Abstract
Bax inhibitor-1 (BI-1), an evolutionarily conserved protein, is a suppressor of cell death induced by the proapoptotic protein Bax and is involved in the response to biotic and abiotic stress in animals, plants and yeast. Rice false smut caused by Ustilaginoidea virens is one of the destructive rice diseases worldwide. Although BI-1 proteins are widely distributed across filamentous fungi, few of them are functionally characterized. In this study, we identified a BI-1 protein in U. virens, UvBI-1, which contains a predicted Bax inhibitor-1-like family domain and could suppress the cell death induced by Bax. By co-transformation of the CRISPR/Cas9 construct along with donor DNA fragment containing the hygromycin resistance gene, we successfully generated Uvbi-1 deletion mutants. The UvBI-1 deletion showed an increase in mycelia vegetative growth and conidiation, suggesting this gene acts as a negative regulator of the growth and conidiation. In addition, the Uvbi-1 mutants exhibited higher sensitivity to osmotic and salt stress, hydrogen peroxide stress, and cell wall or membrane stress than the wild-type strain. Furthermore, UvBI-1 deletion was found to cause increased production of secondary metabolites and loss of pathogenicity of U. virens. Taken together, our results demonstrate that UvBI-1 plays a negative role in mycelial growth and conidiation, and is critical for stress tolerance, cell wall integrity, secondary metabolites production and pathogenicity of U. virens. Therefore, this study provides new evidence on the conserved function of BI-1 among fungal organisms and other species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis is one type of programmed cell death (PCD) that is highly conserved and plays a fundamental role in various biological processes including cell proliferation, generation of developmental patterns, and biotic and abiotic stress responses (Ameisen 2002; Hamann et al. 2008; Heath 1998). The Bcl-2 (B cell lymphoma-2) family proteins are critical apoptotic regulators, and different members have unique functions, with involvement in anti- (e.g., Bcl-2 and Bcl-XL) or proapoptotic functions (e.g., Bax and Bak) by modulating the release of cytochrome c from mitochondria. Bax and Bak-induced apoptosis is actively inhibited by anti-apoptotic Bcl-2 family members (Danial and Korsmeyer 2004; Scorrano and Korsmeyer 2003). Although Bcl-2 family proteins have been described in mammals, worms and insects, they are not encoded in plant and fungal genomes, including that of Saccharomyces cerevisiae (Borner 2003; Danial and Korsmeyer 2004; Sharon et al. 2009). Despite the absence of Bcl-2 homologues, mammalian Bax-induced plant cell death can be suppressed by plant BI-1, indicating that the physiology of cell death is similar in animals and plants (Kawai-Yamada et al. 2001).
Bax inhibitor-1 (BI-1) was initially identified in a screen for human proteins capable of inhibiting Bax-mediated cell death in yeast (Xu and Reed 1998). BI-1 is an endoplasmic reticulum (ER) localized sensor and interacts with Bcl-2 and Bcl-XL but not Bax or Bak to suppress mammalian apoptosis most likely by regulating Ca2+ homeostasis (Chae et al. 2004; Henke et al. 2011; Lisak et al. 2015; Lisbona et al. 2009; Xu and Reed 1998). BI-1 proteins that contain multiple transmembrane domains are evolutionarily conserved multifunctional proteins and present in various organisms including animals, plants, fungi, bacteria, and even viruses (Chae et al. 2003; Chen et al. 2015; Huckelhoven 2004). The expression of BI-1 is upregulated in different human cancers, e.g., breast and prostate cancers, among others (Grzmil et al. 2003, 2006; Robinson et al. 2011), but it is downregulated during the progression of chronic liver disease (Kotsafti et al. 2010). Overexpression of BI-1 in yeast increases the resistance to heat or oxidative stress and provides protection from Bax-induced cell death, whereas deletions of BI-1 abolish an anti-apoptotic function (Cebulski et al. 2011; Chae et al. 2003). BI-1-deficient cells in mice display increased sensitivity to apoptosis induced by ER stress. Conversely, BI-1 overexpression protects against apoptosis induced by ER stress, which correlates with inhibition of Bax activation. BI-1 overexpression also reduces releasable Ca2+ from the ER (Chae et al. 2004). In Arabidopsis, overexpression of AtBI-1 in plants suppresses Bax-, hydrogen peroxide-, and salicylic acid-induced plant cell death and delays methyl jasmonate-induced leaf senescence by suppressing the activation of MAP kinase 6 (Kawai-Yamada et al. 2004; Yue et al. 2012). AtBI-1 is also involved in cell death regulation by interacting with different proteins against pathogen infections (Gaguancela et al. 2016; Matsumura et al. 2003; Weis et al. 2013). In N. benthamiana, silencing of BI-1 reduces the autophagic activity induced by both N gene-mediated resistance to tobacco mosaic virus (TMV) and methyl viologen (MV), whereas overexpression of plant BI-1 increases autophagic activity and notably causes autophagy-dependent cell death (Xu et al. 2017). In a crustacean, Bax inhibitor-1 silencing suppresses white spot syndrome virus replication in the red swamp crayfish, Procambarus clarkia (Du et al. 2013). TaBI-1 is a negative cell death regulator from wheat, which contributes to resistance and plays an important role in the response to biotic stresses (Lu et al. 2018; Wang et al. 2012). Few studies have examined the role of BI-1 in filamentous fungi. MrBI-1, from the insect pathogenic fungi Metarhizium robertsii, was shown to regulate heat tolerance, apoptotic-like cell death, and virulence (Chen et al. 2015). Targeted silencing of Ss-Bi1 in Sclerotinia sclerotiorum resulted in reduced pathogen virulence in host plants and increased sensitivity to heat and ER stresses (Yu et al. 2015b).
During the past decades, rice false smut caused by U. virens has become one of the major fungal diseases with the typical symptom of forming dark green smut balls inside rice seeds (Rush et al. 2000; Sun et al. 2013; Wang et al. 1998; Zhou et al. 2008). In addition to negative impact on rice yield, ustiloxins produced by U. virens are poisonous to humans and animals and can also inhibit the radicle and plumule growth of plant seedlings (Abbas et al. 2014; Meng et al. 2015; Sun et al. 2016; Tsukui et al. 2015). Although the fungal genome has been sequenced, few genes are functionally characterized due to limitation of available molecular genetic tools for U. virens. Mutants with disrupted UvSUN2, Uvt3277, and UvPRO1 genes were generated by random insertional mutagenesis via the Agrobacterium tumefaciens-mediated transformation (ATMT). UvSUN2 contributes to virulence, fungal growth, cell wall construction, and stress responses in U. virens (Yu et al. 2015a). UvPRO1 plays a critical role in hyphal growth and conidiation in stress response and pathogenesis (Lv et al. 2016). By combining T-DNA insertion disruption and RNA interference (RNAi) approaches, the low-affinity iron transport protein Uvt3277 was functional characterized using the mutants showing increased pathogenicity (Zheng et al. 2017). Only one Uvhog1 deletion mutant was identified by targeted gene deletion, and the results suggest that Uvhog1 is conserved in regulating stress responses, hyphal growth, and possibly secondary metabolism (Zheng et al. 2016). A recent study demonstrated that the CRISPR-Cas9 system could be used as an efficient gene replacement or editing tool in U. virens, and CRISPR-edited Uvslt2 mutants show reduction in growth rate and conidiation and an increase in sensitivity to cell wall stresses (Liang et al. 2018). Thus, the CRISPR/Cas9 method is efficient for generating targeted deletion mutants.
In this study, we identified UvBI-1 as a Bax inhibitor-1, which contains six transmembrane and BI-1-like domains and is capable of suppressing the cell death induced by Bax. Using the CRISPR/Cas9 genome editing approach Uvbi-1 deletion mutants were generated to facilitate its functional characterization. We have shown that UvBI-1 was a negative regulator of the mycelia growth, conidiation and secondary metabolites production. The Uvbi-1 mutants were sensitive to stress and almost lost the pathogenicity, indicating the essential role of this gene in U. virens.
Results
Identification of UvBI-1 from U. virens
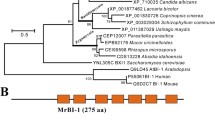
To identify the ortholog of BI-1, we selected Bax inhibitor-1 domain sequence of AtBI-1 as a query to process a BLASTP search in the U. virens genome and two proteins (KDB14758.1 and KDB15472.1) were found to contain Bax inhibitor-1-like domain. We termed the two proteins as UvBI-1 (KDB14758.1) and UvBI-1b (KDB15472.1). The UvBI-1 consisted of a predicted 1028 bp open reading frame interrupted by two introns and encoded a protein of 278 amino acids. The predicted UvBI-1 protein contained six transmembrane and Bax inhibitor-1-like domain (Fig. 1a). Sequence alignment analysis revealed that the BI-1-like domain is well conserved among humans, animals, plants and filamentous fungi (Fig. 1b). The phylogenetic tree revealed that BI-1 protein is most similar to those from Metarhizium (Fig. 1c). Whereas, the UvBI-1b showed high similarity to Ss-Bi-1, and they both show a closer relationship with BI-1 from plant, animal and bacterial. An unrelated protein UvbZIP1 from U.virens was selected for analysis and it was out of the BI-1 group. All this result showed that BI-1 was well conserved, indicating that UvBI-1 might have important functions in U. virens.
Functional domain analysis, sequence alignment and phylogenetic analysis of UvBI-1. a Physical map of UvBI-1 shows a conserved BI-1-like superfamily and six transmembrane domains. b Alignment of the amino acid sequences of UvBI-1 with those from other organisms. The BI-1 proteins from fungi, bacteria, plants and mammals was performed using BioEdit software. The black solid shading show amino acid identity and gray show fifty percent similarity. c Phylogenetic analysis of UvBI-1 with different orthologous. The phylogenetic tree was constructed based on alignment of the full sequences of BI-1 from fungi to mammals by using the Mega5, and UvBI-1b, Ss-Bi-1 and a related protein UvbZIP1 were included. The UvBI-1 was indicated black triangle
UvBI-1 suppresses cell death induced by Bax
Considering UvBI-1 contained a BI-1-like domain, we tested whether UvBI-1 suppressed the cell death induced by Bax. The UvBI-1 gene was coexpressed with Bax in N. benthamiana leaves by agrobacterium-mediated transient expression. For the controls, the GFP was also coexpressed with Bax, and UvBI-1, GFP and Bax were expressed independently. The results showed that no cell death was observed when UvBI-1 and Bax were coexpressed, whereas cell death was observed when Bax was expressed separately or coexpressed with the GFP (Fig. 2). In order to rule out the possibility that no cell death were observed due to the nonexpression of the genes, we detected the expression of genes by reverse transcription (RT)-PCR, and the results showed that all the gene expressed (Fig. S1, see Supporting Information). This result indicated that UvBI-1 suppressed the cell death induced by Bax and therefore had the ability of a Bax inhibitor.
UvBI-1 suppresses the cell death induced by Bax. N. benthamiana leaves were infiltrated with A. tumefaciens cells containing vectors carrying the UvBI-1 gene or control gene GFP, either alone or followed 24 h later with A. tumefaciens cells carrying a mouse Bax. The numbers show the ratio of cell death and the total number of agroinfiltrations. The photograph was taken at 5 days after infiltration
UvBI-1 is highly expressed in the vegetative stage
To gain insights into the possible function of UvBI-1 in U. virens, we examined the gene expression by quantitative real-time polymerase chain reaction (qRT-PCR) during the different stages of the fungal infection. The abundance of UvBI-1 transcripts during vegetative growth in PSB was relatively higher than that invasive growth. During its invasive growth in plants, the transcript level of UvBI-1 decreased almost more than fivefold at 3, 5 and 6 days post-inoculation (dpi) in rice panicles, compared with that of mycelia. However, the transcript abundance began to increase at 7 dpi and almost reached the 60% level similar to that of mycelia (Fig. 3). Considering the special infection characteristic that a lot of the fungal mycelia developed and covered the floral organs after 6 dpi (Song et al. 2016), we concluded that the UvBI-1 was highly expressed during vegetative growth.
The transcriptional pattern of UvBI-1. The expression of UvBI-1 was quantified using qRT-PCR, with the synthesis of cDNA for each sample including mycelia and at 3, 5, 6 and 7 days post inoculation (dpi). Relative transcript level in the mycelia stage was regarded as the reference and normalized to 1. Three independent biological experiments with three replicates in each were performed. Representative results from one of these experiments are shown. The letters represent statistical significance (P < 0.05)
UvBI-1 gene deletion and ΔUvbi-1 mutant complementation
Targeted gene replacement was performed to investigate the function of UvBI-1 in U. virens. The deletion fragments flanking approximately 1.6 kb up- or downstream of the UvBI-1 ORF region were fused partially with the hygromycin B gene. The gRNA spacers of UvBI-1 were cloned into the CRISPR-Cas vector. Uvbi-1 deletion mutants were generated by replacing the endogenous UvBI-1 ORF with the donor template containing hygromycin B gene via the transformation of the strain JZ11-28 protoplasts with linear donor DNA fragments and the CRISPR construct. The hygromycin-resistant transformants were selected on the PSA medium containing hygromycin (100 mg/ml), and the deletion mutants were verified by PCR amplification using specific pairs of primers (Fig. S2, see Supporting Information). Complete inactivation of Uvbi-1 transcription in the deletion mutants was verified by RT-PCR using the RNA samples extracted from the mycelium (Fig. 4). Furthermore, the ORF sequence of UvBI-1 containing the native promoter region was introduced into the deletion strain to create the complemented strains Uvbi-1/BI-1 (Fig. S3, see Supporting Information). There is no difference between deletion mutations and complemented strains except the complemented strains contain the complemented fragment. Three of the deletion mutants and two of complemented strains were selected for the phenotype test, and the results from two deletion mutants and one complemented strain were presented.
Targeted gene replacement and complementation of UvBI-1 in U. virens. a The UvBI-1 target replacement strategy. A 1028 bp fragment of the UvBI-1 coding region was replaced with a 1.4 kb fragment containing the hygromycin B-resistance cassette (HPH) to create the UvBI-1 deletion mutant. b The detection of UvBI-1 expression in the testing strains. The inactivation of UvBI-1 transcription was verified by semiquantitative RT-PCR, using the cDNA from wild-type strain JZ11-28, the deletion mutants, and the complemented strain
UvBI-1 is a negative regulator for vegetative growth
To investigate the role of UvBI-1 in U. virens mycelial growth, we incubated the wild type, mutants and the complemented strain on PSA, YTD and PDA media, respectively. The mutants showed a faster growth on PSA, PDA and YTD media compared with the wild type and complemented strains (Fig. 5a, b). Considering the increased growth rate of the mutants, we investigated whether there was a difference in biomass between the wild type and mutants by culturing the strains in liquid PSB medium and measuring the dry weight 7 days later. The results showed that the dry weight of the Uvbi-1 mutant hyphae was higher than that of the wild type and the complemented strain (Fig. 5c). This result indicated that UvBI-1 was a negative regulator of hyphal growth and biomass in U. virens.
UvBI-1 disruption results in increased vegetative growth. a Mycelia growth of the testing strains on PSA, PDA and YTD media. The wild-type strain (JZ11-28), Uvbi-1 mutants and complemented strain (Uvbi-1/BI-1) were inoculated on PSA, PDA and YTD plates and cultured at 27 °C for 14 days. b The colony diameters of the testing strains. The colony diameter were measured and subjected to statistical analysis. c The dry weight of the tested strains was measured after 7 days of culture in PSB. Three independent biological experiments were performed with three replicates each time, with similar results yielded in each biological experiment. Representative results from one of these experiments are presented. Error bars represent the standard deviation, and asterisks represent a significant difference (P < 0.05)
Deletion of UvBI-1 leads to an increase in conidia formation
Conidia play an important role in the disease infection of rice false smut. Given that the mutants showed increased hyphal growth, we investigated the role of UvBI-1 in conidia formation by examining the canidiation of wild type, Uvbi-1 mutants and the complement strain after 7 days of culture in PSB. The conidiation increased by approximately four-fold in mutants compared with the wild type and complement, with a conidia concentration of approximately 1.6 × 107 conidia/ml (Fig. 6). WA (water agar) and PSA plates were employed to investigate conidia germination, and no difference was detected between the wild type and deletion strains (Fig. S4, see Supporting Information). These results all showed that UvBI-1 played a role as a negative regulator of conidia formation but did not affect conidia germination.
UvBI-1 deletion results in an increase in conidiation. a The conidiation of the testing strains. The JZ11-28, ΔUvbi-1 mutants (ΔUvbi-1-4 and ΔUvbi-1-13) and complemented strain (Uvbi-1/BI-1) were cultured in 50 ml of PSB medium with 160 rpm shaking at 27 °C for 7 days before being photographed. b Statistical analysis of conidia concentration. The conidia concentrations were measured in each independent biological experiment. Three independent biological experiments were performed with three replicates each time, with similar results yielded in each biological experiment. Error bars represent the standard deviation, and asterisks represent a significant difference (P < 0.05). Scale bar = 20 μm
Deletion of UvBI-1 results in defects in response to hyperosmotic and salt stresses
Because BI-1 proteins have an important role in abiotic stress, we investigated whether UvBI-1 contributed to the fungal response to osmotic or salt stress. The wild type, mutants and complemented strain were inoculated on PSA containing 0.2 or 0.4 M NaCl or 0.5, 0.7 or 0.9 M sorbitol. Mycelial growth was observed and inhibitory rates were calculated after incubation for 14 days. Significant differences were observed between the wild type and Uvbi-1 mutants. Compared with the wild type and complemented strain, the mutants showed a higher inhibitory rate under different NaCl and sorbitol concentrations (Fig. 7). This result indicated that UvBI-1 play a positive role in hyperosmotic and salt stress responses in U. virens.
UvBI-1 contributes to the tolerance to hyperosmotic and salt stresses. a The growth of mutants under hyperosmotic and salt stress. The JZ11-28, ΔUvBI-1 mutants (ΔUvbi-1-4 and ΔUvbi-1-13) and complemented strain (Uvbi-1/BI-1) were incubated on PSA supplemented with 0.2 and 0.4 M NaCl or 0.5, 0.7 and 0.9 M sorbitol at 27 °C for 14 days before being photographed. b Statistical analysis of mycelial growth under hyperosmotic and salt stresses. Colony diameters were measured in each independent biological experiment. Measurements of growth inhibition rate are relative to the growth rate of each untreated control. Three independent biological experiments were performed with three replicates each time, with similar results yielded in each biological experiment. Error bars represent the standard deviation, and asterisks represent a significant difference between the UvBI-1 mutant and JZ11-28 and complemented strain (P < 0.05)
The Uvbi-1 deletion mutants show increased sensitivity to oxidative stress
Reactive oxygen species (ROS) often elevates rapidly at the pathogen infection site and plays an essential role in plant defense responses (Lamb and Dixon 1997; Lehmann et al. 2015; Qi et al. 2017). To investigate whether UvBI-1 was involved in the oxidative stress response, the wild type, Uvbi-1 mutants and the complemented strain were exposed to different concentrations of H2O2. The mycelia growth of the mutants was affected, leading to an average of approximately 70% (0.01%), 80% (0.03%) and 100% (0.05%) inhibition of the growth rate, whereas the wild-type strain showed less inhibition (Fig. 8). As confirmation, the complemented strain was as resistant to H2O2 as the wild-type strain. This result indicated that UvBI-1 contributed to the oxidative stress response in U. virens.
UvBI-1 contributes to the tolerance to oxidative stress. a The growth of mutants under oxidative stress. The JZ11-28, ΔUvBI-1 mutants (ΔUvbi-1-4 and ΔUvbi-1-13) and complemented strain (Uvbi-1/BI-1) were incubated on PSA supplemented with 0.01, 0.03 and 0.05% H2O2 at 27 °C for 14 days before being photographed. b Statistical analysis of mycelial growth in response to oxidative stress. Measurements of growth inhibition rate are relative to the growth rate of each untreated control. Three independent biological experiments were performed with three replicates each time, with similar results yielded in each biological experiment. Error bars represent the standard deviation, and asterisks represent a significant difference (P < 0.05)
Deletion of UvBI-1 results in deficiency in cell wall integrity
The fungal cell wall has an essential role in maintaining hyphal morphology and adaptation to the environment. To further investigate the cause of defects resulting from loss of UvBI-1, the integrity of cell walls and membranes of the Uvbi-1 mutants was examined. The mycelial growth was measured on PSA medium containing the various cell wall stressors of sodium dodecyl sulfate (SDS), Calcofluor White (CFW) and Congo Red (CR). Because SDS is a detergent that reduces membrane stability, any cell wall defects will lead to increased sensitivity (Bickle et al. 1998). The CFW binds to chitin to interfere with its polymerization, whereas CR binds to the cell wall component β-1,4-glucan; both stressors are commonly used to detect cell integrity (Roncero and Duran 1985; Wood and Fulcher 1983). The Uvbi-1 mutant colonies showed smaller diameter on the SDS-containing PSA, and the inhibitory rate was significantly higher than that of the wild type and complemented strain. Similarly, UvBI-1 mutants exhibited less resistance to CFW and CR than that of the wild type and the complemented strain, indicating that UvBI-1 was involved in maintaining the integrity of the cell wall (Fig. 9). Additionally, unlike the wild type, a degradation halo of CR by the Uvbi-1 mutants was observed, indicating an excess of CR-degrading activity in the Uvbi-1 mutants (Fig. 9a). These results all demonstrated that the Uvbi-1 mutants had increased sensitivity to membrane and cell wall stress agents, suggesting that the UvBI-1 was involved in regulating the fungal responses to membrane and cell wall stresses.
UvBI-1 is involved in the tolerance of the cell wall or membrane to stress inducers. a The growth of mutants under cell wall or membrane stress. The JZ11-28, ΔUvBI-1 mutants (ΔUvbi-1-4 and ΔUvbi-1-13) and complemented strain (Uvbi-1/BI-1) were incubated on PSA supplemented with 0.015 and 0.03% SDS, 60 and 120 μg/ml CFW and 60 and 120 μg/ml CR. The photos were taken after 14 days of incubation. b Statistical analysis of mycelial growth in response to cell wall or membrane stress. Measurements of growth inhibition rate are relative to the growth rate of each untreated control. Three independent biological experiments were performed with three replicates each time, with similar results yielded in each biological experiment. Error bars represent the standard deviation, and asterisks represent a significant difference (P < 0.05)
Culture filtrates of the Uvbi-1 mutants inhibit rice seed elongation
In addition to causing rice yield losses, the ustiloxins produced by U. virens poses a considerable safety risk. Until now, most of the uxtiloxin were identified from rice false smut balls. Nonetheless, some secondary metabolites from mycelium may be also toxic to rice seeds. To determine whether deletion of UvBI-1 affected the production of secondary metabolites compounds, culture filtrates separated from PSB cultures of 5-day-old wild-type, mutants and complemented strain and were used to soak rice seeds for this test. The rice shoot growth was significantly stunted in samples treated with culture filtrates of the Uvbi-1 mutants, whereas increases in shoot lengths were observed for the treatments of the wild type and complemented strain (Fig. 10). This result showed that more secondary metabolites may be produced in the mutants than wild type and complemented strain, and UvBI-1 most likely involved in secondary metabolites production that would be toxic to rice seeds.
Culture filtrates of the ΔUvBI-1 mutants show increased inhibition of rice seed elongation. a The growth of rice shoots treated with testing culture filtrates. The rice seeds were soaked in 25 ml of culture filtrates in 100 ml conical flasks at 27 °C for 7 days before being photographed. b Statistical analysis of rice shoots. The length of shoots was measured after the incubation. Three independent biological experiments were performed with three replicates each time, with similar results yielded in each biological experiment. Error bars represent the standard deviation, and asterisks represent a significant difference (P < 0.05)
Uvbi-1 mutants show attenuation of pathogenicity on host plants
To evaluate the role of UvBI-1 in the fungal virulence and pathogenicity, a mixture of crushed mycelia and conidia was inoculated into rice panicles. At 21 days after inoculation, typical rice false smut balls had fully developed in the rice panicles inoculated with the wild type, whereas false smut balls were rarely observed for the Uvbi-1 mutants (Fig. 11). The attenuated pathogenicity of the Uvbi-1 mutant strain could be complemented and full virulence restored by reintroducing the UvBI-1 gene. These data showed that UvBI-1 was required for full virulence of U. virens.
UvBI-1 contributes to the pathogenicity of U. virens. a Disease symptoms caused by the testing JZ11-28, deletion mutation and complemented strains. The conidia and hyphal mixed suspension was injected into a single rice panicle from the middle to upper portion at the late booting stage, and plants were maintained in a greenhouse at 27 °C with 90–100% relative humidity for 7 days. The plants were then placed at 80% relative humidity until rice false smut symptoms appeared. The photos were taken 21 days after inoculation. b Statistical analysis of the number of smut balls. The number of smut balls was measured for a single rice panicle. At least three independent biological experiments were performed with nine rice panicles each time, with similar results yielded in each biological experiment. Error bars represent the standard deviation, and asterisks represent a significant difference (P < 0.05)
Discussion
In this study, we characterized UvBI-1 in the rice false smut fungus U. virens, which encoded a Bax Inhibitor-1-like family protein. The results of transient expression in N. benthamiana leaves showed that UvBI-1 has the ability to suppress the cell death induced by Bax, indicating its function as a Bax inhibitor. Deletion of UvBI-1 increase the hyphal growth, conidiation and secondary metabolites production, but decrease stress tolerance and cell wall integrity, and almost completely abolished the fungal pathogenicity. Our results demonstrate that UvBI-1 plays an essential role in U. virens and shares the conserved function of BI-1 among different fungal species and other organisms.
It is difficult to generate gene deletion in U. virens based on the conventional gene replacement approach by introducing homology recombination fragments via the PEG-mediated protoplast transformation (Zheng et al. 2016). Therefore, ATMT was used to generate T-DNA insertional mutants. To date, UvSUN2, Uvt3277, and UvPRO1 have been disrupted using insertional mutation (Lv et al. 2016; Yu et al. 2015a; Zheng et al. 2017). Although the gene replacement frequency was less than 0.2% using ATMT, Uvhog1 was deleted by homologous recombination (Zheng et al. 2016). Recently, the CRISPR-Cas9 system was used to significantly increase the efficiency of gene replacement by homologous recombination in U. virens (Liang et al. 2018). In this study, the CRISPR-Cas construct and the replacement fragments were simultaneously introduced into U. virens using PEG-mediated protoplast transformation to generate Uvbi-1 deletion mutants. We were able to readily obtain targeted gene deletion mutants, indicating the high efficiency of this method.
BI-1 is an evolutionarily conserved protein that is involved in growth and development, biotic and abiotic stress responses, and likely plays an indispensable role in cell protection (Huckelhoven 2004; Watanabe and Lam 2009). In M. robertsii, no obvious difference was observed in the growth rate between the MrBI-1 deletion mutant and the wild type (Chen et al. 2015). Nevertheless, mycelial growth and conidia formation were significantly increased in the UvBI-1 deletion mutants, indicating the negative role of UvBI-1 in the development progress. Moreover, the germination of conidia was not affected by UvBI-1 deletion. UvBI-1 showed a high transcriptional level in mycelia at 7 dpi, indicating its important role during the vegetative growth.
Deletion of MrBI-1 did not increase sensitivity to H2O2 (Chen et al. 2015). Nonetheless, Uvbi-1 deletion mutants were sensitive to H2O2, which is consistent with the result for Ss-bi1 in S. sclerotiorum (Yu et al. 2015b). We also found that Ss-Bi1 was orthologous with UvBI-1b that also contained the BI-1-like domain in U. virens. In plants, the generation of ROS is regarded as one of the first responses to fungal invasion (Lehmann et al. 2015). The ROS can induce apoptosis either in the pathogen only or in the host and the pathogen (Greenberg and Yao 2004). In M. oryzae, the deletion of Moatf1 leads to reduced tolerance to oxidative stress and attenuated virulence, and the inhibition of plant ROS generation restores the extension of infection hyphae (Guo et al. 2010). In Arabidopsis, AtBI-1 suppresses Bax-induced cell death downstream of ROS generation (Kawai-Yamada et al. 2004). Although Bax has not been discovered in fungi, these results suggested that BI-1 in fungi most likely had a similar function of inhibiting Bax. Deletion of UvBI-1 most likely resulted in lost ability to suppress the apoptosis induced by ROS. In addition, UvBI-1 deletion mutants were also more sensitive to other stresses. Based on the infect characteristic of U. virens, hyphae invade into the spikelet 2 or 3 days after inoculation (Song et al. 2016). During this stage, there is few nutrition can be obtained from rice, the pathogen also has to confront with the plant defense. Therefore, UvBI-1 is likely involved in the tolerance to the oxidative burst and other stresses from the host plants and contributes to the pathogenicity of U. virens.
In mammals, BI-1 interacts with Bcl-2 and Bcl-XL to suppress apoptosis by regulating ER calcium (Xu and Reed 1998). Additionally, BI-1 is involved in unfolded protein response (UPR) by physically interacting with the ER stress sensor IRE1α (Lisbona et al. 2009). The AtBI-1 associated proteins were identified based on a coimmunoprecipitation approach, and some of the proteins were shown to function in the BI-1-related processes (Weis et al. 2013). In plants, BI-1 interacts with ATG6 to regulate autophagy and programmed cell death (Xu et al. 2017). The wheat Bax inhibitor-1 protein interacts with an aquaporin TaPIP1 and increases disease resistance (Lu et al. 2018). Thus, further research is required to identify the interacting proteins of UvBI-1 in U. virens for elucidating the mechanism by which UvBI-1 regulates the development process, stress tolerance and pathogenicity.
Experimental procedures
Fungal strains, media and growth conditions
The U. virens wild-type JZ11-28 and mutant strains were cultured on potato sucrose agar (PSA) at 27 °C. The strains were cultured on YTD (yeast extract, 1 g; tryptone, 1 g; d-glucose, 10 g; agar, 15 g) and PDA (potato, 200 g; d-glucose, 20 g; agar 15 g) media for the growth assessment. For conidiation, the strains were cultured in PSB medium with shaking at 160 rpm at 27 °C for 7 days. Then, the cultured mixtures were filtrated, and the concentrations of conidia were measured using a hemocytometer. For the biomass test, six plugs of fungi 5 mm in diameter were cultured in 50 ml of PSB medium with 160 rpm shaking at 27 °C for 7 days; the hyphae were collected by filtration through two layers of gauze and measured for dry weights after drying at 50 °C in an oven for 3 days. For germination test, the conidia were spread on the plates, three region were selected on one plate and 100 conidia were observed with each region. For different stress tests, vegetative growth was assayed after incubation at 27 °C for 14 days on PSA plates and PSA with 0.2 and 0.4 M NaCl, 0.5, 0.7 and 0.9 M sorbitol, 0.01, 0.03 and 0.05% H2O2, 0.015 and 0.03% SDS (w/v), 60 and 120 μg/ml CFW, and 60 and 120 μg/ml CR. The inhibition rate was calculated as follows: inhibition rate = (average of strain colony diameters on PSA—average of strain colony diameters on PSA with chemical added)/average of strain colony diameters on PSA × 100%, and it should minus the agar diameter when the colony diameter was measured. All experiments were repeated three times with three replicates each time. Mycelia were harvested from 7-day-old cultures grown in liquid PSB and used for genomic DNA and total RNA extractions.
Protein sequence analysis
The domain was analyzed using CD-search from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) and SMART (http://smart.embl-heidelberg.de/) (Letunic et al. 2015; Marchler-Bauer et al. 2017). Sequence alignments were performed using BioEdit, and the calculated phylogenetic tree was viewed using the Mega5 program (Tamura et al. 2013).
Agrobacterium-mediated transient expression in N. benthamiana
The UvBI-1 was amplified from cDNA of U. virens using the indicated primers (Table S1, see Supporting Information) and cloned into the vector pGR107 (Wang et al. 2011) using a ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China). The constructs were confirmed by sequencing and were introduced into A. tumefaciens strain GV3101 by electroporation. For infiltration into leaves, recombinant strains of A. tumefaciens were cultured in LB medium containing 100 µg/ml kanamycin at 30 °C with shaking at 200 rpm for 24 h. The cultures were harvested, washed with 10 mM MgCl2 three times and resuspended in 10 mM MgCl2 to a final OD600 of 0.4. The A. tumefaciens cell suspension was infiltrated into N. benthamiana leaves using a syringe without a needle.
Reverse transcription PCR analysis
Total RNA samples were isolated using Trizol, and the cDNA was synthesized using a Thermo scientific RevertAid First Stand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA USA). The reaction mixtures were diluted 6 times with distilled water and used as templates for quantitative real-time PCR or RT-PCR. Real-time PCR amplifications of genes were performed in a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc., Hercules, CA) using SYBR Green I fluorescent dye. The β-tubulin gene served as the internal control for the expression studies. The RT-PCR was conducted with 27 cycles to confirm the deletion and reintroduction of the UvBI-1 gene with the indicating primers and β-tubulin used as the internal control. All experiments in this section were performed in three independent biological experiments with three replicates in each test.
Targeted gene deletion and complementation of UvBI-1
For the CRISPR/Cas system vector construct, a short cassette was generated by annealing the sense and antisense oligonucleotides BI-1f and BI-1r (Table S1, see Supporting Information) and inserted between the two Esp3I sites of pCas9: tRp-gRNA vector. The UvBI-1 gene replacement was constructed by the double-joint PCR method (Yu et al. 2004). Briefly, fragments of a 2.2 kb upstream flanking sequence and a 2.1 kb downstream flanking sequence were amplified from U. virens genomic DNA using the first-round amplification primers. The 5′ and 3′ partials of the hygromycin B gene were amplified using the primer HYG_F and H3, H2 and HYG_R, respectively. The 1.6 kb upstream flanking sequence and 1.7 kb downstream flanking sequence were joined with the 5′ and 3′ partials of the hygromycin B gene using the third round amplification primers, respectively (Table S1, see Supporting Information). The CRISPR vector and the gene recombinant fragments were introduced into protoplasts using protoplast-mediated transformation as described (Zheng et al. 2016). To generate the complementation of the UvBI-1 mutant, a fragment of the ORF sequence of U. virens and a 1 kb upstream flanking sequence were amplified and inserted into a PCETNS4 vector that contained a geneticin resistance gene using the primer pairs. Then the vector was transformed into the NO. 13 deletion mutant strain by using protoplast-mediated transformation.
Toxicity assays with culture filtrates and pathogenicity assay
After 7 days of culture in 50 ml of PSB, the filtrates were collected and centrifuged at 7000 rpm for 5 min. The supernatant was collected and incubated at 75 °C for 30 min. Seeds of rice cultivar Wanxian 98 were soaked in 0.1% potassium permanganate with 160 rpm shaking at 27 °C for 50 min and then washed 5 times with sterile distilled water. Fifty seeds were soaked in 25 ml of culture filtrate in a 100 ml conical flask, and shoot growth was measured after incubation at 27 °C for 7 days.
For the pathogenicity test, the rice was inoculated with U. virens as described (Jia et al. 2015; Song et al. 2016). The plugs were placed into potato sucrose broth (PSB) and shaken at 160 rpm at 27 °C for 7 days, and the cultured fungi were crushed. The conidia were measured and adjusted to 5 × 106/ml with PSB. Approximately 2 ml of the conidia and hyphal mixed suspension in PSB was injected into a single rice panicle from the middle to upper portion at the late booting stage (3–5 days before heading) using a syringe. Inoculated plants were maintained in a greenhouse at 27 °C with 90–100% relative humidity (RH) for 7 days and then were placed at 27 °C and 80% RH until rice false smut symptoms appeared. The symptoms were observed, and the number of smut balls was measured 21 days after inoculation. All experiments in this section were performed in at least three independent biological experiments with at least three replicates in each test.
References
Abbas H, Shier W, Cartwright R, Sciumbato G (2014) Ustilaginoidea virens infection of rice in arkansas toxicity of false smut galls, their extracts and the ustiloxin fraction. Am J Plant Sci 5:3166–3176
Ameisen JC (2002) On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ 9:367–393
Bickle M, Delley PA, Schmidt A, Hall MN (1998) Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J 17:2235–2245
Borner C (2003) The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol Immunol 39:615–647
Cebulski J, Malouin J, Pinches N, Cascio V, Austriaco N (2011) Yeast Bax inhibitor, Bxi1p, is an ER-localized protein that links the unfolded protein response and programmed cell death in Saccharomyces cerevisiae. Plos One 6(6):e20882
Chae HJ, Ke N, Kim HR, Chen SR, Godzik A, Dickman M, Reed JC (2003) Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene 323:101–113
Chae HJ, Kim HR, Xu CY, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC (2004) BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell 15:355–366
Chen YX, Duan ZB, Chen PL, Shang YF, Wang CS (2015) The Bax inhibitor MrBI-1 regulates heat tolerance, apoptotic-like cell death, and virulence in Metarhizium robertsii. Sci Rep. 5:10625
Danial NN, Korsmeyer SJ (2004) Cell death: critical control points. Cell 116:205–219
Du ZQ, Lan JF, Weng YD, Zhao XF, Wang JX (2013) BAX inhibitor-1 silencing suppresses white spot syndrome virus replication in red swamp crayfish, Procambarus clarkii. Fish Shellfish Immun 35:46–53
Gaguancela OA, Zuniga LP, Arias AV, Halterman D, Flores FJ, Johansen IE, Wang A, Yamaji Y, Verchot J (2016) The IRE1/bZIP60 Pathway and Bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Mol Plant Microbe Interact 29:750–766
Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6:201–211
Grzmil M, Thelen P, Hemmerlein B, Schweyer S, Voigt S, Mury D, Burfeind P (2003) Bax Inhibitor-1 is overexpressed in prostate cancer and its specific down-regulation by rna interference leads to cell death in human prostate carcinoma cells. Am J Pathol 163:543–552
Grzmil M, Kaulfuss S, Thelen P, Hemmerlein B, Schweyer S, Obenauer S, Kang TW, Burfeind P (2006) Expression and functional analysis of Bax inhibitor-I in human breast cancer cells. J Pathol 208:340–349
Guo M, Guo W, Chen Y, Dong S, Zhang X, Zhang H, Song W, Wang W, Wang Q, Lv R, Zhang Z, Wang Y, Zheng X (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 23:1053–1068
Hamann A, Brust D, Osiewacz HD (2008) Apoptosis pathways in fungal growth, development and ageing. Trends Microbiol 16:276–283
Heath MC (1998) Apoptosis, programmed cell death and the hypersensitive response. Eur J Plant Pathol 104:117–124
Henke N, Lisak DA, Schneider L, Habicht J, Pergande M, Methner A (2011) The ancient cell death suppressor BAX inhibitor-1. Cell Calcium 50:251–260
Huckelhoven R (2004) BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis 9:299–307
Jia Q, Lv B, Guo MY, Luo CX, Zheng L, Hsiang T, Huang JB (2015) Effect of rice growth stage, temperature, relative humidity and wetness duration on infection of rice panicles by Villosiclava virens. Eur J Plant Pathol 141:15–25
Kawai-Yamada M, Jin LH, Yoshinaga K, Hirata A, Uchimiya H (2001) Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBl-1). Proc Natl Acad Sci USA 98:12295–12300
Kawai-Yamada M, Ohori Y, Uchimiya H (2004) Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16:21–32
Kotsafti A, Farinati F, Cardin R, Burra P, Bortolami M (2010) Bax Inhibitor-1 down-regulation in the progression of chronic liver diseases. BMC Gastroenterol 10:35
Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Phys 48:251–275
Lehmann S, Serrano M, L’Haridon F, Tjamos SE, Metraux JP (2015) Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 112:54–62
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260
Liang YF, Han Y, Wang CF, Jiang C, Xu JR (2018) Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front Plant Sci 9:699
Lisak DA, Schacht T, Enders V, Habicht J, Kiviluoto S, Schneider J, Henke N, Bultynck G, Methner A (2015) The transmembrane Bax inhibitor motif (TMBIM) containing protein family: tissue expression, intracellular localization and effects on the ER CA2+-filling state. Biochim Biophys Acta Mol Cell Res 1853:2104–2114
Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, Reed JC, Glimcher LH, Hetz C (2009) BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1 alpha. Mol Cell 33:679–691
Lu PP, Yu TF, Zheng WJ, Chen M, Zhou YB, Chen J, Ma YZ, Xi YJ, Xu ZS (2018) The wheat Bax inhibitor-1 protein interacts with an aquaporin TaPIP1 and enhances disease resistance in Arabidopsis. Front Plant Sci 9:20
Lv B, Zheng L, Liu H, Tang JT, Hsiang T, Huang JB (2016) Use of random T-DNA mutagenesis in identification of gene UvPRO1, a regulator of conidiation, stress response, and virulence in Ustilaginoidea virens. Front Microbiol 7:2086
Marchler-Bauer A, Bo Y, Han LY, He JE, Lanczycki CJ, Lu SN, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang ZX, Yamashita RA, Zhang DC, Zheng CJ, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45:D200–D203
Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R (2003) Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J 33:425–434
Meng J, Sun W, Mao Z, Xu D, Wang X, Lu S, Lai D, Liu Y, Zhou L, Zhang G (2015) Main ustilaginoidins and their distribution in rice false smut balls. Toxins 7:4023–4034
Qi JS, Wang JL, Gong ZZ, Zhou JM (2017) Apoplastic ROS signaling in plant immunity. Curr Opin Plant Biol 38:92–100
Robinson KS, Clements A, Williams AC, Berger CN, Frankel G (2011) Bax Inhibitor 1 in apoptosis and disease. Oncogene 30:2391–2400
Roncero C, Duran A (1985) Effect of calcofluor white and congo red on fungal cell-wall morphogenesis—invivo activation of chitin polymerization. J Bacteriol 163:1180–1185
Rush MC, Shahjahan AKM, Jones JP, Groth DE (2000) Outbreak of false smut of rice in Louisiana. Plant Dis 84:100–100
Scorrano L, Korsmeyer SJ (2003) Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochem Bioph Res Co. 304:437–444
Sharon A, Finkelstein A, Shlezinger N, Hatam I (2009) Fungal apoptosis: function, genes and gene function. FEMS Microbiol Rev 33:833–854
Song JH, Wei W, Lv B, Lin Y, Yin WX, Peng YL, Schnabel G, Huang JB, Jiang DH, Luo CX (2016) Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ Microbiol 18:3840–3849
Sun X, Kang S, Zhang Y, Tan X, Yu Y, He H, Zhang X, Liu Y, Wang S, Sun W, Cai L, Li S (2013) Genetic diversity and population structure of rice pathogen Ustilaginoidea virens in China. PLoS One 8:e76879
Sun W, Dong X, Xu D, Meng J, Fu X, Wang X, Lai D, Zhou L, Liu Y (2016) Preparative separation of main ustilaginoidins from rice false smut balls by high-speed counter-current chromatography. Toxins 8(1):20
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tsukui T, Nagano N, Umemura M, Kumagai T, Terai G, Machida M, Asai K (2015) Ustiloxins, fungal cyclic peptides, are ribosomally synthesized in Ustilaginoidea virens. Bioinformatics 31:981–985
Wang S, Bai Y, Zhou Y, Yao J, Bai J (1998) The pathogen of false smut of rice. Acta Phytopathol Sin 28:19–24 (In Chinese, abstract in English)
Wang Q, Han C, Ferreira AO, Yu X, Ye W, Tripathy S, Kale SD, Gu B, Sheng Y, Sui Y, Wang X, Zhang Z, Cheng B, Dong S, Shan W, Zheng X, Dou D, Tyler BM, Wang Y (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23:2064–2086
Wang XJ, Tang CL, Huang XL, Li FF, Chen XM, Zhang G, Sun YF, Han DJ, Kang ZS (2012) Wheat BAX inhibitor-1 contributes to wheat resistance to Puccinia striiformis. J Exp Bot 63:4571–4584
Watanabe N, Lam E (2009) Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int J Mol Sci 10:3149–3167
Weis C, Pfeilmeier S, Glawischnig E, Isono E, Pachl F, Hahne H, Kuster B, Eichmann R, Huckelhoven R (2013) Co-immunoprecipitation-based identification of putative BAX INHIBITOR-1-interacting proteins involved in cell death regulation and plant-powdery mildew interactions. Mol Plant Pathol 14:791–802
Wood PJ, Fulcher RG (1983) Dye interactions—a basis for specific detection and histochemistry of polysaccharides. J Histochem Cytochem 31:823–826
Xu QL, Reed JC (1998) Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell 1:337–346
Xu GY, Wang SS, Han SJ, Xie K, Wang Y, Li JL, Liu YL (2017) Plant Bax inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 13:1161–1175
Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Dominguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981
Yu M, Yu J, Hu J, Huang L, Wang Y, Yin X, Nie Y, Meng X, Wang W, Liu Y (2015a) Identification of pathogenicity-related genes in the rice pathogen Ustilaginoidea virens through random insertional mutagenesis. Fungal Genet Biol 76:10–19
Yu Y, Xiao JF, Yang YH, Bi CW, Qing L, Tan WZ (2015b) Ss-Bi1 encodes a putative BAX inhibitor-1 protein that is required for full virulence of Sclerotinia sclerotiorum. Physiol Mol Plant Pathol 90:115–122
Yue HY, Nie SJ, Xing D (2012) Over-expression of Arabidopsis Bax inhibitor-1 delays methyl jasmonate-induced leaf senescence by suppressing the activation of MAP kinase 6. J Exp Bot 63:4463–4474
Zheng D, Wang Y, Han Y, Xu J-R, Wang C (2016) UvHOG1 is important for hyphal growth and stress responses in the rice false smut fungus Ustilaginoidea virens. Sci Rep 6:24824
Zheng MT, Ding H, Huang L, Wang YH, Yu MN, Zheng R, Yu JJ, Liu YF (2017) Low-affinity iron transport protein Uvt3277 is important for pathogenesis in the rice false smut fungus Ustilaginoidea virens. Curr Genet 63:131–144
Zhou YL, Pan YJ, Xie XW, Zhu LH, Xu JL, Wang S, Li ZK (2008) Genetic diversity of rice false smut fungus, Ustilaginoidea virens and its pronounced differentiation of populations in North China. J Phytopathol 156:559–564
Acknowledgements
We thank JinRong Xu from Northwest A&F University, Yangling, China, and Purdue University, West Lafayette, IN, the United States, for the CRISPR-Cas9 system plasmid, Yuanchao Wang from Nanjing Agricultural University for the pGR107 vector and Bax and Daohong Jiang from Huazhong Agricultural University for the complemented vector PCETNS4. The National Natural Science Foundation of China (No. 31701736), the National Key Research and Development Program (2016YFD0300700) and the Fundamental Research Funds for the Central Universities (No. 2662017JC003 and 2662018JC051) supported this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, S., Wang, Y., Wei, W. et al. The Bax inhibitor UvBI-1, a negative regulator of mycelial growth and conidiation, mediates stress response and is critical for pathogenicity of the rice false smut fungus Ustilaginoidea virens. Curr Genet 65, 1185–1197 (2019). https://doi.org/10.1007/s00294-019-00970-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-019-00970-2