort (C-index = 0.818 vs. 0.828) and the validation cohort (C-index = 0.628 vs. 0.753). The Hosmer-Lemeshow test indicated that the calibration of the radiomics-based model was poorer than that of the metabolism-based model (training cohort: p = 0.853 vs. 0.885; validation cohort: p < 0.05 vs. 0.913). According to the calibration curves, the calibration of the two models was better in the training cohort than that in the validation cohort (Supplementary Figs. S3 and S4).
Discussion
In the present study, we developed R-signatures with moderate predictive ability in a training cohort and a validation cohort (R-signaturePFS: AUC = 0.788 and 0.473, respectively; R-signatureOS: AUC = 0.637 and 0.730, respectively). Although no significant association was found between the R-signaturePFS and PFS in the validation cohort, the R-signatures were associated with PFS in the training cohort and with OS in both cohorts. These results provide evidence that radiomic features extracted from pretreatment 18F-FDG PET images can predict lymphoma outcomes. We further developed radiomics-based models combining R-signatures and clinical variables to predict PFS and OS among patients with ENKTL, and the models were then visualised via nomograms with the aim of identifying patients at a high risk of early progression and death who could be offered an alternative treatment strategy. The radiomics-based models achieved good predictive values (PFS: C-index = 0.811 and 0.588, p = 0.415 and 0.228; OS: C-index = 0.818 and 0.628, p = 0.853 and < 0.05). However, the performance of the radiomics-based model was inferior to that of the metabolism-based model in the two cohorts.
The main goal of radiomics is to build a prediction model for clinical outcomes using selected radiomic features, and integrating radiomic features with traditional prognostic indicators (clinical indicators) in one model can improve the prediction performance of a single prognostic indicator [8, 9, 31, 32]. Traditional PET metabolic parameters have been confirmed to be significant prognostic indicators for outcomes of patients with ENKTL and are widely used in clinical management [4, 5]. However, most radiomic indices extracted from PET images have been considered to be significantly correlated with metabolic parameters, especially MTV and TLG [33, 34], and such correlations are postulated to be inherent to the definitions of the features as opposed to being variable depending on the tumour type or acquisition/reconstruction protocols [11], which were also reported to differ in susceptibility to methodological, biological, and metabolic features [35, 36]. Based on these considerations, we built radiomics-based models to investigate the potential added prognostic value of radiomic features in ENKTL patients by comparing the models with metabolism-based models. Our results suggested that the radiomics-based models may provide limited prognostic information for ENKTL patients compared with the metabolism-based model.
Our findings are in line with those of recent studies indicating that the performance of PET-derived radiomic features for tumour prognosis prediction is poor compared with that of PET-derived metabolic parameters. In a cohort of 82 patients with aggressive B cell lymphoma, MTV was correlated with the response to therapy, but texture features could not predict the therapy response, although several features were correlated with the presence of a residual mass and outcomes [18]. Rogasch et al reported that the asphericity feature could predict the response after chemotherapy in 50 children with paediatric Hodgkin’s lymphoma (HL), although MTV showed a better performance [35]. In contrast, several studies have reported promising results for the utilisation of radiomic features derived from PET to risk-stratify patients with lymphoma. Lue et al found that the intensity nonuniformity of pretreatment PET was a prognostic indicator in 42 patients with HL and may outperform MTV [31]. In a cohort of 17 patients with ENKTL, texture features (dissimilarity and LISZE) extracted from pretreatment PET images were independent predictors of PFS, whereas the SUVmax, MTV, and TLG were not associated with PFS [37]. Wang et al demonstrated a relationship between PET radiomic features and OS in 19 patients with renal/adrenal lymphoma, and MTV was not an independent factor [38].
These differences may be attributed to many sources. First, as mentioned above, several studies incorporated radiomic and metabolic features into one model, which may result in an underlying risk of redundancy and underestimation of the performance of both types of features. Second, a sample size of at least 10 to 15 patients per predictor variable has been proposed to be required to produce valid estimates for multiple regression models [39]. We reduced the number of radiomic features to 4 and 3, which is reasonable for minimising false detection rates. Previous studies with small sample sizes and datasets may have a risk of bias. Additionally, many variables may affect the stability and prognostic value of PET-based radiomic features, such as the scanner, segmentation, reconstruction parameters, formulas used to define the radiomic features, and software [40, 41]. Heterogeneity among different studies may have contributed to controversial results. Lastly, validation analysis has been regarded as an indispensable step in radiomics research to show the potential value of a radiomics model for clinical application [8, 42]. In this study, the performance of the models in the internal validation cohort was inferior to that in the training cohort, suggesting that the stability of the models should be considered with caution. Comparing previous findings directly is difficult considering the limited number of lymphoma radiomics studies that have performed internal and/or external validation.
Segmentation is an important step in radiomics research as edges can substantially affect feature values. No consensus on optimal segmentation is available for lymphoma because lymphoma lesions usually have heterogeneous sizes, shapes, and locations. On PET images, some normal organs with high uptake and physiological FDG uptake and excretion may cause confusion [43]. Therefore, identifying accurate and robust segmentation methods for lymphoma is important. We defined ROIs using a semiautomatic threshold-based method (the 40% threshold segmentation method) and assessed the interobserver repeatability of the segmentation, and the results of the ICC analysis indicated that 27 of 38 radiomic features had good repeatability (ICC ≥ 0.85), which is consistent with previous studies demonstrating that most PET image features exhibited high stability in test-retest and interobserver analyses [40, 44]. Several different segmentation methods have been devised for lymphoma [43, 45, 46]. Hu et al proposed an entropy-based optimisation strategy to detect and segment lymphoma in PET images and reported a good performance [43]. Hu et al proposed an automatic approach for ENKTL segmentation that was more stable than traditional deep-learning segmentation [46].
The limitations of the present study are as follows. First, this was a single-centre, retrospective study, and external validation was not performed, which may have impacted patient selection, the examination protocol, and the radiomic quantification results. Second, we used only 18F-FDG PET images to extract radiomic features. PET images have relatively low spatial resolution and high noise, which may influence lesion identification [9]. A combination of PET and CT images may expand the feature pool and lead to the discovery of more predictive radiomic features. In addition, we extracted only traditional radiomic features, which may contain limited information. Other types of features (model- or transform-based features and deep-learning features) have gained popularity because they are more specific to data and clinical outcomes [47], and such features should be further investigated to better understand the predictive power of radiomics.
Conclusions
In conclusion, a pretreatment 18F-FDG PET radiomics-based model was designed and showed significant stratification power in predicting PFS and OS in ENKTL, but the performance was inferior to that of the metabolism-based model. Therefore, further multi-centre, prospective studies with external validation are required to ensure that the results are reproducible and do not require refinement to achieve a higher level of evidence.
Abbreviations
- AUC:
-
Area under curves
- CI:
-
Confidence interval
- C-index:
-
The Harrell concordance index
- DICOM:
-
Digital Imaging and Communications in Medicine
- ECOG PS:
-
Eastern Cooperative Oncology Group performance status
- ICC:
-
The intraclass correlation coefficients
- IPI:
-
International Prognostic Index
- LASSO:
-
The least absolute shrinkage and selection operator
- LDH:
-
Lactate dehydrogenase
- MTV:
-
Metabolic tumour volume
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- ROIs:
-
Regions of interest
- SUVmax :
-
Standardised uptake value
- TLG:
-
Total lesion glycolysis
References
Tse E, Kwong YL (2017) The diagnosis and management of NK/T-cell lymphomas. J Hematol Oncol 10:85
William BM, Armitage JO (2013) International analysis of the frequency and outcomes of NK/T-cell lymphomas. Best Pract Res Clin Haematol 26:23–32
Yamaguchi M, Miyazaki K (2017) Current treatment approaches for NK/T-cell lymphoma. J Clin Exp Hematop 57:98–108
Ding JJ, Chen YL, Zhou SH, Zhao K (2018) Positron emission tomography/computed tomography in the diagnosis, staging, and prognostic evaluation of natural killer/T-cell lymphoma. J Int Med Res 46:4920–4929
Wang H, Shen G, Jiang C, Li L, Cui F, Tian R (2018) Prognostic value of baseline, interim and end-of-treatment18F-FDG PET/CT parameters in extranodal natural killer/T-cell lymphoma: a meta-analysis. PLoS One 13:1–14
Jiang C, Zhang X, Jiang M et al (2015) Assessment of the prognostic capacity of pretreatment, interim, and post-therapy (18)F-FDG PET/CT in extranodal natural killer/T-cell lymphoma, nasal type. Ann Nucl Med 29:442–451
Yan J, Chu-Shern JL, Loi HY et al (2015) Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med 56:1667–1673
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D (2017) Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging 44:151–165
Davnall F, Yip CSP, Ljungqvist G et al (2012) Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 3:573–589
Orlhac F, Soussan M, Maisonobe JA, Garcia CA, Vanderlinden B, Buvat I (2014) Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med 55:414–422
Lee JW, Lee SM (2018) Radiomics in oncological PET/CT: clinical applications. Nucl Med Mol Imaging 52:170–189
Carvalho S, Leijenaar RTH, Troost EGC et al (2018) 18F-fluorodeoxyglucose positron-emission tomography (FDG-PET)-Radiomics of metastatic lymph nodes and primary tumor in non-small cell lung cancer (NSCLC) - a prospective externally validated study. PLoS One 13:e0192859
Mena E, Sheikhbahaei S, Taghipour M et al (2017) 18F-FDG PET/CT metabolic tumor volume and intratumoral heterogeneity in pancreatic adenocarcinomas: impact of dual-time point and segmentation methods. Clin Nucl Med 42:e16–e21
Nakajo M, Jinguji M, Nakabeppu Y et al (2017) Texture analysis of18F-FDG PET/CT to predict tumour response and prognosis of patients with esophageal cancer treated by chemoradiotherapy. Eur J Nucl Med Mol Imaging 44:206–214
Feliciani G, Fioroni F, Grassi E et al (2018) Radiomic profiling of head and neck cancer: 18F-FDG PET texture analysis as predictor of patient survival. Contrast Media Mol Imaging 2018:3574310
Aide N, Talbot M, Fruchart C, Damaj G, Lasnon C (2018) Diagnostic and prognostic value of baseline FDG PET/CT skeletal textural features in diffuse large B cell lymphoma. Eur J Nucl Med Mol Imaging 45:699–711
Parvez A, Tau N, Hussey D, Maganti M, Metser U (2018) 18F-FDG PET/CT metabolic tumor parameters and radiomics features in aggressive non-Hodgkin’s lymphoma as predictors of treatment outcome and survival. Ann Nucl Med 32:410–416
Zhou Y, Ma XL, Pu LT, Zhou RF, Ou XJ, Tian R (2019) Prediction of overall survival and progression-free survival by the 18F-FDG PET/CT radiomic features in patients with primary gastric diffuse large B-cell lymphoma. Contrast Media Mol Imaging 2019:5963607
Ganeshan B, Miles KA, Babikir S et al (2017) CT-based texture analysis potentially provides prognostic information complementary to interim FDG-PET for patients with Hodgkin’s and aggressive non-Hodgkin’s lymphomas. Eur Radiol 27:1012–1020
Kang D, Park JE, Kim YH et al (2018) Diffusion radiomics as a diagnostic modal for atypical manifestation of primary central nervous system lymphoma: development and multicenter external validation. Neuro Oncol 20:1251–1261
Wang L, Dong T, Xin B et al (2019) Integrative nomogram of CT imaging, clinical, and hematological features for survival prediction of patients with locally advanced non-small cell lung cancer. Eur Radiol 29:2958–2967
Nioche C, Orlhac F, Boughdad S et al (2018) Lifex: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 78:4786–4789
Parmar C, Velazquez ER, Leijenaar R et al (2014) Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 9:e102107
Orlhac F, Nioche C, Soussan M, Buvat I (2017) Understanding changes in tumor texture indices in PET: a comparison between visual assessment and index values in simulated and patient data. J Nucl Med 58:387–392
Weir JP (2005) Quantifying test-retest reliability using the intraclass correlation coefficient and the SEM. J Strength Cond Res 19:231
Sauerbrei W, Royston P, Binder H (2007) Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med 26:5512–5528
Alba AC, Agoritsas T, Walsh M et al (2017) Discrimination and calibration of clinical prediction models: users’ guides to the medical literature. JAMA 318:1377–1384
Zieffler AS, Harring JR, Long JD (2011) Comparing groups. Wiley, Hoboken
Pencina MJ, D’Agostino RB, Steyerberg EW (2011) Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 30:11–21
Lue KH, Wu YF, Liu SH et al (2019) Prognostic value of pretreatment radiomic features of 18F-FDG PET in patients with Hodgkin lymphoma. Clin Nucl Med 44:e559–e565
Desseroit MC, Visvikis D, Tixier F et al (2016) Development of a nomogram combining clinical staging with18F-FDG PET/CT image features in non-small-cell lung cancer stage I–III. Eur J Nucl Med Mol Imaging 43:1477–1485
Brooks FJ, Grigsby PW (2014) The effect of small tumor volumes upon intra-tumoral tracer uptake heterogeneity studies. J Nucl Med 55:37–42
Ha S, Choi H, Paeng JC, Cheon GJ (2019) Radiomics in oncological PET/CT: a methodological overview. Nucl Med Mol Imaging 53:14–29
Rogasch JMM, Hundsdoerfer P, Hofheinz F et al (2018) Pretherapeutic FDG-PET total metabolic tumor volume predicts response to induction therapy in pediatric Hodgkin’s lymphoma. BMC Cancer 18:1–9
Ben Bouallègue F, Tabaa YA, Kafrouni M, Cartron G, Vauchot F, Mariano-Goulart D (2017) Association between textural and morphological tumor indices on baseline PET-CT and early metabolic response on interim PET-CT in bulky malignant lymphomas. Med Phys 44:4608–4619
Ko KY, Liu CJ, Ko CL, Yen RF (2016) Intratumoral heterogeneity of pretreatment 18F-FDG PET images predict disease progression in patients with nasal type extranodal natural killer/T-cell lymphoma. Clin Nucl Med 41:922–926
Wang M, Xu H, Xiao L, Song W, Zhu S, Ma X (2019) Prognostic value of functional parameters of 18 F-FDG-PET images in patients with primary renal/adrenal lymphoma. Contrast Media Mol Imaging 2019:1–9
Chalkidou A, O’Doherty MJ, Marsden PK (2015) False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One 10:1–18
Leijenaar RT, Carvalho S, Velazquez ER et al (2015) Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 52:1391–1397
Zwanenburg A (2019) Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging 46:2638–2655
Liu Z, Wang S, Dong D et al (2019) The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges. Theranostics 9:1303–1322
Hu H, Decazes P, Vera P, Li H, Ruan S (2019) Detection and segmentation of lymphomas in 3D PET images via clustering with entropy-based optimization strategy. Int J Comput Assist Radiol Surg 14:1715–1724
Van Velden FHP, Nissen IA, Jongsma F et al (2014) Test-retest variability of various quantitative measures to characterize tracer uptake and/or tracer uptake heterogeneity in metastasized liver for patients with colorectal carcinoma. Mol Imaging Biol 16:13–18
Camacho MR, Etchebehere E, Tardelli N, et al (2020) Validation of a multi-foci segmentation method for measuring metabolic tumor volume in Hodgkin’s lymphoma. J Nucl Med Technol 48:30–35
Hu X, Guo R, Chen J et al (2020) Coarse-to-fine adversarial networks and zone-based uncertainty analysis for NK/T-cell lymphoma segmentation in CT/PET images. IEEE J Biomed Health Inform. https://doi.org/10.1109/JBHI.2020.2972694
Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö (2019) Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol 25:485–495
Funding
This study has received funding by the Key Projects of the Ministry of Science and Technology (grant 2017YFC0113304).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Rong Tian, PhD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise. No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board because this was a retrospective study.
Ethical approval
The study was approved by the institutional ethics review board of the West China Hospital, Sichuan University.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, H., Zhao, S., Li, L. et al. Development and validation of an 18F-FDG PET radiomic model for prognosis prediction in patients with nasal-type extranodal natural killer/T cell lymphoma. Eur Radiol 30, 5578–5587 (2020). https://doi.org/10.1007/s00330-020-06943-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-06943-1