Abstract
Main conclusion
A WRKY transcription factor encoding NbWRKY79, which was induced by salt and ABA was isolated from Nicotiana benthamiana . Overexpression of NbWRKY79 resulted in enhanced tolerance to salt stress.
In plants, there are many families of transcriptional regulators, one of which is WRKY transcription factors, which have a significant effect on the adaptation to abiotic stress. Nevertheless, most of the mechanisms in plant to which WRKY genes are concerned to tolerate salinity are still undiscovered. In this study, a gene from Nicotiana benthamiana, NbWRKY79, was isolated and characterized. NbWRKY79 contains one WRKY domain and localizes in the nucleus. NbWRKY79 was induced after the plant was exposed to salinity and abscisic acid (ABA). The overexpression of NbWRKY79 remarkably enhanced the tolerance of tobacco plant to salinity, which was confirmed when the plant growth, root growth and chlorophyll content were studied through physiological analyses. The sensitivity to ABA-mediated seed germination and seedling root growth of NbWRKY79 transgenic lines were increasing. In addition, the reduced accumulation of reactive oxygen species and malondialdehyde content as well as an increase in proline content and the activity of antioxidant enzymes such as superoxide dismutase, guaiacol peroxidase, catalase and ascorbate peroxidase during salt treatment were found, and these indicated that the transgenic plants enhanced tolerance to oxidative stress when comparing to the wild-type plants. Furthermore, it was found that ABA content and transcript levels of ABA-inducible genes, including NbAREB, NbDREB and NbNCED, were significantly increased in the salt stress conditions. These results recommend that NbWRKY79 holds the key to salt stress response.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stress, one of which is salinity, is the principal cause of crop yield loss worldwide and adversely affects plant growth and productivity. Plants are sessile, so they must continually exhibit high-level tolerance against stresses from the environmental surroundings (Mittler 2006). To survive and grow under fluctuating and stressful environmental conditions, plants have evolved intricate mechanisms to recognize external signaling networks and to be evidence of adaptive responses at the physiological, biochemical, and molecular levels. In these responses, many genes involved in stress response are regulated to not only protect plant cells against environmental stress via the generation of important proteins and enzymes of metabolism but also control signaling pathways and gene expression. Transcription factors (TFs), key components of the signaling pathways, are present at the upstream region, which can regulate downstream genes that respond to stress and play an important role in supporting stress tolerances (Nakashima et al. 2009; Rushton et al. 2010). WRKY proteins are one of the largest families of plant transcription factors (Ülker and Somssich 2004). The WRKY transcription factors are named after the term WRKY domain, which has a highly conserved amino acid sequence, WRKYGQK, at the N terminus and an atypical zinc-finger structure at the C terminus (Eulgem et al. 2000; Eulgem and Somssich 2007; Rushton et al. 2010). According to the phylogenetic data of WRKY domains and the pattern of the zinc-finger-like motif, WRKY proteins are grouped into I, II, and III (Eulgem et al. 2000). Group II is subdivided into IIa, IIb, IIc, IId, and IIe (Rushton et al. 2010). There have been 104 and 74 WRKY genes identified in rice and Arabidopsis thaliana, respectively (Eulgem et al. 2000). All WRKY proteins that were characterized have one or two WRKY domains. The plant responses to biotic and abiotic stresses were controlled by WRKY proteins attaching to W-box elements (TTGACC/T) in the target gene promoters to up- or down-regulate them at the transcription level (Eulgem et al. 2000; Eulgem and Somssich 2007; Ren et al. 2010; Rushton et al. 1995). Many studies concentrated on defense-responsive WRKY proteins (Birkenbihl et al. 2012; Journot-Catalino et al. 2006; Li et al. 2006; Matsushita et al. 2013; Qiu and Yu 2009; Xu et al. 2006); however, a few studies have verified the roles of these proteins in abiotic stress responses (Yan et al. 2014). Recent evidence suggests that WRKY genes possibly participate in the positive and negative regulation of hormones and the abiotic stress response such as salinity, drought, osmotic, and heat stress (Chen et al. 2012; Li et al. 2013; Liu et al. 2014; Miller et al. 2008; Ren et al. 2010; Rushton et al. 2012). Nevertheless, which of the specific WRKY genes are specifically associated with abiotic stress tolerance are still unclear.
In this study, we isolated a novel group II WRKY gene, NbWRKY79, from Nicotiana benthamiana plant and characterized its function in response to salt stress. In comparison with wild-type plants, NbWRKY79 transgenic plants exhibited enhanced salt tolerance with remarkably increased levels of proline content, reduced levels of MDA, lower ROS content, higher activity of ROS-scavenging isoenzymes, and higher ABA content.
Materials and methods
Plant material, growth conditions, and treatments
Wild-type and transgenic N. benthamiana seeds were surface sterilized with a solution of 20% commercial bleach (2% sodium hypochlorite) for 10 min, and rinsed three times with sterile distilled water. For root growth measurements, 5-day-old seedlings cultured on one-half strength solid MS medium (Duchefa Biochemie, Haarlem, Netherlands) were transferred to one-half strength MS medium supplemented with different concentrations of NaCl (0, 200 and 400 mM) or ABA (0, 25 and 50 μM). The tobacco seedlings were cultured at 25 ± 2 °C with cool white fluorescent light (100 μE s−1 m2 light intensity) under long-day condition (16 h light/8 h dark). Plates were oriented vertically with seedlings kept upside down. Salt stress was imposed on the soil-grown plants by treating with 100 mM NaCl daily. Three replicates were performed for each experiment.
Expression constructs
Homology search using an N. benthamiana Blast tool at the database Sol genomics network Boyce Thompson Institute for Plant Research (SGN, http://solgenomics.net/tools/blast/index.pl) was performed for isolation of the full-length cDNA sequence of NbWRKY79. The expressed sequence tags (ESTs) with accession number Niben.v0.4.2.Scf781 in N. benthamiana were obtained using Nicotiana tabacum NtWRKY79 as the query nucleotide sequence. The design of the forward and reverse primers spanning the open reading frame of NbWRKY79 was based on the ESTs. PCR amplifications were performed with pfu polymerase (Promega, Madison, WI, USA) in 1× reaction buffer, 0.1 mM dNTPs, 0.2 μM each of primers, and templates, under the following conditions: 2 min at 95 °C, followed by 30 cycles of 95 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min, and a final polymerization at 72 °C for 10 min. The resulting full-length NbWRKY79 was cloned into the pENTR-D TOPO (Invitrogen, Carlsbad, CA, USA) and subcloned into plant destination vector pH7WG2 (Plant Systems Biology, Ghent, Belgium) by LR recombination reaction (Invitrogen, Carlsbad, CA, USA). The integrity of the constructs was confirmed by sequence analysis.
For subcellular localization assay, NbWRKY79 in pENTR-D TOPO (Invitrogen, Carlsbad, CA, USA) was subcloned into plant destination vector pSITEII-4C1 (Martin et al. 2009) by LR recombination reaction (Invitrogen, Carlsbad, CA, USA) to generate Venus fusion proteins.
For promoter analysis, promoter region (2.0 kb) upstream the start codon of NbWRKY79 gene was amplified using N. benthamiana genomic DNA as template and the following primers: forward, 5′-CCAACCACTGGAAGCTTTCA-3′ and reverse, 5′- TCCGAGGAAATATATATTGAAG-3′. The NbWRKY79 promoter fragment was cloned into the pENTR-D TOPO (Invitrogen, Carlsbad, CA, USA) and subcloned into plant destination vector pHGWFS7 (Plant Systems Biology, Ghent, Belgium) to generate ProNbWRKY79-β-glucuronidase (GUS) reporter construct. The transgenic lines were confirmed by PCR using promoter-specific primers.
Transient expression of fluorescent proteins in tobacco leaves
The Agrobacterium strain GV3011 harboring the YFP:NbWRKY79-derived construct was used for the transient experiment as described (Bhaskar et al. 2009). The Agrobacterium were grown at 28 °C in Luria–Bertani medium containing 50 mg l−1 of kanamycin. Overnight cultures were centrifuged and the cell pellets were reconstituted in infiltration medium (10 mM MgCl2, 10 mM MES and 200 μM acetosyringone) to an optical density at 600 nm of 2, and then left at room temperature for 3–4 h. The cell suspensions of Agrobacterium strains harboring the YFP:NbWRKY79 were infiltrated into leaves of 2–4-week-old N. benthamiana plants using a needleless syringe. Four days after the infiltration, the abaxial epidermis of infiltrated tobacco leaves was assayed for fluorescence through confocal laser-scanning microscopy (LSM780, Carl Zeiss, Germany). Nuclei were stained by adding one drop of 4′,6-diamidino-2-phenylindole (DAPI) staining solution (1 μg ml−1) to epidermal cells. Epidermal cells were set in the dark condition for 15 min, and DAPI fluorescence was visualized by UV illumination.
Plant transformation
ProNbWRKY79-β-glucuronidase (GUS) reporter and NbWRKY79 constructs were introduced into A. tumefaciens strain GV 3101 by electroporation. N. benthamiana transformation was carried out using the leaf-dish method described by Horsch et al. (1985). The kanamycin-resistant plantlets regenerated from transformed callus were transferred to Vriezenveen 70L substrates (Potgrond Vriezenveen bv, Westerhaar, Netherlands) and grown in a growth chamber at 27 °C under a 16-h light and 8-h dark regime. Young plantlets in the pots were covered with clear plastic cups to retain moisture, and hardened off gradually by removing the cups. The seeds of 20 independent lines were harvested from these primary transformants. Ten T1 transgenic lines were obtained using kanamycin selection and PCR. The transgenic T2 lines were retained for further experiments.
Isolation and purification of total RNA
Total RNA was isolated from leaf tissues of 3-week-old N. benthamiana plants using a RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany), treated with DNase I (QIAGEN) to remove genomic DNA contamination, then purified and concentrated with a RNeasy MinElute Cleanup Kit (QIAGEN, Hilden, Germany). The yield and purity of total RNA was measured spectrophotometrically (NanodropND 2000, Nanodrop technologies, Wilmington, DE, USA). RNA samples of 2 µg µl−1 with high purity (OD260/280 and OD260/230 >2) were employed for further analysis.
Semi-quantitative PCR analysis
To assess the expression of NbWRKY79 under salt stress or exposure to ABA, 3-week-old tobacco seedlings were treated with 200 mM NaCl or 50 μM ABA. The total RNA from stressed samples was isolated at five different time points and up to a maximum time of 48 h after imposition of salt or ABA treatment. Then cDNA was prepared. Semi-qPCR analysis was performed using NbWRKY79 forward primer 5′-CTCCTAACGGTTCAGATGATGG-3′ and NbWRKY79 reverse primer 5′-GATGCAGAGGATGTTCTGTCC-3′. The N. benthamiana elongation factor 1-α (NbEF1-α) gene was used as an internal loading control. Biological and technical replicates of each sample were applied for analysis.
The expression levels of combination of native endogenous NbWRKY79 and overexpressing NbWRKY79 in the wild-type plants or the transgenic lines were identified through semi-qPCR using the following primers: forward, 5′-CTCCTAACGGTTCAGATGATGG-3′ and reverse, 5′-GATGCAGAGGATGTTCTGTCC-3′. The forward primer 5′-CTCCTAACGGTTCAGATGATGG-3′ and the reverse primer 5′-CATGTACACACACACAACGGG-3′, which were derived from the 3′-untranslated region of the NbWRKY79 mRNA, were used to analyze the expression level of the native endogenous NbWRKY79 mRNA in the transgenic lines.
Real-time PCR analysis
The cDNA was the result of the reverse transcription using 1 µg of total RNA in 20 µl of reaction according to the manufacturer’s instructions (Promega, Madison, WI, USA). Quantitative real-time PCR (qRT-PCR) reactions were conducted to quantify the transcript levels of the selected genes using the StepOnePlus Real Time PCR system (Applied Biosystems, Foster City, CA, USA) and iQ™ SYBR® Green Supermix (Bio-RAD, Hercules, CA, USA). The NbAREB1 (ABA-responsive element binding), NbDREB (dehydration-responsive element binding), and NbNCED (nine-cis-epoxycarotenoid dioxygenase) primers were used to determine NbAREB, NbDREB, and NbNCED expression levels under salt stress in this study by following the method of Yan et al. (2014) (Table S). The thermal cycles consisted of an initial 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Following PCR, a melting curve analysis was accomplished. Relative quantification of specific mRNA levels was analyzed using the cycle threshold (Ct) 2−ΔΔCt method (Livak and Schmittgen 2001). All reactions were made in triplicates of three independent samples. To normalize the relative expression levels, Nicotiana benthamiana β-actin genes were used as the reference gene. All statistical analyses to identify the significant difference of relative expression of individual genes between control and NaCl-treated samples were made by one-way analysis of variance (ANOVA) followed by Duncan’s Multiple Range test, using the SPSS software (version 11.5 for Windows; SPSS, Inc., Chicago, IL, USA). The different letters were the means and comparisons with p < 0.05 were considered significantly different.
Histochemical detection of GUS activity
Expression using GUS assays in transgenic N. benthamiana plants containing reporter constructs was first examined in 20 independent T1 plants to determine lines with a consistent expression pattern. The T2 generation was analyzed in detail. For histochemical detection of GUS activity, seedlings or explants were treated as described by Jefferson et al. (1987). Plant tissues were cleaned up after GUS staining with ethanol and acetic acid (3:1) treatment to remove chlorophyll before visual examination using a Leica MZ12.5 stereomicroscope (Leica, Wetzlar, Germany).
Measurements of photosynthesis parameter, proline content and MDA content
The Portable Photosynthesis System CIRAS-2 was used to determine the photosynthetic rate of the wild-type plant and the transgenic lines as described by Li et al. (2014b). For the measurement of proline and MDA content, leaves of the wild-type plant and the transgenic lines were collected after 4 weeks of salt treatment. MDA content determination was done using the thiobarbituric acid (TBA) reaction as described by Ma et al. (2013). Proline content measurement was performed by the ninhydrin reaction method as described by Bates et al. (1973). All the assays were conducted at least three times per sample. The results were subjected to ANOVA using SPSS version 11.5 (SPSS, Inc., Chicago, IL, USA) and comparisons with p < 0.05 were considered significantly different.
In situ detection of reactive oxygen species
Superoxide anion and H2O2 levels in N. benthamiana leaves treated with 200 mM NaCl were visually detected by treating with nitroblue tetrazolium (NBT; Sigma, St. Louis, MI, USA) and 3,3′-diaminobenzidine (DAB; Sigma, St. Louis, MI, USA), respectively (Thordal-Christensen et al. 1997).
Leaf discs salt stress bioassay and estimation of chlorophyll
For the salt treatment, leaf discs (9 mm in diameter) were segregated from healthy, fully expanded leaves of 3-week-old wild-type plants and NbWRKY79-overexpressing lines. The discs were floated in liquid MS medium supplied with different concentrations of NaCl (0, 100, 200 and 400 mM) for 4 days in a growth chamber. Growth chamber conditions were maintained at 27 °C and 70% humidity under long-day regime (16 h white light/8 h dark). The effect of salt stress on leaf discs was estimated by quantifying the chlorophyll content from 10 leaf discs per wild-type plants and NbWRKY79-overexpressing lines. The leaf discs were submerged into liquid nitrogen and ground using the TissueLyser LT (QIAGEN, Hilden, Germany). The extract of chlorophyll was collected from plant materials by homogenizing leaf discs in 1-ml aliquots of 80% (v/v) chilled acetone, and the absorbance of the extract was measured. The significant difference between the samples at p < 0.05 was subjected to ANOVA using SPSS version 11.5 (SPSS, Inc., Chicago, IL, USA).
Protein extraction and antioxidant enzyme activity assays
After treatment with 200 mM NaCl, protein of tobacco leaves was extracted. Approximately 1 gam of tobacco leaves was ground in protein extract buffer (50 mM Tris–HCl, pH 7.4, 250 mM sucrose, 10 mM NaF, 10 mM Na3VO4, 1 mM sodium-tartrate, 10% v/v glycerol, 50 mM Na2S2O5, 1% SDS, 1 mM PMSF). Homogenized leaf tissue was centrifuged at 15,700g for 10 min at room temperature and the supernatant was collected. Protein content was identified by the BioRad Dc Protein Assay (Bio-Rad, Hercules, CA, USA) at OD750.
Isoenzyme of SOD, CAT and POD were indicated as described by Wang and Yang (2005). APX in-gel activity was assayed using the method of Mittler and Zilinskas (1993). POD activity in the leaves was assayed according to Chen et al. (2002). The antioxidant enzyme activities of SOD, CAT and APX were determined as per the method of Kumar et al. (2008). Each assay was replicated at least three times per sample.
Quantification of endogenous ABA content
To determine the ABA accumulation under salt stress, 3-week-old tobacco plants were treated with or without 200 mM NaCl for one week. After treatment, the leaf samples were collected, weighed and ground in liquid nitrogen and homogenized in 80% (v/v) methanol containing 1 mM butylated hydroxytoluene overnight at 4 °C to extract the ABA. After centrifugation at 4000 g for 20 min at 4 °C, the supernatant was collected and then loaded onto a Sep-Pak C18 cartridge (Waters, Milford, MA, USA) and dried in N2. The eluate containing ABA was dissolved in 0.01 M phosphate-buffered saline, pH 7.5. The endogenous ABA content was analyzed using an ABA ELISA Kit (Mybiosource, San Diego, CA, USA) according to the manufacturer’s instructions.
Results
Isolation of a full-length cDNA encoding NbWRKY79 from N. benthamiana
A full-length cDNA clone of 744 nt was isolated and termed as NbWRKY79. The nucleotide sequence of the NbWRKY79 cDNA contained an open reading frame, which encodes for a protein of 247 amino acids. The molecular mass of the predicted polypeptide was 27.68 kDa. The NbWRKY79 protein sequence showed significant homology to Nicotiana tabacum NtWRKY79, Solanum tuberosum StWRKY2-like, and Nicotiana tomentosiformis NtoWRKY33, with 88, 54, and 81% identity, respectively. Due to the high degree of homology with Nicotiana tabacum NtWRKY79, this new WRKY protein was designated as NbWRKY79. The gene sequence encoding NbWRKY79 was deposited in GenBank (accession no. KR559678). Sequence analysis indicated that the deduced protein contains a single WRKY DNA-binding domain (WRKYGQK) and a zinc-finger-like motif C2-H2 (C-X4-C-X23-H-X-H) (Fig. S1), which categorized this protein as a member of group II WRKY superfamily (Eulgem et al. 2000).
Expression of NbWRKY79 is induced by salt stress and ABA
To analyze the expression profile of NbWRKY79, semi-quantitative RT-PCR was used. NbWRKY79 was found to be up-regulated under salt and ABA treatments (Fig. 2). For salt stress, NbWRKY79 transcription was induced within 6 h of NaCl treatment. The transcript level was significantly increased after 12 h of NaCl stress to seedlings and the level was continuously increased until 48 h (Fig. 1a). In the case of ABA treatment, the expression of NbWRKY79 was increased and the highest level was reached at 6 h, we thus speculated that NbWRKY79 is an ABA-sensitive gene (Fig. 1b). According to the result above, NbWRKY79 may be concerned to salt stress and ABA adaptation.
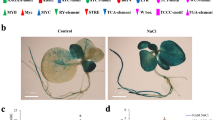
Localization of NbWRKY79 in N. benthamiana epidermal cells and tissue-specific expression patterns of N. benthamiana plants transformed with an NbWRKY79 promoter driven GUS reporter gene (pNbWRKY79::GUS construct) in response to salt stress. a YFP:NbWRKY79 constructs were transiently expressed in epidermal cells of N. benthamiana by agro-infiltration. Epidermal cells were observed by CLSM. Merged, merging of YFP and bright field imaged; YFP, representing of YFP signals; DAPI, staining with DAPI indicated the localization of cell nuclei. White bars, 10 µm. GUS expression patterns in uninduced transgenic seedlings (b) and in transgenic seedlings treated with NaCl (c). Five-day-old transgenic seedlings were treated without salt stress (Murashige and Skoog medium) or for 24 h with 200 mM NaCl. In untreated transgenic seedlings, GUS activity was found in root tip, lateral root and along vascular bundles. In transgenic seedlings treated with NaCl, GUS activity was presented in cotyledons, root (root tip, lateral root and along vascular bundles). Ten seedlings at least were examined and typical results are presented. d GUS activity in sepals, petals, anthers, stigma, ovary and leaf of transgenic plants treated with NaCl. Ten samples at least were examined and typical results are presented
Subcellular localization of NbWRKY79
To identify the subcellular localization of NbWRKY79 protein, we fused YFP with NbWRKY79 under regulation of the Cauliflower mosaic virus 35S promoter (Fig. S2). In transient expression assay, YFP-NbWRKY79 fusions and the control empty-vectors were introduced into leaf epidermal cells of N. benthamiana by Agrobacterium-mediated infiltration. The cells were dyed by DAPI to reveal nuclei and then investigated by a fluorescence microscopy. As a result, YFP-NbWRKY79 was shown to localize in the nucleus whereas the control YFP was distributed throughout epidermal cells (Fig. 3a). These findings suggest that NbWRKY79 is a nuclear protein.
Increased salt tolerance in transgenic lines overexpressing NbWRKY79. Five-day-old wild-type and NbWRKY79-overexpressing (O.E) lines geminated on one-half strength MS medium were transferred to one-half strength MS medium supplemented with 0 mM to 400 mM NaCl and solidified with agar. Seedlings were kept in a growth chamber at 25 °C for 5 days under 16 h of light (50 - 100 µmol m−2 s−1) and 8 h of darkness daily. a Comparison of primary root length of the wild-type seedlings and NbWRKY79-overexpressing lines under salt stress. The plants grown on MS medium and MS medium supplemented with NaCl, and root lengths were after 5 days. White bar, 2 cm. b Comparison of root measurements. Data are the means of analyses of five individual seedlings from each line ± SD. c Phenotype of wild-type plant and NbWRKY79-overexpressing (O.E) N. benthamiana lines under salt stress. Soil-grown plants were irrigated with a 200 mM NaCl solution daily. Photos were taken 4 weeks after the plants were treated. d Leaf surface area. e Photosynthesis rate. f Proline content. g MDA content. The graphs represent the mean and SD over three replicates. The different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test
Expression analysis using NbWRKY79 promoter–GUS fusions in transgenic plants
To further elucidate whether the different roles of NbWRKY79 in stress response correlate with differential expression patterns, GUS reporter gene fusions (ProNbWRKY79:GUS) were constructed using approximately 2.0 kb of the upstream regulatory region for NbWRKY79 gene (Fig. S3). We introduced the construct harboring the NbWRKY79 promoter fused to the GUS reporter gene into the wild-type N. benthamiana plants. Non-transformed plants were also applied as a negative control. In consequence, no expression of GUS gene was observed in any tissues of non-transformed plants (Fig. S4) but the expression was detected in primary root and lateral root of plants transformed with ProNbWRKY79:GUS (Fig. 3b). Next, we analyzed the inducible patterns of GUS expression in N. benthamiana plants in response to salt stress. Salt treatments strongly induced GUS activity in primary roots, lateral roots, cotyledons, true leaves, and shoot apical meristem (Fig. 3c). In addition, GUS activity was detected in petals, sepals, anthers, stigma, and ovary in the transformed plants under salt stress (Fig. 3d). These results indicate that NbWRKY79 has differential salt stress responses.
Overexpression of NbWRKY79 exhibits enhanced tolerance to salt stress
Transgenic plants overexpressing NbWRKY79 were produced to further examine the role of NbWRKY79. Two representative lines, O.E1 and O.E2, were selected from ten T1 transgenic lines for further analyses by semi-quantitative PCR (Fig. S5a). Semi-quantitative RT-PCR results made manifest that all transgenic plants expressed a high level of NbWRKY79 mRNA compared with the wild-type plant in normal condition (Fig. S5b). The expression level of native endogenous NbWRKY79 gene remained unaltered in the wild-type plants and the transgenic lines. All transgenic lines were indistinguishable from the wild-type plants in both morphology and development throughout their life cycle (Fig. S4). Then their T2 homozygous line was selected for further analyses.
To ascertain the possible effects of NbWRKY79 overexpression on salt tolerance, 5-day-old wild-type and NbWRKY79-overexpressing seedlings cultivated on MS medium were transferred to MS medium supplemented with 0 to 400 mM of NaCl. The growth of seedlings was monitored by measuring their root length after 5 days (Fig. 3a, b). Under control conditions, in the absence of both salt treatments, roots of wild-type seedlings and NbWRKY79-overexpressing lines grew at the same rate. At 200 mM NaCl, the root growth rate of wild-type plants and NbWRKY79-overexpressing lines was slower than the rate of those in the absence of NaCl. In comparison, seedlings of NbWRKY79-overexpressing lines grew faster than wild-type seedlings and had longer primary roots. The growth of roots in NbWRKY79-overexpressing lines was inhibited at 400 mM NaCl. The wild-type plants did not grow at this concentration.
To further examine the effect of NbWRKY79 overexpression on vegetable growth stage, soil-grown wild-type plants and NbWRKY79-overexpressing lines were exposed to long-term high salinity. Three-week-old plants were irrigated with 200 mM NaCl solution for 4 weeks and physiological parameters were measured. The growth of the wild-type plants was more deferred than the growth of NbWRKY79-overexpressing lines (Fig. 3c). The wild-type lines had a severe signs of stress, particularly serious chlorosis and wilting, whereas NbWRKY79-overexpressing lines displayed mild symptoms. In addition, the transgenic lines had greater leaf area and photosynthesis rate than the wild-type plants (Fig. 3d, e). Compared with the wild-type plants, the transgenic lines exhibited remarkably higher levels of proline content (Fig. 3f) but lower levels of MDA content (Fig. 3g) under salt condition.
The salt stress tolerance of NbWRKY79-overexpressing lines was further investigated using leaf-disc assay. Leaf discs from 3-week-old wild-type plants and NbWRKY79-overexpressing lines were soaked in the MS solutions including various NaCl concentrations, ranging from 0 to 400 mM, for 4 days. As shown in Fig. 4a, bleaching was perceived to a more severe extent in the wild-type plants than in the transgenic lines under salt treatment. This result was further confirmed through measurement of the chlorophyll content in the leaf discs after NaCl treatment (Fig. 4b).
Leaf disc assay and ROS generation in salt-stressed N. benthamiana leaves. a Leaf discs from the wild-type plants and NbWRKY79-overexpressing (O.E1 and O.E2) lines were treated with different concentrations of NaCl (0, 100, 200, and 400 mM). Black bar, 1 cm. b Quantification of chlorophyll content from the leaf disc assay. The graphs represent the mean and SD over three replicates. Wild-type and NbWRKY79-overexpressing (O.E1 and O.E2) leaves were taken from the plants, which were treated with 200 mM NaCl for two days. The tobacco leaves were stained with DAB (c) and NTB (d) to detect ROS accumulation in shoots and roots. A representative experiment from three with similar results is shown. The different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test
NbWRKY79 overexpression decreases salt stress-induced ROS production
So as to further research the effect of salt stress on NbWRKY79-overexpressing lines, accumulation of ROS, \({\text{O}}_{ 2}^{ \cdot - }\) and H2O2, leaves of the wild-type plants and NbWRKY79-overexpressing lines during salt treatment was tested by ROS staining using NBT and DAB, respectively. High amounts of ROS were observed in the wild-type leaves but not in the transgenic lines after 48-h exposure to 200 mM NaCl (Fig. 4c, d).
Analysis of antioxidant enzyme activity in NbWRKY79-overexpressing plants
To look into the elemental mechanism of NbWRKY79 that caused decreased ROS accumulation, and thus enhanced the stress tolerance, we surveyed the total activities of ROS-scavenging enzymes (SOD, POD, CAT and APX). Salt stress induced the ROS-scavenging enzyme activity in both the wild-type plants and the transgenic lines. Under normal growth conditions, the activities of SOD, POD, CAT and APX in the transgenic lines were 1.3-, 1.8-, 1.4- and 1.9- fold higher than wild-type plants, respectively (Fig. 5a–d). The total activities of antioxidant enzymes in the transgenic lines exposed to salt treatment were increased approximately 1.5-, 2.2-, 2.1- and 2.7-fold for SOD, POD, CAT, and APX, respectively (Fig. 5a–d). Activity in gel assay showed that the increased antioxidant enzyme activities might indicate the decreased oxidative damage in transgenic lines under salt stress. This observation suggests that NbWRKY79 may be associated with ROS signaling (Fig. 5e–h).
Relative activity of antioxidant enzymes, SOD—superoxide dismutase (a), POD—peroxidase (b), CAT—catalase (c), APX—ascorbate peroxidase (d) and on gel-activity of antioxidant enzymes SOD—superoxide dismutase (e), POD—peroxidase (f), CAT—catalase (g) and APX—ascorbate peroxidase (h) in leaves of NbWRKY79 overexpressing (O.E1 and O.E2) lines to these in leaves of wild-type plants. Protein was extracted from the leaves, which were treated with 200 mM NaCl for 2 weeks. The activity of the antioxidant enzymes was quantified. The graphs represent the mean and SD over three replicates. The different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test. For the on gel-activity of antioxidant enzymes, protein was extracted from leaves of NbWRKY79-overexpressing lines, which were treated with 200 mM NaCl for 0-24 h. Native PAGE of leaf extracts containing 200 mg protein. The experiments were repeated with three independent samples and similar results were obtained
Overexpression of NbWRKY79 exhibits enhanced sensitivity to ABA
The increased transcription levels of NbWWRKY79 in response to ABA treatment suggested that NbWWRKY79 might play significant part in the ABA signaling pathway. In the absence of exogenously supplied ABA, germination patterns of the transgenic lines and the wild-type plants were similar (Fig. 6a). However, NbWRKY79-overexpressing lines showed lower germination than the wild-type plants in medium supplemented with different concentrations of ABA (25 and 50 μM) (Fig. 6a). In addition, during the post-germination growth stage, the root growth of NbWRKY79-overexpressing lines and wild-type seedlings was relatively similar in the absence of exogenously applied ABA (Fig. 6b). At 25 and 50 μM ABA, the root growth of transgenic lines was significantly inhibited and the inhibition level was more remarkable than that in the wild-type seedlings (Fig. 6b). The results indicated that overexpression of NbWRKY79 increased the sensitivity of transgenic lines to ABA.
ABA accumulation and expression of ABA-inducible genes
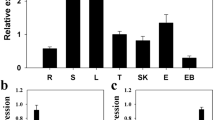
The phytohormone ABA plays crucial parts in the response of plants to abiotic stresses such as high salinity (Cutler et al. 2010; Finkelstein et al. 2002). Salt stress-induced ABA accumulation was reported in many previous studies (Barrero et al. 2006; Jia et al. 2002; Ruiz-Sola et al. 2014). In this study, to determine whether salt stress tolerance in NbWRKY79-overexpressing lines is concerned with the ABA signal pathway, the endogenous ABA contents were assayed. All plants were initially cultivated under normal irrigating conditions and then treated with 200 mM NaCl for a week. Under normal conditions, the ABA level in NbWRKY79-overexpressing lines was close to that in the wild-type plants. Under salt stress, the ABA level was significantly increased in both the wild-type and the transgenic lines. The accumulated ABA in NbWRKY79-overexpressing O.E1 and O.E2 lines was 1.63- and 1.71-fold higher than in the wild-type plants, respectively (Fig. 7a). Furthermore, expressions of ABA-inducible genes, including NbAREB1 (ABA-responsive element binding), NbDREB (dehydration-responsive element binding) and NbNCED (nine-cis-epoxycarotenoid dioxygenase) (Luo et al. 2012), were analyzed under salt stress. Figure 7b–d showed that the expression levels of ABA-inducible genes in the transgenic lines were higher than those in the wild-type plants under NaCl treatment. The results suggest that the endogenous ABA accumulation and the expression of some ABA-responsive genes might be altered by overexpression of NbWRKY79.
ABA accumulation and ABA-inducible gene expressions in the wild-type and NbWRKY79 (O.E)-overexpressing lines. a Endogenous ABA levels in the leaves of the wild-type plants and NbWRKY79 transgenic lines were determined under normal condition and salt treatment with 200 mM NaCl. The expression levels of ABA-inducible genes, NbAREB (b), NbDERB (c) and NbNCED (d) in the wild-type plants and transgenic lines during the salt stress responses as determined using qPCR. The values represent the mean ± SE of three independent experiments. The different letters above the columns indicate significant differences (P < 0.05) according to Duncan’s multiple range test
Discussion
WRKY proteins are one of the largest families of plant transcription factors and have approximately 100 members in Arabidopsis and rice (Eulgem et al. 2000). The WRKY gene family has been indicated to hold the key to regulation of the transcriptional reprogramming in plant stress responses (Chen et al. 2012; Rushton et al. 2010). Preceding studies have indicated that a huge number of genes encoding WRKY transcription factors in plants are induced under biotic stresses. In B. cinerea, some WRKY genes including WRKY70 and WRKY33, which are similar to genes in Arabidopsis, are responsive to necrotrophic fungal pathogens (AbuQamar et al. 2006; Zheng et al. 2006). However, it is still unknown regarding the function of WRKY proteins in abiotic-stress conditions. Changes in the expression patterns of WRKY genes impact various signaling pathways and regulatory networks. WRKY proteins have numerous potential transcriptional activation and repression domains in their structure; therefore, they can activate or repress gene transcription (Rushton et al. 2010). In addition, preceding studies have reflected that WRKY proteins are concerned with plant ABA signaling. Hence, WRKY proteins may function as activators or repressors in ABA signaling (Chen et al. 2012; Miller et al. 2008; Wang et al. 2009; Xie et al. 2005; Zou et al. 2004). A novel WRKY gene termed NbWWKY79 from Nicotiana benthamiana was isolated and characterized in this study. Overexpression of NbWRKY79 exhibited a significant increase in plant tolerance to salt stress (Figs. 3, 4). The transcription of GhWRKY17 and GhWRKY25 (overexpressing in N. benthamiana) belonging to group I and II WRKY genes was also strongly induced during the early development period in cotton (Gossypium hirsutum) after exposure to salt and ABA treatment (Liu et al. 2015; Yan et al. 2014). Further analysis showed that NbWRKY79 associated with plant salt stress response through ABA signaling pathway and the regulation of ROS accumulation. These results transpired the participation of NbWRKY79 genes in plant tolerate to salinity.
As a crucial phytohormone, plant development and various stress responses were related to ABA, activating the gene expression in response to stresses (Chinnusamy et al. 2004). That the changes of ABA at the cellular and whole-plant levels increase ABA content is helpful to plants after exposure to stress conditions (Xiong and Zhu 2003). Salt stress can induce ABA accumulation when ABA synthesis is activated and ABA degradation is inhibited. The accumulation of ABA was set off by salt stress, which in turn induces the expression of stress-related genes through ABA-dependent and -independent regulatory systems (Zhu 2002). In this study, expression of NbWRKY79 was up-regulated by exogenous ABA. The NbWRKY79-overexpressing lines were significantly more sensitive to exogenous ABA than those in wild-type plants (Fig. 6). The NbWRKY79-overexpressed plants exhibited more accumulation of ABA under salt stress compared with wild-type plants (Fig. 7). In addition, the expression of NbAREB, NbDREB and NbNCED, reported to have function in the ABA-dependent and -independent pathways, was up-regulated under salt stress in the transgenic lines (Fig. 7). These findings suggest that NbWRKY79 is concerned with the ABA-dependent signaling pathways under salt stress conditions.
Salt stress can induce the accumulation of ROS, which causes damage to cellular membrane, mediates lipid peroxidation, and leads to oxidative stress (Banu et al. 2009; Bowler et al. 1992; Leshem et al. 2007; Mazel et al. 2004). It is critical to regulate ROS accumulation under salt stress. Prior studies have shown that ROS accumulation is modulated through the ABA-triggered regulatory genes involved in ROS production and ROS scavenging (Mittler et al. 2004). However, the regulatory mechanisms of ABA-dependent ROS signaling under salt stress are still unclear (Yan et al. 2014). In the present study, accumulation of ROS in the wild-type plants and the transgenic lines in response to high-salt stress was determined by histochemical staining of DAB and NBT. Our study provided the evidence that NbWRKY79-overexpressed plants exhibited a lower ROS accumulation and MDA content in the transgenic lines compared to wild-type plants under salt stress (Fig. 3). Furthermore, this study revealed that the activity of ROS-scavenging enzymes including SOD, POD, CAT, and APX in the transgenic lines was higher than that in the wild-type plants in both normal and salt stress conditions (Fig. 5). Another study has reported that overexpression of a gene from cotton plants (Gossypium hirsutum), named GhWKRY44, increases ability to scavenge reactive oxygen species (ROS) (Li et al. 2014a). Once the activities of ROS-scavenging enzymes are up-regulated, they support plants from oxidative damage (Apel and Hirt 2004; Suzuki et al. 2012). These results indicate that NbWRKY79 is concerned with the regulation of SOD, POD, CAT, and APX activities, which resulted in the suppression of ROS accumulation so as to endure less from oxidative damage under salt conditions.
In this study, NbWRKY79 gene encoding for a WKRY transcription factor whose transcription was induced by salt and ABA was extracted from N. benthamiana. NbWRKY79 exhibits important physiological functions in salt stress response. Overexpression of NbWRKY79 caused enhanced tolerance to salt stress, which reflected a partial correlation between the activation of ROS-related antioxidant enzymes and reduced accumulation of ROS under salt stress. Our study also indicated that salt stress tolerance in the transgenic plants might involve ABA signaling pathway. Importantly, overexpression of NbWRKY79 was achieved without having an effect on their phenotypes under normal conditions (Fig. S6). Overall, our findings identified NbWRKY79 as a potential genetic resource for ameliorating salt tolerance in plants.
Author contribution statement
TNN and LHT conceived and designed the research. TNN and DSM conducted experiments. TNN and NVT analyzed data. TNN and LHT wrote the manuscript. All authors read and approved the manuscript.
Abbreviations
- ABA:
-
Abscisic acid
- APX:
-
Ascorbate peroxidase
- CAT:
-
Catalase
- DAB:
-
3,3′-Diaminobenzidine
- GUS:
-
β-Glucuronidase
- MDA:
-
Malondialdehyde
- NBT:
-
Nitroblue tetrazolium
- POD:
-
Guaiacol peroxidase
- qPCR:
-
Quantitative-PCR
- ROS:
-
Reactive oxygen species
- Semi qPCR:
-
Semi-quantitative-PCR
- SOD:
-
Superoxide dismutase
- YFP:
-
Yellow fluorescent protein
References
AbuQamar S et al (2006) Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant J 48:28–44. doi:10.1111/j.1365-313X.2006.02849.x
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399. doi:10.1146/annurev.arplant.55.031903.141701
Banu MNA, Hoque MA, Watanabe-Sugimoto M, Matsuoka K, Nakamura Y, Shimoishi Y, Murata Y (2009) Proline and glycine betaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J Plant Physiol 166:146–156. doi:10.1016/j.jplph.2008.03.002
Barrero JM, Rodríguez PL, Quesada V, Piqueras P, Ponce MR, Micol JL (2006) Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ 29:2000–2008. doi:10.1111/j.1365-3040.2006.01576.x
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207. doi:10.1007/BF00018060
Bhaskar PB, Venkateshwaran M, Wu L, Ané JM, Jiang J (2009) Agrobacterium-mediated transient gene expression and silencing: a rapid tool for functional gene assay in potato. PLoS One 4:e5812. doi:10.1371/journal.pone.0005812
Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea Infection. Plant Physiol 159:266–285. doi:10.1104/pp.111.192641
Bowler C, Montagu MV, Inze D (1992) Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 43:83–116. doi:10.1146/annurev.pp.43.060192.000503
Chen Y-A, Shin J-W, Liu Z-H (2002) Effect of light on peroxidase and lignin synthesis in mungbean hypocotyls. Plant Physiol Biochem 40:33–39. doi:10.1016/S0981-9428(01)01345-6
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA) Gene Regul Mech 1819:120–128. doi:10.1016/j.bbagrm.2011.09.002
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55:225–236. doi:10.1093/jxb/erh005
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic Acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679. doi:10.1146/annurev-arplant-042809-112122
Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10:366–371. doi:10.1016/j.pbi.2007.04.020
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206. doi:10.1016/S1360-1385(00)01600-9
Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14:S15–S45. doi:10.1105/tpc.010441
Horsch RB, Fry JE, HoVmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231. doi:10.1126/science.227.4691.1229
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jia W, Wang Y, Zhang S, Zhang J (2002) Salt stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J Exp Bot 53:2201–2206. doi:10.1093/jxb/erf079
Journot-Catalino N, Somssich IE, Roby D, Kroj T (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18:3289–3302. doi:10.1105/tpc.106.044149
Kumar P, Tewari R, Sharma P (2008) Modulation of copper toxicity-induced oxidative damage by excess supply of iron in maize plants. Plant Cell Rep 27:399–409. doi:10.1007/s00299-007-0453-1
Leshem Y, Seri L, Levine A (2007) Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J 51:185–197. doi:10.1111/j.1365-313X.2007.03134.x
Li J, Brader G, Kariola T, Tapio Palva E (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46:477–491. doi:10.1111/j.1365-313X.2006.02712.x
Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET (2013) Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol 200:457–472. doi:10.1111/nph.12378
Li J, Wang J, Wang N, Guo X, Gao Z (2014a) GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tissue Organ Cult (PCTOC) 121:127–140. doi:10.1007/s11240-014-0688-9
Li J-b, Luan Y-s, Yin Y-l (2014b) SpMYB overexpression in tobacco plants leads to altered abiotic and biotic stress responses. Gene 547:145–151. doi:10.1016/j.gene.2014.06.049
Liu Q-L, Xu K-D, Pan Y-Z, Jiang B-B, Liu G-L, Jia Y, Zhang H-Q (2014) Functional analysis of a novel Chrysanthemum WRKY transcription factor gene involved in salt tolerance. Plant Mol Biol Rep 32:282–289. doi:10.1007/s11105-013-0639-3
Liu X, Song Y, Xing F, Wang N, Wen F, Zhu C (2015) GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma. doi:10.1007/s00709-015-0885-3
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Luo X et al (2012) Over-expression of GsZFP1, an ABA-responsive C2H2-type zinc finger protein lacking a QALGGH motif, reduces ABA sensitivity and decreases stomata size. J Plant Physiol 169:1192–1202. doi:10.1016/j.jplph.2012.03.019
Ma N-N, Zuo Y-Q, Liang X-Q, Yin B, Wang G-D, Meng Q-W (2013) The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato. Physiol Plant 149:474–486. doi:10.1111/ppl.12049
Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59:150–162. doi:10.1111/j.1365-313X.2009.03850.x
Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H (2013) Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 73:302–313. doi:10.1111/tpj.12035
Mazel A, Leshem Y, Tiwari BS, Levine A (2004) Induction of salt and osmotic stress tolerance by overexpression of an intracellular vesicle trafficking protein AtRab7 (AtRabG3e). Plant Physiol 134:118–128. doi:10.1104/pp.103.025379
Miller G, Shulaev V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133:481–489. doi:10.1111/j.1399-3054.2008.01090.x
Mittler R (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci 11:15–19. doi:10.1016/j.tplants.2005.11.002
Mittler R, Zilinskas BA (1993) Detection of ascorbate peroxidase activity in native gels by inhibition of the ascorbate-dependent reduction of nitroblue tetrazolium. Anal Biochem 212:540–546. doi:10.1006/abio.1993.1366
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498. doi:10.1016/j.tplants.2004.08.009
Nakashima K et al (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50:1345–1363. doi:10.1093/pcp/pcp083
Qiu Y, Yu D (2009) Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot 65:35–47. doi:10.1016/j.envexpbot.2008.07.002
Ren X et al (2010) ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J 63:417–429. doi:10.1111/j.1365-313X.2010.04248.x
Ruiz-Sola MÁ, Arbona V, Gómez-Cadenas A, Rodríguez-Concepción M, Rodríguez-Villalón A (2014) A root specific induction of carotenoid biosynthesis contributes to ABA production upon salt stress in Arabidopsis. PLoS One. doi:10.1371/journal.pone.0090765
Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol Biol 29:691–702. doi:10.1007/BF00041160
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258. doi:10.1016/j.tplants.2010.02.006
Rushton DL et al (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J 10:2–11. doi:10.1111/j.1467-7652.2011.00634.x
Suzuki N, Koussevitzky S, Mittler RON, Miller GAD (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35:259–270. doi:10.1111/j.1365-3040.2011.02336.x
Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—powdery mildew interaction. Plant J 11:1187–1194. doi:10.1046/j.1365-313X.1997.11061187.x
Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498. doi:10.1016/j.pbi.2004.07.012
Wang Y-S, Yang Z-M (2005) Nitric oxide reduces aluminum toxicity by preventing oxidative stress in the roots of Cassia tora L. Plant Cell Physiol 46:1915–1923. doi:10.1093/pcp/pci202
Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X (2009) A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230:1155–1166. doi:10.1007/s00425-009-1014-3
Xie Z, Ruas P, Shen QJ (2005) Regulatory networks of the phytohormone abscisic acid. In: Gerald L (ed) Vitamins & hormones, vol 72. Academic Press, San Diego, California, USA, pp 235–269. doi:10.1016/S0083-6729(05)72007-0
Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133:29–36. doi:10.1104/pp.103.025395
Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18:1310–1326. doi:10.1105/tpc.105.037523
Yan H, Jia H, Chen X, Hao L, An H, Guo X (2014) The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol 55:2060–2076. doi:10.1093/pcp/pcu133
Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48:592–605. doi:10.1111/j.1365-313X.2006.02901.x
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273. doi:10.1146/annurev.arplant.53.091401.143329
Zou X, Seemann JR, Neuman D, Shen QJ (2004) A WRKY gene from creosote bush encodes an activator of the abscisic acid signaling pathway. J Biol Chem 279:55770–55779. doi:10.1074/jbc.M408536200
Acknowledgements
We would like to thank Dr. Chi Wen-Chang, Department of Life sciences, National Cheng Kung University, Taiwan, for the kind gift of Gateway vectors. We are grateful to Professor Nguyen Duc Quang (Corvinus University of Budapest, Hungary) and Dr. Malik A. Hussain (The University of Melbourne, Australia) for their careful review of the manuscript and providing helpful comments. Furthermore, we thank Dr. Vu Thanh Ngoc (Industrial University of Ho Chi Minh City, Vietnam) for English editing.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Z.-L. Zhang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2017_2423_MOESM1_ESM.tif
Figure S1 Comparison of the predicted N. benthamiana NbWRKY79 amino acid sequence with those of its homologs from other plants. WRKY amino acid sequences from Nicotiana tabacum, Solanum tuberosum and Nicotiana tomentosiformis were aligned (Genbank accessions: N. tabacum, NtWRKY79, TOBFAC, ET047018; S. tuberosum, StWRKY2-like, XP_006350428.1; N. tomentosiformis, NtoWRKY33, XP_009598832.1; N. benthamiana, NbWRKY79, KR559678. The black boxes indicate identical residues, and gray boxes indicate conservative substitutions. Hyphens indicate gaps introduced to optimize alignments. The red box indicate WRKY domain. The blue boxes indicate zinc-finger-like motif C2-H2. The alignment was done with ClustalW program (http://www.ch.embnet.org/software/ClustalW.html) and BoxShade program (http://www.ch.embnet.org/software/BOX_form.html) (TIFF 5790 kb)
11738_2017_2423_MOESM2_ESM.tif
Figure S2 A schematic diagram interpreting the constructs utilized for transient transformation of N. benthamiana leaf epidermal cells. The full-length NbWRKY79 cDNA sequences were cloned into the pSITEII-4C1 vector (Martin et al. 2009) to generate a Venus fusion proteins. 2X35S, tandem CaMV 35S promoter; TL, TEV leader; DEST, destination cassette; TER, CaMV 35S transcriptional terminator (TIFF 351 kb)
11738_2017_2423_MOESM3_ESM.tif
Figure S3 NbWRKY79 promoter sequence (2.0 kb). The nucleotides in bold big-letters indicate start- and stop-codon of NbWRKY79 ORF, respectively (TIFF 3038 kb)
11738_2017_2423_MOESM5_ESM.tif
Figure S5 Identification of T1 (a) and T2 transgenic lines (b) and expression of native endogenous NbWRKY79. The transcript level of NbWRKY79 was determined in wild-type plants and T2 NbWRKY79 overexpressing lines. The NbEF1-α was included as an internal control (TIFF 1132 kb)
Rights and permissions
About this article
Cite this article
Nam, T.N., Thia, L.H., Mai, D.S. et al. Overexpression of NbWRKY79 enhances salt stress tolerance in Nicotiana benthamiana . Acta Physiol Plant 39, 121 (2017). https://doi.org/10.1007/s11738-017-2423-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2423-8