Abstract
Objective
To develop and validate a radiomics-based nomogram for preoperatively predicting grade 1 and grade 2/3 tumors in patients with pancreatic neuroendocrine tumors (PNETs).
Methods
One hundred thirty-eight patients derived from two institutions with pathologically confirmed PNETs (104 in the training cohort and 34 in the validation cohort) were included in this retrospective study. A total of 853 radiomic features were extracted from arterial and portal venous phase CT images respectively. Minimum redundancy maximum relevance and random forest methods were adopted for the significant radiomic feature selection and radiomic signature construction. A fusion radiomic signature was generated by combining both the single-phase signatures. The nomogram based on a comprehensive model incorporating the clinical risk factors and the fusion radiomic signature was established, and decision curve analysis was applied for clinical use.
Results
The fusion radiomic signature has significant association with histologic grade (p < 0.001). The nomogram integrating independent clinical risk factor tumor margin and fusion radiomic signature showed strong discrimination with an area under the curve (AUC) of 0.974 (95% CI 0.950–0.998) in the training cohort and 0.902 (95% CI 0.798–1.000) in the validation cohort with good calibration. Decision curve analysis verified the clinical usefulness of the predictive nomogram.
Conclusion
We proposed a comprehensive nomogram consisting of tumor margin and fusion radiomic signature as a powerful tool to predict grade 1 and grade 2/3 PNET preoperatively and assist the clinical decision-making for PNET patients.
Key Points
• Radiomic signature has strong discriminatory ability for the histologic grade of PNETs.
• Arterial and portal venous phase CT imaging are complementary for the prediction of PNET grading.
• The comprehensive nomogram outperformed clinical factors in assisting therapy strategy in PNET patients.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Pancreatic neuroendocrine tumors (PNETs) account for 2% and 10% of all pancreatic tumors [1], which now have an increasing diagnosed incidence probably due to the improvement of medical imaging in the past few years [2]. The 2010 WHO classification categorizes PNETs into three grades on the basis of the Ki-67 proliferation index and mitotic count [3]. All PNETs have potential to be malignant tumors according to the new classification; furthermore, PNETs with different histologic grades usually indicate a varied biological aggressiveness and have a significant correlation with prognosis [4,5,6].
As the WHO 2010 grading has shown a strong guidance for therapeutic decision-making in previous clinical practice [7, 8], the accurate assessment of grade is critical for the PNET patient. However, the grade can only be obtained through histopathological exams after surgery. The endoscopic ultrasonography-guided fine-needle aspiration before surgery had an efficient diagnosis of PNETs, but showed poor performance for differentiating histologic grade of PNETs and inevitable invasiveness [9, 10]. Many studies have tried to identify preoperative imaging biomarkers to predict tumor grade of PNETs using computed tomography (CT) and magnetic resonance (MR) [11,12,13,14,15,16,17,18,19,20,21,22,23]; however, limited accuracy or insufficient validation cannot fulfill the clinical requirements.
An emerging technique, termed as radiomics, provides a new way to solve this problem. Based on computer vision technology, radiomics automatically extracts imaging features from encrypted medical images [24,25,26]. The most correlated features with the clinical target would be selected using machine learning methods based on big medical data, thus generating a stable imaging marker–radiomic signature to realize the prediction. It has been successfully applied in cancer screening, diagnosis, and treatment evaluation [27,28,29,30,31,32]. In terms of grading classification, previous studies revealed the validity of radiomics on high-grade and low-grade discrimination in clear cell renal cell carcinoma, colorectal adenocarcinoma, and gliomas [33,32,35]. However, regarding PNET, research on histologic grade classification using the radiomics method is relatively limited. CT textural analysis was recently explored for PNET grading, but the results lacked either accuracy or essential external validation [18, 19]. These studies enlightened the work to solve the PNET grading using a quantitative imaging approach.
Thus, in this multicenter study, we build a radiomic-based predictive model to noninvasively and operatively achieve PNET grading using CT images. Meanwhile, we would also explore the predictive value of clinical and radiological variables, as comparisons with the radiomic signature. A final combined model integrating both radiomic and clinical factors is expected to accurately classify PNET grading.
Materials and methods
Workflow
The workflow of the radiomics analysis included image segmentation, feature extraction, radiomics signature modeling. and model analysis (Fig. 1).
Patients
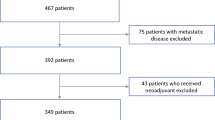
Both of the institutional review boards of the participating centers approved the retrospective study, and the requirement of the informed consent for patients was waived. According to the inclusion and exclusion criteria (Fig. 2), a total of 138 patients derived from two Chinese hospitals in different regions with pathologically confirmed PNET were recruited in our study. The patients from the two institutions were considered as training and validation cohorts respectively. The training cohort consisted of 104 patients (47 males and 57 females; mean age 52.4 ± 11.6 years; range 18–77 years) between April 2009 and November 2017 from Zhongshan Hospital of Fudan University, while the validation cohort contained 34 patients (15 males and 19 females; mean age 55.6 ± 12.5 years; range 19–82 years) between September 2009 and December 2017 from the Affiliated Hospital of Qingdao University. Clinical and radiological characteristics used in this study and the pathology analysis of the patients are described in Supplementary Information 1.
CT image acquisition, segmentation, and feature extraction
All arterial and portal venous phase CT images were retrieved from a picture archiving and communication system (PACS) for further analysis. The procedure of CT image acquisition, segmentation, reproducibility analysis, and feature extraction are described in Supplementary Information 2.
Clinical risk factors
Univariable analysis was used to assess the association between clinicopathological/radiological factors and PNET grading in the training and validation cohorts. The t test or Mann–Whitney U test were performed for quantitative variables, while the chi-square test or Fisher’s exact test was executed for qualitative variables to assess their differences between grade 1 and grade 2/3 groups. Those significant factors with a p value < 0.05 from the univariable analysis in the training cohort were entered into a stepwise multivariable logistic regression analysis. Variables with a p value < 0.05 from the multivariate analysis were identified as potential clinical risk factors related to the histologic grade and were included for clinical model building with multivariable logistic regression.
Radiomic feature reduction and selection
All of the radiomic features were standardized into a normal distribution with z scores. The intra- and inter-class correlation coefficients (ICCs), which were calculated from 15 segmented lesions in the CT images, were used to determine the intra- and inter-observer reproducibility of the radiomic features. Features with the ICCs higher than 0.80 were obtained in the following analysis. The univariable analysis was performed on the reproducible radiomic features using a t test. Variables with a p value < 0.1 were considered to be correlated to the PNET grading and were adopted for further analysis. In order to reduce the redundancy and unnecessary complexity for the computation and modeling, minimum redundancy maximum relevance (MRMR) was used for feature selection [36]. The algorithm generated an importance score for each feature, and then the top-ranked 20 features were entered into the fivefold cross-validation for the selection of the best feature number. The association between all of the selected features and grades was evaluated by a univariable analysis, and the predictive performance for a single feature was also assessed. All of the processes of the feature analysis were performed on the training cohort and validated on the validation cohort in both arterial and portal venous phase CT images.
Construction of the radiomic signature
A widely used machine learning classifier of the random forest (RF) was trained for the radiomic signature construction. Considering the potential interactions between features, we selected the best feature subset using the “wrapper” method [37] with the top-ranked 20 MRMR features. Specifically, the forward feature selection was done step by step according to the feature score with the fivefold cross-validation in the RF model. We repeated the process in the training cohort which was randomly divided into five folds 100 times; then, the optimal feature number and parameters for the RF model which had the maximum mean cross-validation area under the curve (AUC) for the classification of the grade were adopted for the single radiomic signature building. Furthermore, the fusion radiomic signature was developed by combining the two signatures of the arterial and portal venous phases in the multivariable logistic regression. The radiomic score was calculated for each patient to show the prediction risk of grade 2/3 via the radiomic signature. The association between the radiomic score and grade was explored by a t test.
Development of the nomogram
To test whether the radiomic signature and clinical factors were complementary for the prediction of grade, a comprehensive model incorporating significant clinical factors and the fusion radiomic signature with multivariable logistic regression analysis was also constructed on the training cohort. Next, a nomogram was developed based on the proposed comprehensive model as a graphical presentation. The calibration curves were created to identify the agreement of nomogram-predicted probability and the actual rate for grade 2/3 in both the training and validation cohorts with a Hosmer–Lemeshow test [38]. Decision curve analysis, considering the true-positive and false-positive rates synthetically, was conducted to validate the clinical utility of the nomogram by estimating the net benefits at a range of threshold probabilities [39].
Model evaluation and comparison
We compared the clinical model, radiomic signatures, and the comprehensive nomogram with a receiver operating characteristic (ROC) analysis. The ROC curves were plotted and AUCs were used to quantify the discriminative ability of each model. The performances of different ROC curves were compared by the Delong test. According to the radiomic score of each patient, we divided all of the patients into grade 1 and grade 2/3 by the optimal cutoff value. Thus, several evaluation indices including AUC with a 95% confidence interval (95% CI), accuracy (ACC), sensitivity (SENS), specificity (SPEC), positive predictive value (PPV), and negative predictive value (NPV) were calculated. We further validated the predictive nomogram using a stratification analysis in patients grouped by age and gender.
Statistical analysis
All of the statistical analyses were performed using SPSS (version 20.0) and R software (version 3.4.1). A p value less than 0.05 was considered statistically significant with a two-tailed test. The detailed R packages used in the study are listed in Supplementary Information 3.
Radiomic quality score
To assess the quality of our radiomics study, we calculated the radiomic quality score at http://www.radiomics.world/ [24].
Results
Patient characteristics
A total of 138 patients were contained in our study, among which 57 (41.3%) patients were categorized as grade 1, 69 (50%) patients were grade 2, and 12 (8.7%) patients were grade 3. No significant differences were found in age, gender, and histologic grade between the training and validation cohorts (Supplementary Table S1).
Clinical modeling
All of the basic clinicopathological and radiological characteristics were compared between grade 1 and grade 2/3 groups in both the training and validation cohorts using a univariable analysis, and the results are listed in Table 1. Univariate analysis showed that the tumor margin, carcinoembryonic antigen, preoperative liver metastasis, dilatation of the main pancreatic duct, pancreatic atrophy, and protrusion from the outline of the pancreas were significantly related to grade (p < 0.05). Examples of typical radiological characteristics are illustrated in Supplementary Fig. S1. At the multivariable logistic regression analysis, only the tumor margin (OR 0.091; 95% CI 0.032–0.231; p < 0.001) was considered an independent predictor. The final clinical model with a single factor tumor margin yielded an AUC of 0.764 (95% CI 0.681–0.847) in the training cohort and 0.625 (95% CI 0.500–0.750) in the validation cohort.
Radiomic feature selection
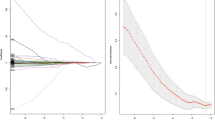
After the reproducibility analysis, 283 and 173 features were included for arterial and portal venous phase CT images separately. The distributions of the radiomic feature reproducibility in both phases are presented in Supplementary Fig. S2. Next, 205 and 82 features in the two phases were found to be significantly related to grade with a univariable analysis. The results of mean AUCs with 100 times fivefold cross-validation in the RF classifier and feature numbers are shown in Fig. 3. It can be inferred from the curves that the RF classifier obtained the best discrimination performance with 15 and 10 features in the arterial and portal venous phases respectively. The number of radiomic features after these feature selection methods is described in Table 2. All of the selected features of the double phases and predictive performance in the training and validation cohorts are shown in Supplementary Table S2.
The mean AUC for 100 times the fivefold cross-validation in the random forest classifier constructed with different feature subset sizes. The optimal feature subset sizes for the arterial and portal venous phases were 15 and 10 with the best cross-validation AUC of 0.795 and 0.684 in the training cohort
Performance of the radiomic signature
Either of the radiomic signatures of the arterial and portal venous phases yielded satisfying predictive performance with AUCs of 0.929 (95% CI 0.868–0.989) and 0.913 (95% CI 0.856–0.971) in the training cohort, and with AUCs of 0.877 (95% CI 0.764–0.991) and 0.879 (95% CI 0.735–1) in the validation cohort. The fusion radiomic signature gained optimal performance in both the training (AUC 0.970; 95% CI 0.943–0.997) and validation (AUC 0.881; 95% CI 0.760–1) cohorts. The best cutoff value for the grade classification of the fusion radiomic score was − 0.171. Significant difference of the fusion radiomic score in grade 1 and grade 2/3 groups was found in both the training (2.726 ± 1.183 vs. − 4.259 ± 4.096) and validation (1.660 ± 1.583 vs. − 1.150 ± 1.754) cohorts with a p value < 0.001 (Supplementary Fig. S3).
Development and validation of the nomogram
A comprehensive model integrating the tumor margin and fusion radiomic signature was established. Performances of the clinical model, radiomic signatures, and the comprehensive model are presented in Table 3. ROC curves of these models are shown in Fig. 4. The comprehensive model displayed the best predictive performance with AUCs of 0.974 (95% CI 0.950–0.998) and 0.902 (95% CI 0.798–1) in the training and validation cohorts respectively. The AUCs were much better than in the clinical model alone (p < 0.001), which indicated a powerful incremental value of the radiomic signature. Additionally, the patient characteristics and the performance of the proposed models with training and validation cohorts composed with each different period of two hospitals are presented in Supplementary Table S3 and S4. The performance of the proposed models after training and validation cohorts randomly divided is presented in Supplementary Table S5. Stratification analysis showed outstanding performance in both subgroups of age and gender (Table 4).
Comparison of receiver operating characteristics (ROC) curves for prediction of the histologic grade. ROC curves of the clinical model, radiomic signature for the arterial phase, portal venous phase, fusion radiomic signature, and the comprehensive model incorporating clinical risk factors and the fusion radiomic signature in the training (a) and validation (b) cohorts
A nomogram based on the comprehensive model which can provide the probability of grade 2/3 for PNETs is given in Fig. 5a. The best cutoff value for the predicted probability to classify grade 1 and grade 2/3 was 0.731. Calibration curves of the nomogram are shown in Fig. 5b with good agreement in both the training (p = 0.732) and validation (p = 0.119) cohorts. Decision curve analysis was adopted for the assessment of grade diagnosis (Fig. 6). The predictive nomogram obtained more net benefits than the clinical model with “treat-all” and “treat-none” strategies when the probability threshold was greater than 1% in the training cohort and 2% in the validation cohort.
(a) The nomogram based on the comprehensive model incorporating the clinical risk factor tumor margin and the fusion radiomic signature. To use the nomogram, locate the margin according to the patient information and draw a line straight up to the points axis to obtain the score associated with the margin. Repeat for the radiomics signature. By summing the scores of each point and locating it on the total points and drawing a line straight down to the bottom axis, the estimated probability of grade 2/3 could be determined. (b) Calibration curves for the training and validation cohorts. The y-axis represents the actual rate of grade 2/3 in the patients; the x-axis represents the nomogram-predicted probability of grade 2/3. The black diagonal dashed line means an ideal agreement fitted by a perfect model. Hosmer–Lemeshow test showed good agreement of the nomogram with p values > 0.05 in both the training and validation cohorts
Decision curves for the comprehensive model and clinical model in the training and validation cohorts. The y-axis represents the net benefit. The benefit was calculated by summing the benefits (true-positive results) and subtracting the harms (false-positive results), weighting by the relative harm of an undetected cancer compared with the harm of an unnecessary treatment. The relative harm was calculated with the “threshold probability p” by formula “p/(1 − p).” The threshold probability is where the expected benefit of treatment and the expected benefit of avoiding treatment are equal [37]. The orange line measures the benefit obtained from the comprehensive model, and the green line measures the benefit of the clinical model. The gray line represents the assumption that all patients were grade 2/3 (“treat all”); the black line represents the assumption that no patients were grade 2/3 (“treat none”). The comprehensive model shows the optimal net benefit in both the training (a) and validation (b) cohorts in most of the ranges of the threshold probability
Radiomic quality score
In addition to the prominent predictive performance, our study got a satisfactory score of 57.7% according to the estimation of the radiomic quality score.
Discussion
In this multicenter study, we proposed an optimal model integrating clinical risk factors and the fusion radiomic signature from the arterial and portal venous phase CT images for the preoperative prediction of histologic grade of PNETs. Firstly, the radiomic analysis showed that both the single-phase radiomic signatures could discriminate grade 2/3 from grade 1 with high accuracy. Secondly, the statistical analysis indicated that the clinical factor mostly related to the grade was tumor margin; however, the clinical model building with it had limited performance. Therefore, we developed the comprehensive model to test whether the radiomics signatures and clinical factor were complementary. The nomogram based on the comprehensive model obtained the most ideal performance with AUC 0.974 and 0.902 in the training and validation cohorts respectively; thereby, we considered it as a powerful tool for the prediction of PNET grading and clinical decision-making.
Previous studies have investigated the relationship between the tumor imaging characteristic and PNET grading [10,11,12,13,14,15,16,17,18,19,20,21,22,23]. These studies were commonly based on small sample size, utilized subjective semi-quantitative imaging descriptors, or lacked reliable external validation. Shigeru et al concluded that the CT ratio in dynamic CT could predict pathological grade 3 disease in PNET with high sensitivity, specificity, and diagnostic accuracy [10]. They did not validate the performance independently and had a poor accuracy of 47% for all grades. Belousova et al found that tumor size, arterial enhancement ratio, and contrast enhancement pattern in multidetector CT showed an accuracy of 74.7%, 79.5%, and 74.4%, respectively, for PNET grading prediction [15]. However, their study lacked the validation for the combined model and had less predictive performance. Two recent studies investigated whether the CT findings and CT texture analysis have predictive performance of PNET grading without independent validation [18, 19]; similarly, they both obtained AUCs less than 0.8, which was much lower than our independent external validation results. To our knowledge, this is the first multicenter study to assess the association between histologic grade and radiomic features in PNET patients in a single center and to validate the consequence in another center.
For the significant predictor in the clinical model, the ill-defined margin mostly existed in grade 2/3 in our study. Namely, the tumor in PNET patients with higher grade was more likely to have an ill-defined margin, which was consistently proven in previous studies [11, 12, 18, 20, 21]. A possible explanation is that higher-grade tumors tend to show more infiltration into the surrounding tissue than lower-grade tumors [40, 41].
For the radiomic signature development, the best features subset was selected using the MRMR feature selection algorithm and was subsequently applied to the RF classifier. MRMR was previously proven to be an effective and reliable feature selection method for radiomics [42]. The RF classifier was also given a stable and satisfying performance in a radiomic analysis [43]. Consistently with previous work, our radiomic model building with MRMR and RF showed great AUC with robust features.
For the fusion radiomic signature, we build a multivariable logistic regression model using the two single-phase radiomic signatures. The fusion radiomic signature outperformed either of the single-phase radiomic signatures. Potential reasons for this finding may be that the combination of the two phases could show the vascularity of PNETs more accurately than only one phase [44]. The fusion signature could also provide more textural information in the tumor microenvironments since the most effective features from the two phases in this study were texture features.
To explore clinical use, we established a nomogram based on the comprehensive model as an individualized tool to predict the risk of grade 2/3 for each PNET patient. Though the variability of CT image acquisition was found between the training and validation cohorts of the two institutions, the predictive nomogram had strong results with a higher AUC, which indicated a great capacity for prediction and generalization of the model. Furthermore, more net benefits of the model for the majority of the threshold probabilities could be derived from the decision curve analysis, meaning that using our nomogram for therapy strategy would get better clinical outcome. Thus, in patients who were diagnosed with PNET preoperatively [45], our nomogram provided a promising tool to assist radiologists and oncologists in diagnosis of grade and treatment. Specifically, with the stratified result of grade 1 and grade 2/3, different treatment strategies could be applied. Patients with grade 1 could undergo treatment of parenchyma-sparing pancreatic resection. And for patients with grade 2/3, the comprehensive treatment planning including surgical resection and systematic therapy would be recommended [46].
This study had several limitations. Firstly, as a retrospective study standing on multicenter cohorts, we used CT imaging with heterogeneous scanning parameters in various companies, which may cause potential noise in the distribution of images in the two cohorts. Though resampling and normalization were applied to all of the voxels in CT images in our study, further standardization for the preprocessing of the CT images in the radiomic analysis could be investigated. Secondly, due to limited sample size, the number of patients with grade 3 PNETs was very small. Therefore, we divided the patients into grade 1 and grade 2/3 PNETs. In the future, a larger sample size could be done for the prediction of three grades in PNETs. Thirdly, manual tumor segmentation was time-consuming. A more stable and efficient method such as automatic segmentation with deep learning could be applied to the radiomic analysis.
In conclusion, the proposed nomogram integrating the clinical predictor tumor margin and fusion radiomic signature had a powerful predictive capability for grade 1 and grade 2/3 in PNET patients. We presented it as a noninvasive and practical method to assist in the clinical diagnosis and decision-making of PNET patients.
Abbreviations
- ACC:
-
Accuracy
- AFP:
-
α-Fetoprotein
- AUC:
-
Area under the curve
- CA199:
-
Carbohydrate antigen 19-9
- CEA:
-
Carcinoembryonic antigen
- CI:
-
Confidence interval
- CT:
-
Computed tomography
- DMPD:
-
Dilatation of the main pancreatic duct
- GLCM:
-
Gray level co-occurrence matrix
- GLDM:
-
Gray level dependence matrix
- GLRLM:
-
Gray level run length matrix
- GLSZM:
-
Gray level size zone matrix
- ICCs:
-
Intra- and inter-class correlation coefficient
- MR:
-
Magnetic resonance
- MRMR:
-
Minimum redundancy maximum relevance
- NGTDM:
-
Neighboring gray tone difference matrix
- NPV:
-
Negative predictive value
- PA:
-
Pancreatic atrophy
- PACS:
-
Picture archiving and communication system
- PBG:
-
Preoperative blood glucose
- PFP:
-
Protrusion from the outline of the pancreas
- PLM:
-
Preoperative liver metastasis
- PNETs:
-
Pancreatic neuroendocrine tumors
- PPV:
-
Positive predictive value
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristics
- ROI:
-
Region of interest
- SENS:
-
Sensitivity
- SPEC:
-
Specificity
- WHO:
-
World Health Organization
References
Yao JC, Hassan M, Phan A et al (2008) One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol 26:3063–3072
Ohmoto A, Rokutan H, Yachida S (2017) Pancreatic neuroendocrine neoplasms: basic biology, current treatment strategies and prospects for the future. Int J Mol Sci 18:143
Bosman FT, Carneiro F, Hruban RH, Theise ND (2010) WHO classification of tumours of the digestive system, 4th edn. International Agency for Research on Cancer, Lyon
Reid MD, Balci S, Saka B, Adsay NV (2014) Neuroendocrine tumors of the pancreas: current concepts and controversies. Endocr Pathol 25:65–79
Klimstra DS, Modlin IR, Coppola D, Lloyd RV, Suster S (2010) The pathologic classification of neuroendocrine tumors: a review of nomenclature, grading, and staging systems. Pancreas 39:707–712
Scarpa A, Mantovani W, Capelli P et al (2010) Pancreatic endocrine tumors: improved TNM staging and histopathological grading permit a clinically efficient prognostic stratification of patients. Mod Pathol 23:824–833
Zhou C, Zhang J, Zheng Y, Zhu Z (2012) Pancreatic neuroendocrine tumors: a comprehensive review. Int J Cancer 131:1013–1022
Öberg K (2012) Neuroendocrine tumors of the digestive tract: impact of new classifications and new agents on therapeutic approaches. Curr Opin Oncol 24:433–440
Larghi A, Capurso G, Carnuccio A et al (2012) Ki-67 grading of nonfunctioning pancreatic neuroendocrine tumors on histologic samples obtained by EUS-guided fine-needle tissue acquisition: a prospective study. Gastrointest Endosc 76:570–577
Horiguchi S, Kato H, Shiraha H et al (2017) Dynamic computed tomography is useful for prediction of pathological grade in pancreatic neuroendocrine neoplasm. J Gastroenterol Hepatol 32:925–931
De Robertis R, Cingarlini S, Martini PT et al (2017) Pancreatic neuroendocrine neoplasms: magnetic resonance imaging features according to grade and stage. World J Gastroenterol 23:275–285
Kim JH, Eun HW, Kim YJ, Han JK, Choi BI (2013) Staging accuracy of MR for pancreatic neuroendocrine tumor and imaging findings according to the tumor grade. Abdom Imaging 38:1106–1114
Jang KM, Kim SH, Lee SJ, Choi D (2014) The value of gadoxetic acid-enhanced and diffusion-weighted MRI for prediction of grading of pancreatic neuroendocrine tumors. Acta Radiol 55:140–148
Besa C, Ward S, Cui Y, Jajamovich G, Kim M, Taouli B (2016) Neuroendocrine liver metastases: value of apparent diffusion coefficient and enhancement ratios for characterization of histopathologic grade. J Magn Reson Imaging 44:1432–1441
Belousova E, Karmazanovsky G, Kriger A et al (2017) Contrast-enhanced MDCT in patients with pancreatic neuroendocrine tumours: correlation with histological findings and diagnostic performance in differentiation between tumour grades. Clin Radiol 72:150–158
Min JH, Kang TW, Kim YK et al (2018) Hepatic neuroendocrine tumour: apparent diffusion coefficient as a potential marker of prognosis associated with tumour grade and overall survival. Eur Radiol 28:2561–2571
Hwang EJ, Lee JM, Yoon JH et al (2014) Intravoxel incoherent motion diffusion-weighted imaging of pancreatic neuroendocrine tumors: prediction of the histologic grade using pure diffusion coefficient and tumor size. Invest Radiol 49:396–402
Choi TW, Kim JH, Yu MH, Park SJ, Han JK (2018) Pancreatic neuroendocrine tumor: prediction of the tumor grade using CT findings and computerized texture analysis. Acta Radiol 59:383–392
Canellas R, Burk KS, Parakh A, Sahani DV (2018) Prediction of pancreatic neuroendocrine tumor grade based on CT features and texture analysis. AJR Am J Roentgenol 210:341–346
Guo C, Chen X, Xiao W, Wang Q, Sun K, Wang Z (2017) Pancreatic neuroendocrine neoplasms at magnetic resonance imaging: comparison between grade 3 and grade 1/2 tumors. Onco Targets Ther 10:1465
Kim DW, Kim HJ, Kim KW et al (2015) Neuroendocrine neoplasms of the pancreas at dynamic enhanced CT: comparison between grade 3 neuroendocrine carcinoma and grade 1/2 neuroendocrine tumour. Eur Radiol 25:1375–1383
Kang J, Ryu JK, Son JH et al (2018) Association between pathologic grade and multiphase computed tomography enhancement in pancreatic neuroendocrine neoplasm. J Gastroenterol Hepatol 33:1677–1682
Zhao W, Quan Z, Huang X et al (2018) Grading of pancreatic neuroendocrine neoplasms using pharmacokinetic parameters derived from dynamic contrast-enhanced MRI. Oncol Lett 15:8349–8356
Lambin P, Leijenaar R, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Chen J, Tian J (2009) Real-time multi-modal rigid registration based on a novel symmetric-SIFT descriptor. Progress in Natural Science-Materials International 19:643-651
Cameron A, Khalvati F, Haider MA, Wong A (2016) MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng 63:1145–1156
Huang YQ, Liang CH, He L et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157–2164
Liu Z, Zhang XY, Shi YJ et al (2017) Radiomics analysis for evaluation of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Clin Cancer Res 23:7253–7262
Elhalawani H, Kanwar A, Mohamed ASR et al (2018) Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci Rep 8:13
Ger RB, Cardenas CE, Anderson BM, Yang J, Mackin DS, Zhang L (2018) Guidelines and experience using imaging biomarker explorer (IBEX) for Radiomics. J Vis Exp:e57132
Zhang B, Tian J, Dong D et al (2017) Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin Cancer Res 23:4259–4269
Ding JL, Xing ZY, Jiang ZX et al (2018) CT-based radiomic model predicts high grade of clear cell renal cell carcinoma. Eur J Radiol 103:51–56
Huang X, Cheng Z, Huang Y et al (2018) CT-based radiomics signature to discriminate high-grade from Low-grade colorectal adenocarcinoma. Acad Radiol. https://doi.org/10.1016/j.acra.2018.01.020
Tian Q, Yan LF, Zhang X et al (2018) Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging. https://doi.org/10.1002/jmri.26010
Peng H, Long F, Ding C (2005) Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27:1226–1238
Phuong TM, Lin Z, Altman RB (2005) Choosing SNPs using feature selection. Computational Systems Bioinformatics Conference 301–309
Kramer AA, Zimmerman JE (2007) Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med 35:2052–2056
Vickers AJ, Elkin EB (2006) Decision curve analysis: a novel method for evaluating prediction models. Med Decis Mak 26:565–574
Hu Y, Rao S, Xu X, Tang Y, Zeng M (2018) Grade 2 pancreatic neuroendocrine tumors: overbroad scope of Ki-67 index according to MRI features. Abdom Radiol (NY). https://doi.org/10.1007/s00261-018-1573-5
Kasajima A, Yazdani S, Sasano H (2015) Pathology diagnosis of pancreatic neuroendocrine tumors. J Hepatobiliary Pancreat Sci 22:586–593
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087
Breiman L (2001) Random forests. Mach Learn 45:5–32
Horton KM, Hruban RH, Yeo C, Fishman EK (2006) Multi–detector row CT of pancreatic islet cell tumors. Radiographics 26:453–464
Low G, Panu A, Millo N, Leen E (2011) Multimodality imaging of neoplastic and non-neoplastic solid lesions of the pancreas. Radiographics 31:993–1015
Valle JW, Eatock M, Clueit B, Gabriel Z, Ferdinand R, Mitchell S (2014) "A systematic review of non-surgical treatments for pancreatic neuroendocrine tumours" (vol 40, pg 376, 2014). Cancer Treat Rev 40:1037–1037
Funding
This study has received funding by the National Natural Science Foundation of China (No. 81227901, 81527805, 61231004, 81771924, 81501616), National Key Research and Development Program of China (2017YFA0205200, 2017YFC1308700), and the Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-SW-STS-160).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jie Tian.
Conflict of interest
The authors declare that they have no competing interests.
Statistics and biometry
Dr. Jingwei Wei from the University of Chinese Academy of Sciences, who is one of the authors, has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board of Zhongshan Hospital Affiliated to Shanghai Fudan University and Affiliated Hospital (Laoshan hospital) of Qingdao University.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1238 kb)
Rights and permissions
About this article
Cite this article
Gu, D., Hu, Y., Ding, H. et al. CT radiomics may predict the grade of pancreatic neuroendocrine tumors: a multicenter study. Eur Radiol 29, 6880–6890 (2019). https://doi.org/10.1007/s00330-019-06176-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-019-06176-x