Abstract
The genotyping of Campylobacter coli was done using three methods, pulsed-field gel electrophoresis (PFGE), Sau-polymerase chain reaction (Sau-PCR), and denaturing gradient gel electrophoresis assay of flagellin gene (fla-DGGE) and the characteristics of these assays were compared. The results showed that a total of 53 strains of C. coli were isolated from chicken and duck samples in three markets. All isolates were clustered into 31, 33, and 15 different patterns with Simpson’s index of diversity (SID) values of 0.972, 0.974, and 0.919, respectively. Sau-PCR assay was simpler, more rapid, and had higher discriminatory power than PFGE assay. Fla-DGGE assay could detect and illustrate the number of contamination types of C. jejuni and C. coli without cultivation, which saved more time and cost than Sau-PCR and PFGE assays. Therefore, Sau-PCR and fla-DGGE assays are both rapid, economical, and easy to perform, which have the potential to be promising and accessible for primary laboratories in genotyping C. coli strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Campylobacter coli is one of the most prevalent organisms and a major cause of human acute enteritis, which is associated with foodborne illness. Large numbers of Campylobacteriosis cases occur every year, with approximately 200,000 cases in the European Union and 800,000 in the USA [1, 2]. Human Campylobacteriosis are often caused by the species of Campylobacter jejuni and C. coli. In general, the main symptoms of C. coli that affect most people include diarrhea, fever, and abdominal cramps, which may cause chronic sequelae in severe cases [3]. Although most Campylobacter infections are caused by C. jejuni, the occurring rate of C. coli infections has increased significantly from approximately 5% to 15% since 2000 [4, 5]. The main source of human infections is considered to be the consumption of contaminated poultry meat [6, 7]. Many studies have reported a higher rate of C. coli isolated from chicken than that of C. jejuni in China [8, 9].
Molecular typing methods are essential tools for the epidemiological investigation of C. coli. Pulsed-field gel electrophoresis (PFGE) is regarded as a “gold standard” fingerprinting method, but it also has disadvantages due to being arduous [10]. Moreover, several studies have reported some rapid genotyping methods of C. coli strains, including multilocus sequence typing (MLST) [11], automated repetitive extragenic palindromic polymerase chain reaction (REP-PCR) [12], and whole-genome sequencing (WGS) [13]. However, these methods have certain limitations, such as the requirement of technical expertise and are expensive.
Sau-polymerase chain reaction (Sau-PCR) is based on a genomic DNA enzymatic digestion with the restriction endonuclease Sau3AI of amplified fragment length polymorphism (AFLP) and subsequent amplification of randomly amplified polymorphic DNA (RAPD), which can be considered for DNA fingerprinting-based analyses with lower costs of time and equipment [14]. Nowadays, it has been applied to many bacterial species; however, there is no report on the application of Sau-PCR to genotype C. coli isolates. Denaturing gradient gel electrophoresis assay of flagellin gene (Fla-DGGE) which is based on the 5’ and 3’ regions of flaA and flaB genes being highly conserved within different strains, can separate DNA fragments of equal length with different base composition regardless of regions in between flaA and flaB varying significantly [15, 16]. It has been applied for subtyping C. jejuni and C. coli in cecal samples of broilers without cultivation [17]. Thus, this study aimed to apply PFGE, Sau-PCR, and fla-DGGE for genotyping C. coli strains from poultry meat, and compare their characteristics.
Material and Methods
Identification of Campylobacter coli
A total of 81 fresh slaughtered poultry samples were collected from three retail markets (A, B, and C) in Guangzhou, China. All samples were transported to the laboratory within 2 h on ice packs and then processed immediately. Briefly, the epidermis was removed with a sterile knife and the subcutaneous muscle was taken to exclude environmental contamination. Ten grams of samples and 10 mL of 0.1% peptone water were placed in a sterile filter stomacher bag and subjected to 30 s of homogenization with a BagMixer lab blender 400 (Bio-Rad, USA), then 1 mL filtered liquid was transferred into 4 mL Bolton broth enrichment (Oxoid, UK) and 4 mL Preston broth enrichment (Oxoid, UK), respectively, and incubated at 37 ℃ for 24 h. After that, a loopful of the enrichment was streaked onto modified charcoal cefoperazone deoxycholate agar (mCCDA) (Oxoid, UK) and Skirrow selective medium (Oxoid, UK) plate, respectively, then cultured at 37 ℃ for 48 h under microaerophilic conditions (85% N2, 10% CO2, and 5% O2). Two presumptive colonies in each plate were picked to grow at 25 ℃ and 42 ℃, then tested for indole acetate, hippuric salt, oxidase, catalase and Gram stained. The isolates with typical colony characteristics were subjected to a Campylobacter PCR test kit (TaKaRa, Japan). Based on the PFGE results, if there were typologically consistent isolates in each sample, only one of them was retained. The identified C. coli isolates were stored at − 80 ℃.
PFGE Analysis
The isolates were subjected to molecular typing by PFGE, which was performed using the standardized protocol described by PulseNet (http://www.cdc.gov/pulsenet/protocols.htm). Briefly, all isolates were grown on Mueller–Hinton agar (Oxoid, UK) with 5% laked horse blood, then diluted to the required concentration and made agarose-embedded plugs, then digested with SmaI restriction enzyme. The digested plugs were run in Seakem agarose gel (Bio-Rad, USA) with 0.5 × Tris–Borate EDTA (TBE) buffer to separate the bands with a CHEF Mapper PFGE system (Bio-Rad, USA) by running for 18 h at 14 ℃ switching directions every 6.76 s and ending with 35.38 s. Gels were stained with ethidium bromide solution and destained with distilled water, then DNA bands were visualized under UV illumination.
Sau-PCR Analysis
For Sau-PCR, the assay was designed and modified as previously described in Corich et al. [14]. The DNA (200 ng) was subjected to digestion at 37 ℃ for 5 h with 5 U Sau3AI in a final volume of 20 μL. Primers SAUA, SAUT, SAG, STG, and SGAG were used for preliminary examination (Supplementary Table S1). The amplification reaction was performed in a 25 μL reaction volume containing 2.5 μL of 10 × buffer (1.5 mM MgCl2), 0.5 μL dNTP (10 mM), 5 μL primer SAG (10 μM), 0.5 U Taq polymerase, 20 ng template DNA, and ddH2O. PCR reactions were carried out with an iCycler Thermal Cycler (Bio-Rad, USA) using the following amplification conditions: 25 ℃ for 5 s, ramp to 60 ℃ at 0.1 ℃/s, 60 ℃ for 30 s, 2 cycles of 94 ℃ for 1 min, 50 ℃ for 15 s, ramp to 25 ℃ at 0.1 ℃/s, ramp to 50 ℃ at 0.1 ℃/s, 50 ℃ for 30 s, 35 cycles of 94 ℃ for 15 s, 48 ℃ for 1 min, 65 ℃ for 2 min, and the final extension at 65 ℃ for 5 min, then stored at 4 ℃. The amplification products were subjected to 2% agarose gel and detected by staining with ethidium bromide, then photographed and analyzed with a Universal Hood II system.
Fla-DGGE Analysis
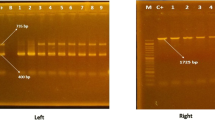
The experimental design and modifications for the fla-DGGE assay were based on prior work done by Yu and Morrison [18]. Primers CF03 and CF02 were used in this assay (Supplementary Table S1). The amplification reaction was performed in a 50 μL reaction volume containing 1.25 U Taq polymerase, 5 μL of 10 × PCR buffer, 1 μL dNTP (10 mM), 20 ng template DNA, 3 μL of each primer (10 μM), and ddH2O. PCR reactions were carried out with a PCR amplifier using the following amplification conditions: 5 min at 94 ℃ (initial step), 35 cycles of 30 s at 94 ℃, 30 s at 56 ℃, 30 s at 72 ℃ and a final extension of 7 min at 72 ℃. The gradient of the chemical denaturant was adjusted to 5%–35%. Amplicons were separated in 8% polyacrylamide gel in 1 × Tris–Acetate-EDTA (TAE) with a Dcode apparatus (Bio-Rad, USA), and the operating conditions were 20 V at 60 ℃ for 15 min and then 200 V at 60 ℃ for 3 h. The amplification products were subjected to 2% agarose gel and detected by staining with ethidium bromide, then photographed and analyzed with a Universal Hood II system (Bio-Rad, USA).
Data Analysis
For the data, Chi-Square test, t-tests, and ANOVA were employed to assess the significant differences of C. coli in chickens and ducks sampled in different markets using SPSS 26.0 software, and P value less than 0.05 was considered a statistically significant difference. The dendrograms were drawn with the Numerical Taxonomy System of Statistic (NTSYS) software, using the Dice correlation coefficient and the unweighted pair group mathematical average (UPGMA) clustering algorithm to determine the phylogenetic relationships of strains. The single numerical index of Simpson’s index of diversity (SID) was used to compare the typing methods [19]. The formula for calculating SID is:
where ‘N’ is the total number of isolates, ‘s’ is the total number of patterns described, and ‘nj’ is the number of isolates belonging to type j. In addition, the concordance between the typing methods was assessed using the adjusted Rand index (AR) and adjusted Wallace (AW) coefficient. The AR shows the proportion of agreement, whereas the AW indicates the probability that two isolates assigned to the same type by one method will also be classified as the same type when using the other method [20,21,22]. The calculation of SID, AR, and AW coefficients was performed using the online tool at the Comparing Partitions website (http://www.comparingpartitions.info/).
Results
Isolation of C. coli
In this study, 81 samples of fresh poultry meat from three markets were tested. The results showed that 44/81 (54.3%) of the samples were positive for C. coli, and a total of 53 C. coli strains were isolated. The prevalence of C. coli in poultry meat from 3 markets is shown in Table 1. The overall proportion of positive samples in poultry meat was 54.6% (95% CI 44.5–64.6), with a higher proportion of duck samples (60.6%) than the chicken samples (50.0%), and a higher proportion in market A (63.4%) than B (48.8%) and C (51.6%). The overall prevalence of C. coli was 49.8% (95% CI 26.6–73.0) and 59.4% (95% CI 38.2–80.6) in chicken and duck samples, respectively. There was no significant difference in the prevalence of C. coli between chicken and duck samples (χ2 = 0.887, P > 0.05). The mean and standard deviation of C. coli from three markets were as follows: market A (8.5 ± 0.707), B (7.0 ± 4.243), and C (6.5 ± 2.121). There was no significant difference in the overall mean of C. coli between chicken and duck samples (t = 0.658, P = 0.547 > 0.05), and no significant difference was found in the overall mean of C. coli from three markets (F = 0.283, P = 0.772 > 0.05). In addition, the overall separation rate of C. coli isolates was 50.0% (95% CI 29.8–70.2), with the highest rate recovered from chicken samples in market B, and the lowest rate recovered from duck samples in market B.
PFGE Analysis
A total of 53 C. coli isolates were classified into 31 pulsotypes with similarity ranging from 60 to 100% (Fig. 1). The SID value of PFGE was 0.972. Groups P4, P8, and P31 were the three predominant ones, each group consisted of 4 or 5 isolates, and those isolates were all recovered from market A. The number of types varied in different markets, A 10/22 (45.5%), B 14/16 (87.5%), and C 9/15 (60.0%). Some isolates of the same type originated from different sources, such as P1 included 1 isolate from chicken and 2 isolates from duck in market A, P4 included 2 isolates from chicken and 2 isolates from duck in market A, and P31 included 1 isolate from chicken and 3 isolates from duck in market A. Some isolates of the same type originated from different markets, for example, P10 included 2 isolates of chicken from markets B and C. In addition, P28 originated from different sources and markets, which included 1 isolate of chicken from market B and 2 isolates of duck from market C.
Sau-PCR Analysis
In the preliminary examination of Sau-PCR, the results showed that primer SAG generated more and clearer bands than primers SAUA, SAUT, STG, and SGAG (data not shown). The results of three repeated experiments were consistent. Sau-PCR analysis of 53 isolates yielded between 4 and 10 DNA fragments, and they were classified into 33 pulsotypes with similarity ranging from 54 to 100% (Fig. 2). The calculated SID value of Sau-PCR was 0.974. Group S8 was the predominant one, which consisted of 6 isolates. The number of types varied in different markets, A 13/22 (59.1%), B 13/16 (81.3%), and C 10/15 (66.7%). Some isolates of the same type originated from different sources, such as S1 included 1 isolate from chicken and 2 isolates from duck in market A, and S13 included 2 isolates from chicken and duck in market B. Some isolates of the same type originated from different markets, for example, S33 included 2 isolates of chicken from markets B and C. In addition, some isolates of the same type originated from different sources and markets, such as S8 included 2 isolates of chicken and 2 isolates of duck from market A, and 2 isolates of duck from market C, S9 included 1 isolate of duck from market A and 2 isolates of chicken from market B.
Fla-DGGE Analysis
A total of 53 isolates were classified into 15 subtypes by fla-DGGE, and the SID value was 0.919. Fla-DGGE analysis of each isolate yielded one band, and the electrophoretic bands of 15 different types of some C. coli isolates by fla-DGGE are shown in Fig. 3. The number of types varied in different markets, A 9/22 (41.0%), B 8/16 (50.0%), and C 7/15 (46.7%). Groups F1, F9, and F10 were the predominant ones. Some isolates of the same type originated from different sources, for example, F1 included 3 isolates from duck and 6 isolates from chicken in market A. In addition, some isolates of the same type originated from different sources and markets, such as F8 included 3 isolates of chicken from market C and 1 isolate of chicken from market B and 2 isolates of duck from market A, F9 included 2 isolates of duck from market C and 3 isolates of chicken from market B and 1 isolate of duck from market B, F10 included 1 isolate of chicken from market A and 3 isolates of chicken from market B and 1 isolate of duck from market B and 2 isolates of duck from market C.
Comparison of Three Methods
Genotyping was performed by PFGE, Sau-PCR, and fla-DGGE, a total of 53 isolates were clustered into 31, 33, and 15 different patterns with SID values of 0.972, 0.974, and 0.919, respectively. The results of the analysis of the 17 isolates by the three typing methods were highly consistent (Fig. 4). For example, the subtyping results of strain CYC1, CYC3, and CYC4 were all in the same group by three methods, CYC47, CYC48, and CYC49 were all in another identical group. Using the Sau-PCR assay, PFGE groups P4, P8, P22, P28, and P31, comprising 2–5 isolates, were further divided into 2 to 3 Sau-PCR types, respectively. Using the fla-DGGE assay, PFGE groups P4, P6, P8, P10, P11, P28, and P31, comprising 2–5 isolates, were further divided into 2 to 3 fla-DGGE types, respectively.
The congruence among methods was determined by comparing the calculated AR and AW coefficients. The AR coefficient between PFGE and Sau-PCR showed a moderate congruence (AR = 0.500), AWSau-PCR→PFGE = 0.487, AWPFGE→Sau-PCR = 0.514, which meant if two isolates were in the same type by PFGE, they had about 48.7% chance of being identified as the same Sau-PCR type, while conversely, the chance was 51.4%. In addition, the AR coefficient between PFGE and fla-DGGE was 0.207, and between Sau-PCR and fla-DGGE was 0.142. The AW coefficient among methods is shown in Table 2.
Discussion
According to previous studies, the prevalence of C. coli in poultry or raw poultry meat ranged from 7% to 87.8% in China [[22] (n markets = 3); [23] (n markets = 25); [24] (n markets = 14); [25] (n markets = 5)]. In this study, the prevalence of C. coli in poultry samples ranged from 41.7% to 66.7%. The incidence of C. coli was higher in duck (60.6%) than in chicken (50.0%), but there was no significant difference in the prevalence between chicken and duck, which was similar to our previous study [22]. The results showed that 44 (54.3%) samples (n = 81) were positive for C. coli and 53 strains were isolated, which was less than the previous study where 53 (60.2%) samples (n = 88) were positive for C. coli and 68 isolates were detected [22]. There were 20.5% (9/44) of the poultry samples contaminated by 2 types of C. coli in our study. Due to only two suspicious colonies in each sample being picked for identification in this study, the actual contamination rate of C. coli in these samples may be underestimated. In addition, three typing method results all showed that some isolates of the same type originated from different sources and markets, which indicated the samples may be cross-contaminated through multiple pathways. Markets A, B, and C are located in the same district and close to each other. Poultry in the same market may have been transported from the same source or raised on the same farm before being slaughtered. The association between Campylobacter outbreaks and poultry products has been reported in several studies. The contaminated poultry products may lead the Campylobacter outbreaks [6, 26, 27]. People are likely to get illnesses such as diarrhea, fever, and abdominal pain from contaminated poultry products. Therefore, more interventions should be implemented to prevent the occurrence of poultry contamination.
Any typing method that produces a SID value higher than 0.900 is appropriate, while methods that produce a SID value of 0.950 can be considered as more or less ‘ideal’ [12, 28]. In the present study, the SID values of PFGE, Sau-PCR, and fla-DGGE were 0.972, 0.974, and 0.919, respectively, indicating that the three methods were all appropriate for genotyping C. coli. However, discriminatory power is not the only criterion for judging the usefulness of a technique. Optimal typing methods need to have appropriate discriminatory power and must be assessed for the turnaround time, throughput, cost, and technical difficulty of the typing methods [29].
It is commonly accepted that PFGE is one of the most easily accessible methods for typing C. coli. Previously, a study genotyped C. coli strains that were isolated from live broilers and retail broiler meat by PFGE, and the SID value was 0.910 [12]. Other studies have shown the SID value of PFGE in C. coli reached 0.980 [22, 30]. The SID value of PFGE in the current study was within the range of those previously observed. However, due to the complex and time-consuming operation procedures, it is undesirable to genotype large numbers of samples by PFGE. Moreover, compared to other methods, the apparatus and reagents, such as proteinase K and SmaI restriction enzyme used in PFGE were expensive. When using PFGE for genotyping C. jejuni, some strains may still not be typeable due to genetic variations [31].
Sau-PCR fingerprints are created based on the restriction sites of the Sau3AI enzyme in the bacterial genome, which means that Sau-PCR is a simple and fast technique. To the best of our knowledge, the Sau-PCR assay has been applied to genotyping of 10 bacterial species since 2005, such as Listeria monocytogenes, Burkholderia cepacian, and others [32, 33]. A study showed Sau-PCR was useful in investigating the distribution of Lactococcus garvieae strains in the environment, and the discriminatory power was 0.798, which was lower than our study [34]. Another study genotyped Legionella strains in a hospital in Italy, which described Sau-PCR showed greater discriminative power than PFGE [35]. Sau-PCR was used to subtype Burkholderia cepacia and the fingerprinting results were reproducible [33]. Sau-PCR was also applied to Starmerella bacillaris and Staphylococcus xylosus [36, 37]. However, there is no report on the application of Sau-PCR for genotyping C. coli isolates, and only a few studies on Sau-PCR have been reported so far. In the present study, the Sau-PCR assay was used to genotype C. coli, and the results of the DNA fingerprint were reproducible. The SID value of Sau-PCR was 0.974, which was slightly higher than PFGE. Additionally, the results of C. coli genotyping could be generated within one day because of its rapid and simple operating procedure.
The principle of DGGE is that the double-stranded DNA molecules have different denaturation temperatures during gel electrophoresis and remain in different positions on the gel, forming separate bands [38]. It is possible to separate DNA fragments of the same size and different base compositions, even with only one base difference [39]. The 5ʹ and 3ʹ regions of flaA and flaB genes are highly conserved within different strains, while the sequences in between can vary significantly [15]. Primers CF02 and CF03 were designed according to these features, which were favorable for fla-DGGE typing. Fla-DGGE can detect C. coli and C. jejuni without needing the pathogen to be cultured, which can save time and be cost-effective as microaerobic strains are difficult to culture. In addition, fla-DGGE can illustrate the number of contaminants of C. coli and C. jejuni without needing to culture pathogens, which is one of the most important differences as compared to other methods. The SID value of fla-DGGE was lower than Sau-PCR and PFGE, however, its discriminatory power could be improved if PCR products were sequenced.
To evaluate the reliability of the typing methods, several characteristics were assessed to compare the analysis results obtained by the three methods. Among all AR and AW coefficients, between PFGE and Sau-PCR were the highest (AR = 0.500, AWSau-PCR→PFGE = 0.487, AWPFGE→Sau-PCR = 0.514), suggesting that Sau-PCR had a moderate level of agreement with PFGE. The AR and AW coefficients between fla-DGGE and other methods were low, indicating that fla-DGGE had a low level of agreement with other methods. The degree of congruence between the results of two or more typing methods varies with sample size, even for different samples from the same population [40, 41]. Although the present results may differ from a similar analysis using different strains of C. coli, the high congruence between PFGE and Sau-PCR data may be reproducible in other studies.
Conclusion
The present study found that the proportion of C. coli positive samples from poultry meat was high. PFGE, Sau-PCR, and fla-DGGE were able to genotype C. coli strains. Sau-PCR assay was simpler, more rapid, and had higher discriminatory power than PFGE assay. Fla-DGGE assay could detect and illustrate the number of contamination types of C. jejuni and C. coli without cultivation, which saved more time and cost than Sau-PCR and PFGE assays. Therefore, Sau-PCR and fla-DGGE assays are both rapid, economic, and easy to perform, which have the potential to be promising and accessible for primary laboratories in genotyping C. coli strains.
Data Availability
Data available on request from the corresponding author.
Code Availability
Not applicable.
References
European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC) (2018) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. Eur Food Saf Auth 16(12):e5500. https://doi.org/10.2903/j.efsa.2018.5500
Scallan E, Griffin PM, Angulo FJ et al (2011) Foodborne illness acquired in the United states-unspecified agents. Emerg Infect Dis 17(1):16–22. https://doi.org/10.3201/eid1701.P21101
Keithlin J, Sargeant J, Thomas MK et al (2014) Systematic review and meta-analysis of the proportion of Campylobacter cases that develop chronic sequelae. BMC Public Health 14:1203. https://doi.org/10.1186/1471-2458-14-1203
Li Y, Gu Y, Lv J et al (2020) Laboratory study on the gastroenteritis outbreak caused by a multidrug-resistant Campylobacter coli in China. Foodborne Pathog Dis 17(3):187–193. https://doi.org/10.1089/fpd.2019.2681
Zhang P, Zhang X, Liu Y et al (2020) Multilocus sequence types and antimicrobial resistance of Campylobacter jejuni and C. coli isolates of human patients from Beijing, China, 2017–2018. Front Microbiol 11:554784. https://doi.org/10.3389/fmicb.2020.554784
Rosner BM, Schielke A, Didelot X et al (2017) A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci Rep 7(1):5139. https://doi.org/10.1038/s41598-017-05227-x
Zhang P, Zhang X, Liu Y et al (2022) Genomic insights into the increased occurrence of Campylobacteriosis caused by antimicrobial-resistant Campylobacter coli. MBio 13(6):e0283522. https://doi.org/10.1128/mbio.02835-22
Wang Y, Dong Y, Deng F et al (2016) Species shift and multidrug resistance of Campylobacter from chicken and swine, China, 2008–14. J Antimicrob Chemother 71(3):666–669. https://doi.org/10.1093/jac/dkv382
Liu D, Yang D, Liu X et al (2020) Detection of the enterococcal oxazolidinone/phenicol resistance gene optrA in Campylobacter coli. Vet Microbiol 246:108731. https://doi.org/10.1016/j.vetmic.2020.108731
Ribot EM, Swaminathan B, Taskforce P (2006) PulseNet USA: a five-year update. Foodborne Pathog Dis 3(1):9–19. https://doi.org/10.1089/fpd.2006.3.9
Tedersoo T, Roasto M, Mäesaar M et al (2022) The prevalence, counts, and MLST genotypes of Campylobacter in poultry meat and genomic comparison with clinical isolates. Poult Sci 101(4):101703. https://doi.org/10.1016/j.psj.2022.101703
Behringer M, Miller WG, Oyarzabal OA (2011) Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J Microbiol Methods 84(2):194–201. https://doi.org/10.1016/j.mimet.2010.11.016
Whitehouse CA, Young S, Li C et al (2018) Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol 73:122–128. https://doi.org/10.1016/j.fm.2018.01.018
Corich V, Mattiazzi A, Soldati E et al (2005) Sau-PCR, a novel amplification technique for genetic fingerprinting of microorganisms. Appl Environ Microbiol 71(10):6401–6406. https://doi.org/10.1128/AEM.71.10.6401-6406.2005
Wegmuller B, Luthy J, Candrian U (1993) Direct polymerase chain reaction detection of Campylobacter jejuni and Campylobacter coli in raw milk and dairy products. Appl Environ Microbiol 59(7):2161–2165. https://doi.org/10.1128/aem.59.7.2161-2165.1993
Hein I, Mach RL, Farnleitner AH et al (2003) Application of single-strand conformation polymorphism and denaturing gradient gel electrophoresis for fla sequence typing of Campylobacter jejuni. J Microbiol Methods 52(3):305–313. https://doi.org/10.1016/S0167-7012(02)00183-5
Najdenski H, Heyndrickx M, Herman L et al (2008) Fla-DGGE analysis of Campylobacter jejuni and Campylobacter coli in cecal samples of broilers without cultivation. Vet Microbiol 127(1–2):196–202. https://doi.org/10.1016/j.vetmic.2007.08.002
Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36(5):808–812. https://doi.org/10.2144/04365st04
Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 26(11):2465–2466. https://doi.org/10.1128/jcm.26.11.2465-2466.1988
Carriço JA, Silva-Costa C, Melo-Cristino J et al (2006) Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol 44(7):2524–2532. https://doi.org/10.1128/JCM.02536-05
Duarte A, Seliwiorstow T, Miller WG et al (2016) Discriminative power of Campylobacter phenotypic and genotypic typing methods. J Microbiol Methods 125:33–39. https://doi.org/10.1016/j.mimet.2016.03.004
Kong N, Luo Y, Liu J et al (2023) Subtyping of Campylobacter coli isolated from raw poultry meat in retail markets using amplified intergenic locus polymorphism—a novel rapid subtyping method. J Microbiol Methods 204:106662. https://doi.org/10.1016/j.mimet.2022.106662
Tang M, Zhou Q, Zhang X et al (2020) Antibiotic resistance profiles and molecular mechanisms of Campylobacter from chicken and pig in China. Front Microbiol 11:592496. https://doi.org/10.3389/fmicb.2020.592496
Zhu J, Yao B, Song X et al (2017) Prevalence and quantification of Campylobacter contamination on raw chicken carcasses for retail sale in China. Food Control 75:196–202. https://doi.org/10.1016/j.foodcont.2016.12.007
Li B, Ma L, Li Y et al (2017) Antimicrobial resistance of Campylobacter species isolated from broilers in live bird markets in shanghai, China. Foodborne Pathog Dis 14(2):96–102. https://doi.org/10.1089/fpd.2016.2186
Jehanne Q, Pascoe B, Bénéjat L et al (2020) Genome-wide identification of host-segregating single-nucleotide polymorphisms for source attribution of clinical Campylobacter coli isolates. Appl Environ Microbiol 86(24):e01787-e1820. https://doi.org/10.1128/aem.01787-20
Rodrigues CS, Armendaris PM, de Sá CVGC et al (2021) Prevalence of Campylobacter spp. in chicken carcasses in slaughterhouses from south of Brazil. Curr Microbiol 78(6):2242–2250. https://doi.org/10.1007/s00284-021-02478-w
Van Belkum A, Tassios PT, Dijkshoorn L et al (2007) Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect 13:1–46. https://doi.org/10.1111/j.1469-0691.2007.01786.x
Clark CG, Taboada E, Grant CCR et al (2012) Comparison of molecular typing methods useful for detecting clusters of Campylobacter jejuni and C. coli isolates through routine surveillance. J Clin Microbiol 50(3):798–809. https://doi.org/10.1128/JCM.05733-11
Thakur S, White DG, McDermott PF et al (2009) Genotyping of Campylobacter coli isolated from humans and retail meats using multilocus sequence typing and pulsed-field gel electrophoresis. J Appl Microbiol 106(5):1722–1733. https://doi.org/10.1111/j.1365-2672.2008.04142.x
Oyarzabal OA, Backert S, Williams LL et al (2008) Molecular typing, serotyping and cytotoxicity testing of Campylobacter jejuni strains isolated from commercial broilers in Puerto Rico. J Appl Microbiol 105(3):800–812. https://doi.org/10.1111/j.1365-2672.2008.03809.x
Cocolin L, Stella S, Nappi R et al (2005) Analysis of PCR-based methods for characterization of Listeria monocytogenes strains isolated from different sources. Int J Food Microbiol 103(2):167–178. https://doi.org/10.1016/j.ijfoodmicro.2004.12.027
Yan H, Shi L, Alam MJ et al (2008) Usefulness of Sau-PCR for molecular epidemiology of nosocomial outbreaks due to Burkholderia cepacia which occurred in a local hospital in Guangzhou, China. Microbiol Immunol 52(5):283–286. https://doi.org/10.1111/j.1348-0421.2008.00035.x
Foschino R, Nucera D, Volponi G et al (2008) Comparison of Lactococcus garvieae strains isolated in northern Italy from dairy products and fishes through molecular typing. J App Microbiol 105(3):652–662. https://doi.org/10.1111/j.1365-2672.2008.03780.x
De Giglio O, D’Ambrosio M, Spagnuolo V et al (2023) Legionella anisa or Legionella bozemanii? Traditional and molecular techniques as support in the environmental surveillance of a hospital water network. Environ Monit Assess 195(4):496. https://doi.org/10.1007/s10661-023-11078-z
Lemos WJ, Bovo B, Nadai C et al (2016) Biocontrol ability and action mechanism of Starmerella bacillaris (synonym Candida zemplinina) isolated from wine musts against gray mold disease agent Botrytis cinerea on grape and their effects on alcoholic fermentation. Front Microbiol 7:1249. https://doi.org/10.3389/fmicb.2016.01249
Iacumin L, Comi G, Cantoni C et al (2006) Molecular and technological characterization of Staphylococcus xylosus isolated from naturally fermented Italian sausages by RAPD, Rep-PCR and Sau-PCR analysis. Meat Sci 74(2):281–288. https://doi.org/10.1016/j.meatsci.2006.03.020
Fischer SG, Lerman LS (1983) DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A 80(6):1579–1583. https://doi.org/10.1073/pnas.80.6.1579
Myers RM, Fischer SG, Lerman LS et al (1985) Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res 13(9):3131–3145. https://doi.org/10.1093/nar/13.9.3131
Pinto FR, Melo-Cristino J, Ramirez M (2008) A confidence interval for the Wallace coefficient of concordance and its application to microbial typing methods. PLoS ONE 3(11):e3696. https://doi.org/10.1371/journal.pone.0003696
Liu JH, Lan CL, Yao GF et al (2023) Comparison of pulsed-field gel electrophoresis and a novel amplified intergenic locus polymorphism method for molecular typing of Campylobacter jejuni. Arch Microbiol 205(1):49. https://doi.org/10.1007/s00203-022-03392-w
Acknowledgements
Great appreciation is given to all members of our laboratory for their enthusiastic participation in the research.
Funding
The authors would like to acknowledge financial support from the Special Projects in Key Areas of Colleges and Universities in Guangdong Province (2023ZDZX2031), and the Natural Science Foundation of Guangdong Province (No. 2018A030313163).
Author information
Authors and Affiliations
Contributions
G-FY: writing—original draft, formal analysis, visualization and conceptualization. Y-LH: formal analysis and writing—review and editing. J-HL, N-QK and C-YL: writing—review and editing. Y-WL and S-LB: project administration, supervision and funding acquisition. All authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical Approval
Not applicable.
Consent to Participation
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yao, GF., Hu, YL., Kong, NQ. et al. Rapid Genotyping of Campylobacter coli Strains from Poultry Meat by PFGE, Sau-PCR, and fla-DGGE. Curr Microbiol 80, 402 (2023). https://doi.org/10.1007/s00284-023-03517-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03517-4