Abstract
The cervicovaginal microbiota is an essential aspect of women’s reproductive and overall health. In this study, we aimed to evaluate the probiotic properties of a cervicovaginal isolate, obtained from a gynecologically healthy woman and assess its antagonistic effect against various microorganisms isolated from the vagina. Cytological examination was performed using Papanicolaou staining, and the isolated microorganism was identified via 16S Ribosomal RNA Gene Sequence Analysis. Probiotic characteristics were evaluated by determining the tolerance of the isolate to low pH, different NaCl concentrations, and bile salts. Bacterial adherence to stainless steel sheets, antibiotic susceptibility, and antimicrobial activity tests were also conducted and analyzed. Antimicrobial tests and antagonistic activities were assessed through disc diffusion assays. The cervicovaginal isolate was identified as B. velezensis ON116948 and was found to be tolerant to low pH, high NaCl and 0.3% bile salts. Additionally, it exhibited adherence. With the exception of amoxicillin/clavulanic acid (AMC) (30 μg) and oxacillin (OX) (1 μg), this isolate was susceptible to all the antibiotics tested. Candida species did not grow on B. velezensis spread media, while B. velezensis was able to grow on C. albicans, C. glabrata, C. tropicalis, S. condimenti and S. epidermidis spread media with growth zones of 13.7 ± 0.6, 13.3 ± 0.6, 14.2 ± 4.4, 10.5 ± 0.5 and 16.0 ± 1.0 (around discs), respectively. Our findings suggest that the cervicovaginal B. velezensis ON116948 isolate exhibits probiotic properties and antagonistic activity. These results provide important insights into the potential use of this isolate as a probiotic for the prevention of vaginal infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervicovaginal microbiota plays a substantial role in women’s health and reproduction [1]. The balance between the diverse microorganisms that comprise the human vaginal microflora/microecosystem is critical for homeostasis and it has a significant effect on women’s genital system wellbeing [2]. Types, compounds, and interactions between lower genital system microorganisms differ among individuals and the differences occur due to variations of internal (hormonal status, age, pregnancy, ethnicity and immune system) and external factors (smoking, antibiotic, douching, sexual activities and contraceptives) [3, 4]. Lactobacillus species are known for their capacity to colonize epithelial cell surfaces and create antimicrobial compounds such as bacteriocin and hydrogen peroxide to maintain healthy vaginal microflora [2, 5]. Recent studies have demonstrated that women were dominated by different Lactobacillus species and ratios of L. crispatus, L. gasseri, L. iners, L. jensenii, L. acidophilus. Changes in these species are closely related to gynecological problems such as sexually transmitted diseases, bacterial vaginosis (BV) and vulvovaginal candidiasis (VVC) [2, 6, 7] Lactobacilli abundance is known to process glycogen breakdown and produce lactic acid, leading to a low vaginal pH, which is hypothesized to benefit women by reducing diseases such as BV [8]. However, some women with low lactobacilli and a vaginal pH > 4.5 do not experience BV [9]. Similarly, women with a low vaginal pH also do not experience vulvovaginal candidiasis (VVC) by means of some cervicovaginal bacteria displaying anti-Candida activity [10]. Therefore, isolation and identification of new cervicovaginal inhabitants from healthy women would be a very innovative potential to prevent cervicovaginal diseases. Bacillus velezensis (B. velezensis) is an aerobic, gram-positive bacterium and recently has been reported to suppress the growth of microbial pathogens including not only bacteria and fungi but also nematodes [11]. It is also known to promote plant growth and trigger, induced systemic resistance in plants, allowing plants to defend themselves against recurring infections caused by some microorganisms [11]. Nowadays, the usefulness of B. velezensis as a probiotic in various fields is strongly been debated. By this means recent studies focus on the potential of B. velezensis as a probiotic not only in the aquaculture industry but also in husbandry [12].
In this study, we aimed to figure out whether or not a cervicovaginal B. velezensis isolate displays probiotic characteristics and exhibits antagonistic activity against some vaginal opportunistic pathogens belonging to Candida and Staphylococcus species.
Materials and Methods
Cytological Examination of a Cervicovaginal Sample
Cervicovaginal samples were collected from women who underwent examinations at the Gynecology Department of Hacettepe University Hospital (between June 2019 and August 2019). They were taken from the cervix (endocervix and ectocervix) using a speculum and a sterile cytobrush without any contact with the urethra/rectum and external parts of the vagina to avoid any possible contamination. Slides were fixed using 96% ethanol without air-drying and stained according to the Papanicolaou method (Pap) described by Donmez et al. [7]. Briefly, the slides were washed with decreasing concentrations of alcohol solutions (85–50%), followed by a wash with distilled water. After rehydration, the slides were incubated for 2 min with Harri’s Hematoxylin (Merck, Germany) and then rinsed with tap water. Subsequently, the slides were rinsed again with 1% hydrochloric acid (HCl)-alcohol and distilled water, respectively. Then incubated separately for an additional 3 min with Orange G and EA 65 dyes (Merck, Germany). All the slides were rinsed with 95% ethanol to remove any excessive dye. To cover-slip the slides, absolute ethanol, a 50% (v/v) alcohol-xylene solution and pure xylene were applied, followed by the use of Entellan (Merck, Germany) mounting medium. The cytological findings were then investigated by a light microscope and photographed using a camera-attachment (Leica DM 4000B). Following cytological examinations, cervicovaginal smear sample that includes long bacilli in chains was selected for further microbiological experiments.
Microbial Isolation
Cytobrush that has been smeared to a sterile slide, has also been immersed in 10 mL Brain–Heart Infusion (BHI) Broth media (Lab M Ltd, Lancashire, UK) (pH 7) to grow aerobic and facultative anaerobic microorganisms that exist in the cervicovaginal sample and was incubated at 37 °C for 72 h. Following incubation, microbial culture that was grown in BHI was inoculated into BHI agar, by streak plate method in order to obtain different microorganisms as single colonies. Obtained single colonies were Gram-stained and examined under the light microscope (Leica DM 4000B) separately. Microbial cells of a particular single colony, which exhibit the same microbiological morphology as the microbial cells on the cervicovaginal smear sample (long bacilli in chains) were selected to be isolated. The isolated bacterial strain was inoculated into BHI Broth media (pH 7) including 10% glycerol and stored at − 20 °C to be identified and used in further experiments.
Bacterial Identification and Evolutionary Analysis
Bacterial isolate was initially grown in BHI Agar media and colonies belonging to it was examined macroscopically. Afterwards, microscopic examination was applied following Gram staining. Identification was done using the 16S Ribosomal RNA Gene Sequence Analysis (BM Labosis, 2020; Ankara, Turkey). For the identification of bacterial species, the DNA isolation of the samples was performed using the EurX GeneMATRIX Bacterial & Yeast DNA Isolation Kit (Poland). Following DNA isolation, the quantity and purity of the obtained DNA were assessed using a Thermo Scientific Nanodrop 2000 spectrophotometer (USA). In the PCR study, the target gene regions for species identification were amplified using universal primers 27F-1492R and ITS1-ITS4. The primer sequences and PCR conditions used are provided below:
-
27F 5′ AGAGTTTGATCMTGGCTCAG 3′
-
1492R 5′ TACGGYTACCTTGTTACGACTT 3′
-
ITS1 5′ TCCGTAGGTGAACCTGCGG 3′
-
ITS4 5′ TCCTCCGCTTATTGATATGC 3′
The PCR amplification was performed using the kyratec thermocycler under the following conditions: 40 cycles of Denaturation: 95 °C for 45 s, Annealing: 57 °C for 45 s, Extension: 72 °C for 60 s and Final Extension: 72 °C for 5 min. The temperature was lowered to 4 °C and the PCR was completed. The amplification results obtained through PCR were subjected to electrophoresis on a 1.5% agarose gel prepared in 1 × TAE buffer. Electrophoresis was conducted at 100 V for 90 min and the gel was visualized under UV light after staining with ethidium bromide.
A single-step PCR process was performed to amplify the approximately 1470-base pair region. The PCR reaction was carried out using Solis Biodyne (Estonia) FIREPol® DNA Polymerase with Taq polymerase enzyme. After PCR, a single band was obtained on the agarose gel for one of the samples, indicating the successful completion of the PCR process.
During the purification step of PCR products, the obtained single-band samples were purified using the MAGBIO “HighPrep™ PCR Clean-up System” (AC-60005) purification kit following the kit’s procedures. For Sanger sequencing of the sample, the ABI 3730XL Sanger sequencing instrument (Applied Biosystems, Foster City, CA) and the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) were used at the Macrogen Netherlands laboratory. Reads obtained from the 27F and 1492R, and ITS1-ITS4 primers were assembled into contigs to generate a consensus sequence. The BioEdit software was employed for this process, using the CAP contig assembly algorithm.
The 16S Ribosomal RNA Gene Sequence that has been determined has been uploaded to a GenBank and an accession number has been provided. The evolutionary history was inferred using the Maximum Likelihood Method and Tamura-Nei Model [13]. The phylogenetic tree with the highest log likelihood (4478.00) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the BioNJ method to a matrix of pairwise distances estimated using the Tamura-Nei model. This analysis involved 15 nucleotide sequences. There were a total of 1562 positions in the final dataset. Evolutionary analyses were conducted in MEGA X [14].
Microbial Growth Conditions and Harvesting
Bacterial isolate (B. velezensis) was first grown on BHI agar (Lab M Ltd, Lancashire, UK) for 24 h at 37 °C. For pre-culture preparations, single colonies were inoculated into 10 mL BHI broth and incubated overnight at 37 °C. 1.5 mL of this preculture was used to inoculate a second culture of 30 mL BHI broth, which was subsequently grown for 24 h at 37 °C. Cells were harvested by centrifugation at 3220×g for 10 min at 5 °C (Eppendorf 5810R, with an Eppendorf Swing-bucket rotor A-4-62, Hamburg, Germany); washed three times with 10 mM potassium phosphate buffer (pH 7) and was adjusted to 2.0 McFarland optical density standard.
In Vitro Characterization of Probiotic Properties
Tolerance to low pH and high NaCl concentrations, tolerance against bile salts; bacterial adherence to stainless steel plates; antibiotic susceptibility and antimicrobial activity [15, 16] were determined for in vitro evaluation of probiotic properties of cervicovaginal B. velezensis isolate.
Tolerance to Low pH
Since the gastric mucus layer is defined as pH 3, probiotic microorganisms are expected to be tolerant to pH around pH 3 [17]. In vitro assay to determine the resistance of B. velezensis isolate to the acidic condition of the stomach was applied. B. velezensis isolate was grown, harvested, and adjusted to a 2.0 McFarland optical density standard as described previously. . pH tolerance test of B. velezensis isolate was determined by inoculating 0.5 mL of bacterial suspension (adjusted to 2.0 McFarland optical density standard) in to 10 mL of potassium phosphate buffer with pH levels of 2.0, 2.5, 3.0, 3.5 and 7.0 separately (Since tolerance of a potential probiotic microorganism to acidic conditions was tested, pH 7 was used as a control). Bacterial suspensions inside potassium phosphate buffers with different pH levels were incubated at room temperature. Food remains in the stomach for at least 3 h [18]. Therefore, 4 h of incubation was conducted for the pH tolerance test. After 4 h, 100 μL of bacterial suspension was spread on the BHI agar plates and agar plates were incubated at 37 °C for 24 h. After 24 h of incubation, total colonies belong to each pH level were counted to determine the viability and growth pattern.
Tolerance to NaCl
Bacillus velezensis isolate was grown, harvested and adjusted to a 2.0 McFarland optical density standard as described previously . Probiotic strains are expected to withstand high salt concentrations in the human gut. By this means, B. velezensis strain ON116948 was exposed to seven different concentrations of NaCl. In this respect, BHI agar plates were prepared in seven different concentrations of NaCl (0, 0.5, 1, 5, 7.5, 10 and 12.5%). NaCl tolerance test of B. velezensis isolate was determined by spreading 100 μL of bacterial suspension (adjusted to 2.0 McFarland optical density standard) on to the BHI agar plates with seven different NaCl concentrations and incubating plates at 37 °C for 24 h. After incubation, influence of NaCl concentrations on the bacterial growth was evaluated macroscopically.
Tolerance to Bile Salts
The physiological concentration of human bile in the duodenum is defined as around 0.3% [16]. Therefore, tolerance to 0.3% (w/v) bile salt was tested in this part. Bacillus velezensis isolate was grown, harvested, and adjusted to a 2.0 McFarland optical density standard as described previously Bile salts tolerance test of B. velezensis isolate was determined by inoculating 0.5 mL of bacterial suspension in to the 10 mL BHI broth supplemented with 0.3% (w/v) bile salt (sodium deoxycholate) and providing incubation at 37 °C for 24 h. After incubation, influence of bile salts on the bacterial growth was evaluated visually. BHI broth without 0.3% (w/v) bile salts was evaluated as a positive control.
Bacterial Adhesion to Stainless Steel Sheets
Bacillus velezensis isolate was grown, harvested and adjusted to a 2.0 McFarland optical density standard as described previously . Bacterial adhesion to stainless steel sheets was determined by inoculating 1 mL of bacterial suspension in to the sterile petri dishes that include sterile stainless steel sheets and 20 mL BHI broth. After incubation at 37 °C for 24 h, stainless steel sheets were rinsed 3 times with 10 mM potassium phosphate buffer (pH 7) and visually evaluated in terms of bacterial adhesion.
Antibiotic Susceptibility
Antimicrobial susceptibilities of the isolated and identified bacterial strain B. velezensis against different antibiotics including vancomycin (30 μg), gentamicin (10 μg), streptomycin (10 μg), amoxicillin/clavulanic acid (30 μg), penicillin G (10 Units), oxacillin (1 μg), amoxicillin (25 µg), piperacillin (100 μg), norfloxacin (10 μg), nalidixic acid (30 μg) were tested by Kirby-Bauer Disc-Diffusion method according to the recommendation of Clinical and Laboratory Standards Institute (CLSI M100-ED30:2020). Briefly, B. velezensis isolate was grown, harvested and adjusted to a 2.0 McFarland optical density standard as described previously. Standard antibiotic discs were placed onto the surface of BHI plates, previously spread with 100 μL of bacterial suspension and agar plates were incubated at 37 °C for 24 h. After incubation, antibiotic susceptibilities of B. velezensis isolate were determined according to the ZOI values given as below [19]:
-
ZOI ≥ 10.0 mm: Sensitive
-
ZOI < 10.0 mm: Resistant
Antimicrobial Activity
Clinical isolates of Candida albicans V6, Candida tropicalis V89, Proteus mirabilis U15, Staphylococcus epidermidis W17 which were previously isolated and identified [20,21,22] and Candida glabrata V23, Escherichia coli V7 and Staphylococcus condimenti V36 strains which were isolated and identified using the 16S Ribosomal RNA Gene Sequence Analysis (BM Labosis, 2020; Ankara, Turkey) were selected to be tested for this part of the study.
All microorganisms were grown, harvested and adjusted to a 2.0 McFarland optical density standard as described previously. 20 μL of B. velezensis were deposited onto sterile antibiotic assay discs (Whatman, 6MM PK 1000). These discs and 10 μL aliquot of B. velezensis were placed onto the surface of BHI plates, previously spread with 100 μL C. albicans V6, C. glabrata V23, C. tropicalis V89, E. coli V7, P. mirabilis U15, S. condimenti V36 and S. epidermidis W17 clinical strains. Apart from this, 20 μL of C. albicans V6, C. glabrata V23, C. tropicalis V89, E. coli V7, P. mirabilis U15, S. condimenti V36 and S. epidermidis W17 clinical strains were also deposited onto sterile antibiotic assay discs. These discs and 10 μL aliquots of all these seven clinical strains were placed onto the surface of BHI plates, previously spread with 100 μL of B. velezensis isolate. 20 μL of 10 mM potassium phosphate buffer (pH 7) deposited onto sterile antibiotic assay discs and 10 μL aliquots of 10 mM potassium phosphate buffer (pH 7) was used as negative controls. All the BHI plates were incubated for 24 h at 37 °C and the diameters of zone of growths on BHI agar plates were measured using a ruler.
Statistical Analysis
The statistical analysis in this study was performed using Statistical Package for the Social Sciences (SPSS) version 23 (IBM SPSS Statistics Inc., Chicago, IL, USA). The data were described using mean ± standard deviation. To test whether the data follows a normal distribution, the Shapiro–Wilk test was conducted. Since the data was found to deviate from normal distribution, the Mann Whitney U test was used for comparing two groups, while the Kruskal Wallis test was employed for comparisons involving more than two groups. A significance level of P < 0.05 was considered as statistically significant.
Sequencing Data
The bacteria belonging to the only single colony was identified following 16S Ribosomal RNA Gene Sequence Analysis with a GenBank accession number of ON116948.
Results
Cytological Examination of Cervicovaginal Smears
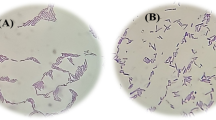
In this study, long bacilli took our attention during the microscopic examination of one of the cervicovaginal smears belonging to a woman (who attended the Gynecology Clinic for the routine Pap-screening program) without any gynecological complaint. Long bacilli colonization on and around the epithelial cells (Fig. 1a, b) was observed. During the microscopic examination, the slide was also examined under a light microscope to identify any potential-specific infections. Bacterial vaginosis (BV) is diagnosed by the presence of clue cells (epithelial cells with adherent bacteria), a decrease in neutrophils and Lactobacilli. The presence of filamentous and yeast forms of fungal microorganisms indicates a fungal infection. Additionally, the detection of motile Trichomonas vaginalis protozoa confirms Trichomoniasis. The observation of characteristic clusters or filaments of Actinomyces species demonstrates Actinomycosis. In the examined sample, there were no indications of specific infections such as BV, fungal infection, Trichomoniasis, or Actinomycosis.
Images of the a Pap staining of the selected cervicovaginal sample smeared slide with a bright field light microscope (400×magnification). This representative image reveals the absence of inflammation and specific infectious agents, as well as the absence of cellular abnormalities. b Pap staining of the selected cervicovaginal sample smeared slide with a bright field light microscope (1000×magnification). Bacterial cells as bacilli indicated by red and black arrows show bacilli colonization on (red arrow) and around (black arrow) cervicovaginal epithelial cells. c Single colonies belong to the bacterial isolate on BHI Agar medium. d Gram-stained bacterial isolate taken with a bright field light microscope 100×magnification (LEICA DM500). Red arrow shows single colony morphology of the isolated bacteria (B. velezensis). The violet color indicates that isolated bacterial cells (B. velezensis) are gram-positive (Color figure online)
Moreover, there was no observed increase in inflammatory cells (neutrophils and macrophages) or any cellular abnormalities associated with inflammation. During the light microscopic examination of the Pap smear, inflammatory cells, including neutrophils and macrophages, can be identified and characterized based on their cytological features. Neutrophils display segmented nuclei divided into 2 to 5 lobes connected by thin chromatin strands, while macrophages can be recognized by their kidney-shaped nuclei and cytoplasmic vacuoles containing digested material. Therefore, the cervicovaginal smear sample without any specific infection or inflammation was selected for further microbiological experiments.
Microbial Isolation, Identification and Evolutionary Analysis
When the microbial growth after the application of the streak plate method was examined, only one type of single colony morphology was observed on the BHI agar (pH 7) plate as seen in Fig. 1c. The microbial cells belonging to that single colony were determined as Gram-positive bacilli (in chains) following Gram staining (Fig. 1d). It was seen that the microscopic morphology of the bacterial cells belonging to only a single colony was the same as the microscopic morphology of the bacterial cells observed after the cytological examination of the selected cervicovaginal sample. Therefore, bacteria belonging to the only single colony were isolated and identified as B. velezensis with 99% identity ratio, following 16S Ribosomal RNA Gene Sequence Analysis with a GenBank accession number of ON116948. The information about the sequence similarity is presented in Fig. 2 with a phylogenetic tree.
In Vitro Characterization of Probiotic Properties
Tolerance to Low pH
Growth of B. velezensis strain ON116948 on BHI agar plates after being incubated inside potassium phosphate buffer adjusted to 2.0, 2.5, 3.0, 3.5 and 7.0 pH levels were evaluated (Fig. 3a). As seen in Fig. 3b, although there became a decrease in the bacterial count in parallel with the decrease in pH level, B. velezensis ON116948 demonstrated growth at each pH level tested in this study (Fig. 3a, b). Thus, it exhibited the potential to survive even in acidic conditions.
a and b Growth of B. velezensis strain ON116948 on plates after being incubated inside potassium phosphate buffer adjusted to different pH levels (For the pH-dependent growth curve analysis of B. velezensis strain ON116948, data from three independent experiments were collected. The results are expressed as mean ± standard deviation. There is a statistically significant difference among the pH values, Kruskal Wallis H test, *P = 0.011). c Growth of B. velezensis strain ON116948 on plates that include different NaCl concentrations. d Growth of B. velezensis strain ON116948 on stainless steel sheets
Tolerance to NaCl
Regarding the tolerance to NaCl, B. velezensis strain ON116948 was able to grow even in the highest concentration of NaCl (12.5%). The bacterial count was decreased in parallel with the increase in NaCl concentration (Fig. 3c).
Tolerance to Bile Salts
Bacillus velezensis strain ON116948 was determined as tolerant to 0.3% bile salt, by exhibiting growth after 24 h of incubation in 0.3% bile salt-supplemented BHI broth media.
Bacterial Adhesion to Stainless Steel Sheets
Bacillus velezensis strain ON116948 was able to attach well to the stainless steel. The adhesion of B. velezensis strain ON116948 is shown in Fig. 3d.
Antibiotic Susceptibility
In Table 1, the antibiotic susceptibility of B. velezensis in terms of the diameter of the zone of inhibition (ZOI) is given. According to our findings, there was not any ZOI after the application of amoxicillin/clavulanic acid (30 μg) and oxacillin (1 μg) antibiotics. However, except for amoxicillin/clavulanic acid (30 μg) and oxacillin (1 μg); B. velezensis strain ON116948 was sensitive to all antibiotics tested.
Antimicrobial Activity
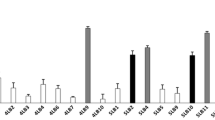
Table 2a presents the growth of tested microorganisms in millimeters (mm) (mean values ± SD, n = 3) on B. velezensis spread media and shows the antagonistic effect of B. velezensis against different microbial species. According to our results, a statistically significant difference was determined among the growth of different tested microorganisms both around discs (P = 0.003) and 10 μL aliquots (P = 0.003) on B. velezensis spread media. This finding indicates that the growth on B. velezensis spread media varied depending on the tested clinical microbial strain. According to our results, there was not any growth zone formed by C. albicans V6, C. glabrata V23, and C. tropicalis V89 strains (both around discs and aliquots) on B. velezensis spread media. However, E. coli V7 and P. mirabilis U15 clinical strains formed growth zones both around discs and 10 μL microbial aliquots (Table 2a, Fig. 4a).
Images of growth of a different clinical isolates on B. velezensis spread media. b B. velezensis on different clinical isolates spread media. Red arrows indicate the zone of growth around discs; black arrows indicate the zone of growth after the addition of 10 μL microbial suspension (Color figure online)
When E. coli V7 and P. mirabilis U15 clinical strains were compared in terms of their growth zone diameters around discs on B. velezensis spread media, growth zone of P. mirabilis U15 was significantly more than the growth zone of E. coli V7 (P = 0.034). Apart from this, no growth zone was observed around the discs where S. condimenti V36 and S. epidermidis W17 strains were deposited. However, a slight growth was observed when 10 μL of S. condimenti V36 and S. epidermidis W17 strains were inoculated on B. velezensis spread media. The difference in the growths between S. condimenti V36 and S. epidermidis W17 when inoculated as 10 μL aliquots was not statistically significant (P = 0.121). (Table 2a, Fig. 4a).
Table 2b presents the growth of B. velezensis strain ON116948 when deposited on discs and when inoculated as 10 μL aliquots on different microbial species spread media. Our findings revealed a statistically significant difference among the growth of all the tested strains both around discs (P = 0.007) and around 10 μL aliquots (P = 0.006), indicating that the growth of B. velezensis varied significantly depending on the specific microorganisms spread to the media. This highlights that B. velezensis has diverse effects on the growth of different microbial species. According to our results, B. velezensis exhibited no growth (both around discs and 10 μL aliquots) on E. coli V7 and P. mirabilis U15 spread media. However, B. velezensis demonstrated growth zones (both around discs and 10 μL aliquots) on media spread with Candida species (C. albicans V6, C. glabrata V23, C. tropicalis V89) and Staphylococcus species (S. condimenti V36 and S. epidermidis W17) (Table 2b, Fig. 4b). While there was a limited statistically significant difference in the growth zones of B. velezensis around discs on S. epidermidis and S. condimenti spread media (P = 0.05), a significant difference was observed in the growth zones around 10 μL aliquots on S. epidermidis and S. condimenti spread media (P = 0.037). Apart from these, no significant difference was observed in the growth zones of B. velezensis both around discs and 10 μL aliquots on C. albicans V6, C. glabrata V23 and C. tropicalis V89 spread media (P = 0.649).
To sum up, B. velezensis strain ON116948 exhibited antagonistic effect against clinical Candida albicans V6, Candida glabrata V23, Candida tropicalis V89, Staphylococcus condimenti V36 and Stapylococcus epidermidis W17 strains. However, it was not effective against two-gram-negative microbial species of E. coli V7 and P. mirabilis U15.
Discussion
The human vaginal micro-ecosystem is composed of several aerobic and anaerobic microorganisms that coexist in a dynamic balance [23]. This micro-ecosystem is the first line of defense against infections by “competitive exclusion” and “direct killing” [24]. Microbial species that exist in this micro-ecosystem are closely related to gynecological problems such as sexually transmitted diseases, bacterial vaginosis (BV) and vulvovaginal candidiasis (VVC) [3, 6, 7, 25, 26]. Previous studies show that various Lactobacillus species are found in the cervix and vagina as Gram-positive bacilli [27] and microorganisms belonging to the Lactobacillus genus are regarded as part of the normal human gastrointestinal and vaginal flora [28].
Additionally, Lactobacilli abundance is known to benefit women by reducing diseases such as BV and VVC [8]. However, in some cases, women without Lactobacilli also do not experience vaginal infections as a result of some other cervicovaginal microorganisms displaying antagonistic activities against various vaginal opportunistic pathogens [10]. Therefore, isolation and identification of new cervicovaginal inhabitants from gynecologically healthy women would be a very innovative solution for the prevention of diverse cervicovaginal infections.
In the first part of this study, long bacilli in chains both in the cervicovaginal smear and in the cervicovaginal culture (grown in BHI Broth, pH 7) of a cervicovaginal sample belonging to a woman who did not have any gynecological complaint was observed (Fig. 1a and b). Following this observation, we isolated and identified that bacteria, to understand whether or not it is a known cervicovaginal inhabitant. The microbial isolate that exhibits long bacilli in chains (Fig. 1a, b and d) was identified as B. velezensis by 16S Ribosomal RNA Gene Sequence Analysis BM Labosis, 2020; Ankara, Turkey) with a GenBank accession number of ON116948 (Fig. 2). To the best of our knowledge, B. velezensis has not been reported to exist in cervicovaginal microbiota before.
In recent years, the isolation, identification, and testing of numerous of B. velezensis strains in various applications within the fields of agriculture, biotechnology and biocontrol have increased. According to these studies, it is seen that the isolated and identified diverse B. velezensis strains exhibit differences in terms of their secondary metabolites, antimicrobial activities, genetic compositions and plant growth-promoting traits [29,30,31,32,33,34].
Bacillus velezensis is a gram-positive bacterium that can be easily isolated and cultured by means of being distributed widely in nature [12]. Due to its wide distribution in nature and enrichment in its metabolites and inhibitory compounds such as surfactin and bacteriocins, B. velezensis is also used as a probiotic both in the aquaculture [35] and in animal nutrition [12]. Moreover, this bacterial species is also reported to inhibit adherence, replication, virulence of pathogens and regulate the immune system of animals [36]. From 2017 to 2020 years, various B. velezensis strains have been reported to be isolated from diverse animal sources such as chicken cecal content, domestic yak, manure of piglets, intestinal tract of healthy grouper, gastrointestinal tract of healthy grass carp, catfish intestine and intestines of free-range Tibetan yaks [12]. Surprisingly, B. velezensis has also been reported to be isolated from human gut microbiota in July 2021 [37].
Probiotics are expected to display tolerance to various adverse conditions such as low pH, high NaCl and 0.3% bile salts [12, 38]. Therefore, in the next part of this study, in vitro characterization of probiotic properties of B. velezensis strain ON116948 was assessed. The gastric mucus layer is defined as pH 3 and probiotic microorganisms are expected to be tolerant to pH 3 [17]. This study reports not only tolerance to pH 3, but also to pH 2 and pH 2.5 (Fig. 3a and b). Apart from this, NaCl is reported to inhibit the growth of certain types of microorganisms in the human gut [39].
Therefore, probiotic strains are expected to withstand high salt concentrations in the human gut. By this means, B. velezensis strain ON116948 was exposed to seven different concentrations of NaCl in this study and demonstrated tolerance not only to 10.0%, but also to 12.5% NaCl concentration (Fig. 3c). The physiological concentration of human bile in the duodenum is defined as around 0.3% [16]. As a result, this concentration is usually selected as an essential bile dose for the selection of probiotic strains [40]. Although the high concentration of bile salts causes disruption of cellular homeostasis, bacterial content leakage, and sometimes cell death [41]. Bacillus velezensis strain ON116948 in the present study showed growth in BHI supplemented with 0.3% bile salts which is in line with the other studies that report probiotic microorganisms having ability to grow in the media supplemented with wide bile concentration (0.05–0.3%) [16]. The outcomes of the tests mentioned above showed that B. velezensis had an abiotic stress tolerance.
Another important characteristics of probiotic bacteria is reported as adhesion to the intestinal flora [15]. According to Darmastuti et al. [42] probiotics with excessive adhesion capacity have high capability to bind host epithelial cell surface especially intestinal epithelial cells. In parallel with this, adhesion-related genes were identified in the genome of B. velezensis R-71003, that play roles on adherence of the strain to the gut [19]. In line with this criterion, B. velezensis strain ON116948 also possessed an in vitro adherence property to the stainless steel surface in this study (Fig. 3d). This finding suggests the colonization potential of B. velezensis not only on GI but also on lower genital tract flora.
Antibiotic resistance poses a threat to both animal and human health [43]. Therefore, absence of antibiotic resistance is known as one of the most important characteristics of a probiotic [19]. Antibiotic resistance of B. velezensis strain ON116948 towards 10 different antibiotics (Vancomycin (30 μg), gentamicin (10 μg), streptomycin (10 μg), amoxicillin/clavulanic acid (30 μg), penicillin G (10 Units), oxacillin (1 μg), amoxicillin (25 µg), piperacillin (100 μg), norfloxacin (10 μg) and nalidixic acid (30 μg)) which were mostly prescribed for bacterial vaginosis or urogenital infections were tested in the next part of this study [44, 45]. In line with characteristics of a probiotic, B. velezensis strain ON116948 was determined as sensitive to all antibiotics tested except amoxicillin/clavulanic acid (30 μg) and oxacillin (1 μg) (Table 1). Several clinical trials support the use of probiotics in the management of acute gastroenteritis and antibiotic-associated diarrhea [46] and it is known that extensive use of some antimicrobials influences the resistance of the gut microbiota [47]. In these situations, probiotics are reported to be directly exposed to antibiotics, sometimes even at a very high concentration. This makes the adaptation of probiotics to antibiotics possible [46]. As an example, one of the recent studies investigates the ecological role of AMC-resistant bifidobacterial strains [47]. Another study emphasizes the mechanism of adaptive gentamycin resistance in a probiotic Lactobacillus casei strain (one of the probiotic species which has been used both as starter cultures and as food additives for improving food texture properties) [46]. Additionally, some of the L. casei strains were determined as AMC-resistant in one of the recent studies [48]. Since the combination of amoxicillin/clavulanic acid is one of the most frequently prescribed antibiotics in the world, it is possible that many probiotics residing in the intestinal microbiota may have developed resistance to it. Since cervicovaginal pathogens such as G. vaginalis strains isolated from vaginal samples diagnosed with BV are reported as AMC-sensitive [49], AMC resistance of B. velezensis strain ON116948 could be a choice to be used to restore the normal cervicovaginal microbiota during the vaginal infections or cervicovaginal cancer [44]. Probiotic bacteria are also known to synthesize some antimicrobial substances against various pathogenic microorganisms [38, 44]. Therefore, probiotics are regarded as ideal alternatives to diverse antibiotics due to their benefits of causing antagonistic and suppressive effect against various pathogens [16]. By this means, in the last part of this study, it is aimed to determine whether or not B. velezensis strain ON116948 demonstrates antagonistic activity against Gram-positive, Gram negative and yeast pathogens (Table 2a and b, Fig. 4a and b). According to our findings, E. coli V7 and P. mirabilis U15 caused an antagonistic activity against B. velezensis isolate. The observed ineffectiveness of B. velezensis against E. coli V7 and P. mirabilis U15, raises the need for further research on the antagonistic activity of B. velezensis strain ON116948 against some other gram-negative bacteria. On the other hand, B. velezensis strain ON116948 suppressed the growth of C. albicans V6, C. glabrata V23, C. tropicalis V89, S. condimenti V36 and S. epidermidis W17 strains. While inhibitory effect of B. velezensis against the microbial growth of some fungal pathogens such as Aspergillus flavus, Cryptococcus neoformans and Fusarium solani has been reported [50], some opportunistic pathogens such as C. albicans, C. glabrata, C. tropicalis, S. condimenti and S. epidermidis have not been reported to be inhibited by B. velezensis until now. Therefore, our findings not only represent a possible probiotic candidate, but also demonstrates a beneficial cervicovaginal colonizer as a result of being able to cause an antagonistic activity against some cervicovaginal opportunistic pathogens belong to Candida and Staphylococcus genus for the first time. However, its safety and efficacy in clinical trials should also be tested in detail before it can be recommended for clinical use.
Conclusion
Based on the findings of this study, it can be concluded that B. velezensis strain ON116948, which was isolated from a cervicovaginal sample of a gynecologically healthy woman, could be a probiotic candidate for the prevention and treatment of vaginal infections. This microorganism demonstrated antagonistic activity against various opportunistic pathogens, including Candida and Staphylococcus species, and its ability to resist AMC might make it a favorable alternative to antibiotics used to treat AMC-sensitive bacterial vaginosis (BV)-associated microorganisms. These results suggest that B. velezensis strain ON116948 has the potential to be used as a novel probiotic in the development of therapeutic strategies for vaginal infections. However, further research is needed to confirm its safety and efficacy in clinical trials before it can be recommended for clinical use.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Anahtar MN, Gootenberg DB, Mitchell CM, Kwon DS (2018) Cervicovaginal microbiota and reproductive health: the virtue of simplicity. Cell Host Microbe 23(2):159–168. https://doi.org/10.1016/j.chom.2018.01.013
Gaspar C, Donders GG, Palmeira-de-Oliveira R, Queiroz JA, Tomaz C, Martinez-de-Oliveira J, Palmeira-de-Oliveira A (2018) Bacteriocin production of the probiotic Lactobacillus acidophilus KS400. AMB Expr. https://doi.org/10.1186/s13568-018-0679-z
Barrientos-Durán A, Fuentes-López A, de Salazar A, Plaza-Díaz J, García F (2020) Reviewing the composition of vaginal microbiota: inclusion of nutrition and probiotic factors in the maintenance of eubiosis. Nutrients 12(2):1–30. https://doi.org/10.3390/nu12020419
Chen X, Huang H, Zhang S, Zhang Y, Jiang J, Qiu Y, Liu J, Wang A (2021) Bacillus velezensis wz-37, a new broad-spectrum biocontrol strain, promotes the growth of tomato seedlings. Agriculture 11(7):1–14. https://doi.org/10.3390/agriculture11070581
Kumherová M, Veselá K, Kosová M, Mašata J, Horáčková Š, Šmidrkal J (2021) Novel potential probiotic Lactobacilli for prevention and treatment of vulvovaginal infections. Probiotics Antimicrob Prot 13(1):163–172. https://doi.org/10.1007/s12602-020-09675-2
Donmez HG, Cagan M, Fadiloglu E, Unal C, Onder SC, Beksac MS (2020) Is bacterial vaginosis associated with autoimmune antibody positivity? Cytopathology 4:298–302. https://doi.org/10.1111/cyt.12846
Donmez HG, Sahal G, Akgor U, Cagan M, Ozgul N, Beksac MS (2020) The relationship between the presence of HPV infection and biofilm formation in cervicovaginal smears. Infection 48(5):735–740. https://doi.org/10.1007/s15010-020-01478-5
Miller EA, Beasley DAE, Dunn RR, Archie EA (2016) Lactobacilli dominance and vaginal pH: why is the human vaginal microbiome unique? Front Microbiol 7:1–13. https://doi.org/10.3389/fmicb.2016.01936
Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ (2011) Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA 108(1):4680–4687. https://doi.org/10.1073/pnas.1002611107
Jang SJ, Lee K, Kwon B, You HJ, Ko GP (2019) Vaginal lactobacilli inhibit growth and hyphae formation of Candida albicans. Sci Rep 9(1):1–9. https://doi.org/10.1038/s41598-019-44579-4
Rabbee MF, Sarafat Ali M, Choi J, Hwang BS, Jeong SC, Baek K (2019) Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules 24(6):1–13. https://doi.org/10.3390/molecules24061046
Khalid F, Khalid A, Fu Y, Hu Q, Zheng Y, Khan S, Wang Z (2021) Potential of Bacillus velezensis as a probiotic in animal feed: a review. J Microbiol 59(7):627–633. https://doi.org/10.1007/s12275-021-1161-1
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10(3):512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Mulaw G, Sisay Tessema T, Muleta D, Tesfaye A (2019) In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int J Microbiol 2019:7179514. https://doi.org/10.1155/2019/7179514
Nath S, Sikidar J, Roy M, Deb B (2020) In vitro screening of probiotic properties of Lactobacillus plantarum isolated from fermented milk product. Food Qual Saf 4(4):213–223. https://doi.org/10.1093/fqsafe/fyaa026
Sanhueza E, Paredes-Osses E, González CL, García A (2015) Effect of PH in the survival of Lactobacillus salivarius strain UCO_979C wild type and the Ph acid acclimated variant. Electron J Biotechnol 18(5):343–346. https://doi.org/10.1016/j.ejbt.2015.06.005
Thakkar P, Modi HA, Prajapati JB (2015) Isolation, characterization and safety assessment of lactic acid bacterial ısolates from fermented food products. Int J Curr Microbiol Appl Sci 4(4):713–725
Kang M, Su X, Yun L, Shen Y, Feng J, Yang G, Meng X, Zhang J, Chang X (2022) Evaluation of probiotic characteristics and whole genome analysis of Bacillus velezensis R-71003 isolated from the intestine of common carp (Cyprinus carpio L.) for its use as a probiotic in aquaculture. Aquacult Rep. https://doi.org/10.1016/j.aqrep.2022.101254
Sahal G, Bilkay IS (2015) Multidrug resistance by biofilm-forming clinical strains of Proteus mirabilis. Asian Biomed 9(4):535–554. https://doi.org/10.5372/1905-7415.0904.424
Sahal G, Bilkay IS (2018) Distribution of clinical isolates of Candida spp. and antifungal susceptibility of high biofilm-forming Candida isolates. Rev Soc Bras Med Trop 51(5):644–650. https://doi.org/10.1590/0037-8682-0136-2018
Sahal G, Bilkay IS (2014) Multi drug resistance in strong biofilm forming clinical isolates of Staphylococcus epidermidis. Braz J Microbiol 45(2):539–544. https://doi.org/10.1590/s1517-83822014005000042
Handalishy II, Behery MA, Elkhouly M (2014) Comparative study between probiotic vaginal tampons and oral metronidazole in treatment of bacterial vaginosis. Al-Azhar Assiut Med j 12:185–203
Lewis FMT, Bernstein KT, Aral SO (2017) Vaginal microbiome and its relationship to behavior, sexual health, and sexually transmitted diseases. Obstet Gynecol 129(4):643–654. https://doi.org/10.1097/AOG.0000000000001932
Freire AD, Custódio AI, Filho JQ, Freitas JC, Gonçalves AK, Cobucci RN (2020) The association between abnormal vaginal flora and cytological evidence of HPV with prematurity in high-risk pregnant women. Gynecol Obstet Reprod Med 26(3):173–178
Tortelli BA, Lewis WG, Allsworth JE, Member-Meneh N, Foster LR, Reno HE, Peipert JF, Fay JC, Lewis AL (2020) Associations between the vaginal microbiome and Candida colonization in women of reproductive age. Am J Obstet Gynecol 222(5):471.e1-471.e9. https://doi.org/10.1016/j.ajog.2019.10.008
Curty G, de Carvalho PS, Soares MA (2020) The role of the cervicovaginal microbiome on the genesis and as a biomarker of premalignant cervical intraepithelial neoplasia and invasive cervical cancer. Int J Mol Sci 21(1):222. https://doi.org/10.3390/ijms21010222
Goldstein EJC, Tyrrell KL, Citron DM (2015) Lactobacillus species: taxonomic complexity and controversial susceptibilities. Clin Infect Dis 60(2):S98–S107. https://doi.org/10.1093/cid/civ072
Fan B, Wang C, Song X, Ding X, Wu L, Wu H, Gao X, Borriss R (2018) Bacillus velezensis FZB42 in 2018 the gram-positive model strain for plant growth promotion and biocontrol. Front Microbiol 9:2491. https://doi.org/10.3389/fmicb.2018.02491
Jiang CH, Liao MJ, Wang HK, Zheng MZ, Xu JJ, Guo JH (2018) Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol Control 12:147–157. https://doi.org/10.1016/j.biocontrol.2018.07.017
Yi Y, Zhang Z, Zhao F, Liu H, Yu L, Zha J, Wang G (2018) Probiotic potential of Bacillus velezensis JW: antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol 78:322–330. https://doi.org/10.1016/j.fsi.2018.04.055
Pandin C, Darsonval M, Mayeur C, Le Coq D, Aymerich S, Briandet R (2019) Biofilm formation and synthesis of Antimicrobial compounds by the biocontrol agent Bacillus velezensis QST713 in an Agaricus bisporus compost micromodel. Appl Environ Microbiol 85(12):e00327-e419. https://doi.org/10.1128/AEM.00327-19
Rabbee MF, Hwang BS, Baek KH (2023) Bacillus velezensis: a beneficial biocontrol agent or facultative phytopathogen for sustainable agriculture. Agronomy 13(3):840. https://doi.org/10.3390/agronomy13030840
Yuan H, Shi B, Wang L, Huang T, Zhou Z, Hou H, Tu H (2022) Isolation and characterization of Bacillus velezensis strain P2–1 for biocontrol of apple postharvest decay caused by Botryosphaeria dothidea. Front Microbiol 12:808938. https://doi.org/10.3389/fmicb.2021.808938
Thurlow CM, Williams MA, Carrias A, Ran C, Newman M, Tweedie J, Allison E, Jescovitch LN, Wilson AE, Terhune JS, Liles MR (2019) Bacillus velezensis AP193 exerts probiotic effects in channel catfish (Ictalurus punctatus) and reduces aquaculture pond eutrophication. Aquaculture 503:347–356. https://doi.org/10.1016/j.aquaculture.2018.11.051
Fooks LJ, Gibson GR (2002) In vitro investigations of the effect of probiotics and prebiotics on selected human intestinal pathogens. FEMS Microbiol Ecol 9(1):67–75. https://doi.org/10.1016/S0168-6496(01)00197-0
Torres-Sánchez A, Pardo-Cacho J, López-Moreno A, Ruiz-Moreno Á, Cerk K, Aguilera M (2021) Antimicrobial effects of potential probiotics of Bacillus spp. isolated from human microbiota: in vitro and in silico methods. Microorganisms 9(8):1615. https://doi.org/10.3390/microorganisms9081615
Borah T, Gogoi B, Khataniar A, Gogoi M, Das A, Borah D (2019) Probiotic characterization of indigenous Bacillus velezensis strain DU14 isolated from Apong, a traditionally fermented rice beer of Assam. Biocatal Agric Biotechnol 18:101008. https://doi.org/10.1016/j.bcab.2019.01.046
Prabhurajeshwar C, Chandrakanth RK (2017) Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: an in vitro validation for the production of inhibitory substances. Biomed J 40(5):270–283. https://doi.org/10.1016/j.bj.2017.06.008
Dunne C, O’Mahony L, Murphy L, Thornton G, Morrissey D, O’Halloran S, Feeney M, Flynn S, Fitzgerald G, Daly C, Kiely B, O’Sullivan GC, Shanahan F, Collins JK (2001) In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am J Clin Nutr 73(2):386S-392S. https://doi.org/10.1093/ajcn/73.2.386s
Mandal S, Puniya AK, Singh K (2006) Effect of alginate concentrations on survival of microencapsulated Lactobacillus casei NCDC-298. Int Dairy J 16(10):1190–1195. https://doi.org/10.1016/j.idairyj.2005.10.005
Darmastuti A, Hasan PN, Wikandari R, Utami T, Rahayu ES, Suroto DA (2021) Adhesion properties of Lactobacillus plantarum Dad-13 and Lactobacillus plantarum Mut-7 on Sprague Dawley rat intestine. Microorganisms 9(11):2336. https://doi.org/10.3390/microorganisms9112336
Feito J, Contente D, Ponce-Alonso M, Díaz-Formoso L, Araújo C, Peña N, Borrero J, Gómez-Sala B, Del Campo R, Muñoz-Atienza E, Hernández PE, Cintas LM (2022) Draft genome sequence of Lactococcus lactis subsp. cremoris WA2–67: a promising Nisin-producing probiotic strain isolated from the rearing environment of a Spanish rainbow trout (Oncorhynchus mykiss, Walbaum) farm. Microorganisms 10(3):521. https://doi.org/10.3390/microorganisms10030521
Kamble A, Naik S, Talathi M, Jadhav D, Pingale S, Kaul-Ghanekar R (2022) Cervicovaginal microbiota isolated from healthy women exhibit probiotic properties and antimicrobial activity against pathogens isolated from cervical cancer patients. Arch Microbiol 204(8):491. https://doi.org/10.1007/s00203-022-03103-5
Ranjit E, Raghubanshi BR, Maskey S, Parajuli P (2018) Prevalence of bacterial vaginosis and its association with risk factors among nonpregnant women: a hospital based study. Int J Microbiol 2018:8349601. https://doi.org/10.1155/2018/8349601
Zhang W, Guo H, Cao C, Li L, Kwok LY, Zhang H, Sun Z (2017) Adaptation of Lactobacillus casei Zhang to gentamycin involves an alkaline shock protein. Front Microbiol 8:2316. https://doi.org/10.3389/fmicb.2017.02316
Mancabelli L, Mancino W, Lugli GA, Argentini C, Longhi G, Milani C, Viappiani A, Anzalone R, Bernasconi S, van Sinderen D, Ventura M, Turroni F (2021) Amoxicillin-clavulanic acid resistance in the genus Bifidobacterium. Appl Environ Microbiol 87(7):e03137-e3220. https://doi.org/10.1128/AEM.03137-20
Duche RT, Singh A, Wandhare AG, Sangwan V, Sihag MK, Nwagu TNT, Panwar H, Ezeogu LI (2023) Antibiotic resistance in potential probiotic lactic acid bacteria of fermented foods and human origin from Nigeria. BMC Microbiol 23:142. https://doi.org/10.1186/s12866-023-02883-0
Ozturk S, Erbas G (2017) Investigation of antibiotic sensivity, isolation and identification of Gardnerella vaginalis collected from Ketem/Aydın Province. Kocatepe Med J 18:61–66
Devi S, Kiesewalter HT, Kovács R, Frisvad JC, Weber T, Larsen TO, Kovács ÁT, Ding L (2019) Depiction of secondary metabolites and antifungal activity of Bacillus velezensis DTU001. Synth Syst Biotechnol 4(3):142–149. https://doi.org/10.1016/j.synbio.2019.08.002
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
Conceptualization: GS, HGD, MSB; Sample collection: HGD and MSB; Investigation: GS and HGD; Data analysis: GS and HGD; Writing—first draft: GS and HGD; Supervision: MSB; Review and editing: MSB. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no relevant financial or non-financial interests to disclose.
Ethical Approval
This study was approved by “Hacettepe University Ethics Committee” (GO19/507).
Informed Consent
The written informed consent was obtained from all individual participants in accordance with the principles of the declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sahal, G., Donmez, H.G. & Beksac, M.S. Cervicovaginal Bacillus velezensis Isolate: A Potential Probiotic and an Antagonist Against Candida and Staphylococcus. Curr Microbiol 80, 332 (2023). https://doi.org/10.1007/s00284-023-03447-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03447-1