Abstract
A novel Gram-stain positive, endospore-forming bacterium, designated SA4T, was isolated from a soil sample collected from an abandoned marine solar saltern at Wendeng, Shandong Province, PR China. Cells were observed to be rod shaped, alginase positive, catalase positive and motile. The strain was found to grow at temperatures ranging from 15 to 40 °C (optimum 35 °C), and pH 5.0–11.0 (optimum pH 8.0) with 0–7.0 % (w/v) NaCl concentration (optimum NaCl 3.0 %). Phylogenetic analysis based on 16S rRNA gene sequences indicated that strain SA4T belongs to the genus Bacillus and exhibits 16S rRNA gene sequence similarities of 96.6, 96.5, 96.3 and 96.2 % with Bacillus horikoshii DSM 8719T, Bacillus acidicola 105-2T, Bacillus shackletonii LMG 18435T and Bacillus pocheonensis Gsoil 420T, respectively. The menaquinone was identified as MK-7 and the major polar lipids were identified as diphosphatidylglycerol, phosphatidylglycerol and phosphatidylethanolamine. The major fatty acids detected were anteiso-C15:0 (22.3 %), iso-C15:0 (22.6 %), iso-C16:0 (14.8 %) and iso-C14:0 (14.7 %). The DNA G+C content was determined to be 42.4 mol %. Phenotypic, chemotaxonomic and genotypic properties clearly indicated that isolate SA4T represents a novel species within the genus Bacillus, for which the name Bacillus mesophius sp. nov. is proposed. The type strain is SA4T (=DSM 101000T=CCTCC AB 2015209T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Bacillus is composed of rod shaped, endospore-forming bacteria that are members of the phylum Firmicutes and are phylogenetically and phenotypically heterogeneous according to their distribution in various environments (Claus and Berkeley 1986). The majority of established species are mesophiles, and prefer neutral pH and low salt concentrations (Kosowski et al. 2014); however, some can also grow in alkaline environments (Denizci et al. 2010; Nielsen et al. 1995). In the course of screening for polysaccharide-degrading bacteria, we isolated a Gram-stain positive, endospore-forming, alkaliphilic strain, designated SA4T, from a soil sample collected from an abandoned marine solar saltern at Wendeng, PR China. This new isolate can degrade alginate, which is the most abundant polysaccharide in brown algae and is expected to become a renewable bioenergy resource (Obataa et al. 2015). Thus, the isolate has potential ability to produce novel alginases that are resistance to the alkaline conditions encountered during the processing of biomass for bioenergy generation from alginates. In this paper, we report the taxonomic characterisation of strain SA4T. On the basis of phenotypic characteristics, phylogenetic analysis and 16S rRNA data, the strain SA4T is considered to represent a novel species of the genus Bacillus.
Materials and methods
Bacterial strains, isolation and cultivation
The strain SA4T was isolated from a soil sample (temperature, pH and salinity of 31.9 °C, 8.01 and 2.71 %, respectively) collected from an abandoned marine solar saltern at Weihai, Shandong Province, PR China (122°0′21.59”E, 36°69′52.82”N) during October 2013. For culture, the soil sample was suspended in sterilised water, serially diluted, spread on marine 2216 agar (MA; Becton–Dickinson) directly without heat shock and incubated at 30 °C for 48 h. Pure cultures were obtained by several successive single colony isolations. The strain was stored both on MA slants at 4 °C and as suspensions in marine 2216 broth (MB; Becton–Dickinson) with 20 % (v/v) glycerol at −80 °C. The reference strains Bacillus horikoshii DSM 8719T, Bacillus acidicola DSM 14745T and Bacillus shackletonii LMG 18435T were obtained from the culture collections and used as controls in the phenotypic tests.
Phenotypic and physiological characterisation
A single colony of each strain was inoculated in 10 ml MB medium and incubated for 3 days at 30 °C before being subjected to phase-contrast light microscopy (Olympus BX-90). Strain SA4T was cultured over the temperature range 5–50 °C (at intervals of 5 °C) and the pH range 5.0–13.0 (at intervals of 1.0 pH unit) in MB. Growth at various NaCl concentrations was tested in NaCl-free artificial seawater medium supplemented with 5.0 g peptone, 1.0 g yeast extract and various concentrations of NaCl (final concentration: 0–10 % (w/v), using increments of 1.0 %) (Yang and Cho 2008). Cell density in MB was determined from the optical density at 600 nm with a spectrophotometer (UV-2550, Shimadzu, Japan) after incubation for up to 3 days. Growth under anaerobic conditions was determined after incubation in an anaerobic jar containing H2/CO2/N2 (5:5:90, by vol.) on MA at 30 °C for 7 days. The Gram staining and the KOH lysis test were carried out according to the methods described by Gregersen (1978) and Smibert and Krieg (1994). Motility was examined on motility agar (Chen et al. 2007). Oxygen requirement, activities of catalase, urease and oxidase, hydrolysis of casein, starch and aesculin, nitrate reduction, the Voges–Proskauer test, indole and H2S production were determined by the conventional methods described by Cowan and Steel (1965) and Smibert and Krieg (1994). Alginate lyase activity was detected by the formation of clear zones around colonies on MA plates containing 1 % (w/v) sodium alginate after flooding with diluted Lugol solution (Akagawa and Yamasato 1989; Schlesner et al. 1990). The utilisation of sole carbon and nitrogen sources was determined according to the method described by Gao et al. (1994). API 20E and API 50CH kits (bioMérieux) were used according to the manufacturer’s instructions except that the NaCl concentration was adjusted to 3.0 % (w/v) for strain SA4T and that the pH was adjusted to 5.0 for Bacillus acidicola DSM 14745T.
Chemotaxonomic characterisation
Biomass of strain SA4T and reference strains was harvested from cultures after incubation on MA medium at 30 °C for 48 h, when the bacteria were in the logarithmic growth phase. For cell wall chemotaxonomy, the peptidoglycan diamino acid test was carried out according to Schleifer (1985). The amino acid composition of the peptidoglycan hydrolysate (4 M HCl, 16 h, 100 °C) was determined by TLC, 2D-TLC and gas chromatography/mass spectrometry GC/MS. Menaquinones were analysed as described by Collins et al. (1977), using reverse-phase HPLC (Groth et al. 1996). Extraction and analysis of polar lipids by 2D-TLC was performed according to Minnikin et al. (1984) by the DSMZ identification service. For determination of cellular fatty acids, the isolate was harvested after cultivation on MA at 30 °C for 48 h, the references strains were cultivated on MA. The experiments were performed according to the standard protocol of the Sherlock Microbial Identification System version 6.0 (MIDI), analysed by GC (model 7890; Agilent) and identified using the TSBA6 database of the Microbial Identification System (Sasser 1990).
Phylogenetic analysis of 16S rRNA gene sequence
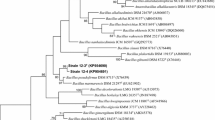
For 16S rRNA gene sequencing and phylogenetic analysis, chromosomal DNA was extracted and purified according to standard methods (Hopwood et al. 1985). The 16S rRNA gene sequences were amplified by PCR with the universal primers 9F (5′-GAGTTTGATCCTGGCTCAG-3′) and 1542R (5′-GGAGAAAGGAGGTGATCCAGCC-3′) as described by Lee et al. (2001). The sequence of the amplified 16S rRNA gene was determined using a DNA sequencer (ABI 310 sequencer; Applied Biosystems). The resultant 16S rRNA gene sequence was compared with that of reference strains with validly published names using the EzTaxon-e server (http://www.eztaxon-e.ezbiocloud.net/; Kim et al. 2012). For determination of pairwise sequence similarity values, sequence data were calculated using the global alignment algorithm obtained through the EzTaxon-e server (Kim et al. 2012). After multiple alignments of data by CLUSTAL_X (Thompson et al. 1997), phylogenetic trees were constructed using the neighbour-joining (NJ) (Saitou and Nei 1987), maximum-parsimony (MP) (Fitch 1971) and maximum-likelihood (ML) (Felsenstein 1981) methods implemented with MEGA version 6 (Tamura et al. 2013). Evolutionary distances were computed according to the Juke–Cantor model (Jukes and Cantor 1969). The reliability of each branch was evaluated by bootstrap analysis based on 1000 replications (Felsenstein 1985). The 16S rRNA gene sequences used for the phylogenetic comparisons are shown in the ML phylogenetic tree with their strain designations and accession numbers.
For determination of DNA G+C content, genomic DNA was prepared using the method described by Hopwood et al. (1985). The G+C content was determined using the HPLC method (Mesbah et al. 1989).
Results and discussion
Phenotypic characteristics
Morphological features showed that cells of the strain SA4T are Gram-positive, motile and rod shaped. Ellipsoidal spore were observed to lie subterminally in unswollen sporangia (supplementary Fig. S1). Growth was observed at temperatures ranging from 15 to 40 °C, and pH 5.0–11.0 with 0–8.0 % (w/v) NaCl concentration. Optimum growth was found at 35 °C with pH 8.0 and 3.0 % (w/v) NaCl concentration. No growth was evident below 15 °C or pH 5.0 or above 9.0 % (w/v) NaCl. Strain SA4T, B. horikoshii DSM 8719T, B. Acidicola DSM 14745Tand B. Shackletonii LMG 18435T were found to be negative for nitrate reduction; all strains were found to produce acid from N-acetylglucosamine and trehalose, but not from d-arabinose, d-fucose, l-fucose or 5-ketogluconate. However, the Voges–Proskauer test was found to be positive for strain SA4T, distinguishing it from the three reference strains. Other detailed physiological and biochemical characteristics of strain SA4T in comparison with its close phylogenetic neighbours are presented in Table 1.
Molecular phylogenetic analysis
An almost complete 16S rRNA gene sequence (1501 bp, GenBank accession number KU324461) of strain SA4T was determined. Pairwise comparisons indicated SA4T to be closely related to B. horikoshii DSM 8719T (96.6 %), followed by B. acidicola 105-2T (96.5 %), B. shackletonii LMG 18435T (96.3 %) and Bacillus pocheonensis Gsoil 420T (96.2 %). In the ML tree, phylogenetic analysis placed strain SA4T in the genus Bacillus although its relationships B. horikoshii DSM 8719T, B. acidicola 105-2T and others was not supported by high bootstrap values (Fig. 1). The topologies of the phylogenetic trees built using NJ and MP methods also support the conclusion that strain SA4T forms a stable clade although its relationships with the reference strains B. horikoshii DSM 8719T, B. acidicola 105-2T and others remains to be clarified (supplementary Figs.S2 and S3). This analysis suggested that strain SA4T represents a distinct species within the genus Bacillus. The genomic DNA G+C content of strain SA4T was determined to be 42.4 mol % whereas that of B. horikoshii DSM 8719T is 41.1 mol % (Nielsen et al. 1995).
Chemotaxonomic characterisation
Analysis of the cell wall peptidoglycan showed that strain SA4T contains meso-diaminopimelic acid as the diagnostic diamino acid. The menaquinone was identified as MK-7. The polar lipids detected were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylcholine, two aminophospholipids (APL) and seven unidentified phospholipids (supplementary Fig. S4). The major fatty acids present in strain SA4T (>10 % of the total fatty acids) were identified as anteiso-C15:0 (22.3 %), iso-C15:0 (22.6 %), iso-C16:0 (14.8 %) and iso-C14:0 (14.7 %). Marginal quantitative differences were observed in the profile of fatty acids compared with the close neighbour, B. horikoshii DSM 8719T (supplementary Table S1). The fatty acid profile of strain SA4T was similar to that of the reference strains tested, although there were differences in the proportions of some fatty acid components. These data are consistent with reports that iso- and anteiso-branched fatty acids of the 14–17 carbon series are typical of those found in the membranes of members of the genus Bacillus (Albert et al. 2005; Kämpfer 1994).
Taxonomic conclusion
The 16S rRNA gene analysis showed that strain SA4T has 96.6 % identity with its close relative B. horikoshii DSM 8719T, which is below the 98.7 % threshold suggested for DNA:DNA hybridization (Tindall et al. 2010; Meier-Kolthoff et al. 2013; Kim et al. 2014). The phenotypic and chemotaxonomic properties of strain SA4T distinguish it from B. horikoshii DSM 8719T. Based on the present polyphasic analysis, strain SA4T is considered to represent a novel species within the genus Bacillus, for which the name Bacillus mesophilus sp. nov. is proposed.
Description of Bacillus mesophilus sp. nov.
Bacillus mesophilus (me.so’phi.lus. Gr. adj. mesos, middle; Gr. n. philos, friend, loving; N.L. masc. adj. mesophilus, a mesophilic bacillus).
Cells are Gram-stain positive motile rods (0.5–0.8 × 2.0–5.0 μm) with a polar flagellum, occurring singly or in short chains. Ellipsoidal endospores are located sub-terminally. Colonies grown at 30 °C on MA for 48 h appear circular, pale pink in colour, flat, about 1–2 mm diameter. Growth occurs at 20–40 °C (optimum 35 °C), pH 5.0–11.0 (optimum pH 8.0), with 0–8.0 % (w/v) NaCl concentration (optimum 3.0 %). Aerobic; catalase and oxidase positive but urease negative. H2S and indole are not produced. Positive for gelatin and alginate hydrolysis, and Voges–Proskauer test. Negative for nitrate reduction, citrate utilisation, β-galactosidase (ONPG), tryptophan deaminase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase. Produces acid from trehalose, sucrose, maltose, glycerol, d-xylose, glucose, fructose, mannose and N-acetylglucosamine, but not from other substrates. The major cellular fatty acids are anteiso-C15:0, iso-C15:0, iso-C16:0 and iso-C14:0. The respiratory quinone is MK-7. The DNA G+C content of the type strain is 42.4 mol %.
The type strain, SA4T (=DSM 101000T=CCTCC AB 2015209T), was isolated from a soil sample collected from a deserted saltworks at Wendeng, Shandong Province, PR China. The GenBank accession number of the 16S rRNA gene sequence of strain SA4T is KU324461.
References
Akagawa M, Yamasato K (1989) Synonymy of Alcafigenes aquamarinus, Alcaligenes faecalis subsp. homari, and Deleya aesta: Deleya aquamarina comb. nov. as the type species of the genus Deleya. Int J Syst Bacteriol 3:462–466
Albert RA, Archambault J, Rosselló-Mora R, Tindall BJ, Matheny M (2005) Bacillus acidicola sp. nov., a novel mesophilic, acidophilic species isolated from acidic Sphagnum peat bogs in Wisconsin. Int J Syst Evol Microbiol 55:2125–2130
Buck JD (1982) Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol 44:992–993
Chen YG, Cui XL, Pukall R, Li HM, Yang YL, Xu LH, Wen ML, Peng Q, Jiang CL (2007) Salinicoccus kunmingensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-west China. Int J Syst Evol Microbiol 57:2327–2332
Claus D, Berkeley RCW (1986) Genus Bacillus. In: Sneath PH, Mair N, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams, Wilkins, Baltimore, pp 1105–1139
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Cowan ST, Steel KJ (1965) Manual for the identification of medical bacteria. Cambridge University Press, London
Denizci AA, Kazan D, Erarslan A (2010) Bacillus marmarensis sp. nov., an alkaliphilic, protease-producing bacterium isolated from mushroom compost. Int J Syst Evol Microbiol 60:1590–1594
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–789
Fitch WM (1971) Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gao JL, Sun JG, Li Y, Wang ET, Chen WX (1994) Numerical taxonomy and DNA relatedness of tropical rhizobia isolated from Hainan Province, China. Int J Syst Bacteriol 44:151–158
Gregersen T (1978) Rapid method for distinction of Gram-negative from Gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:123–127
Groth I, Schumann P, Weiss N, Martin K, Rainey FA (1996) Agrococcus jenensis gen. nov., sp. nov., a new genus of actinomycetes with diaminobutyric acid in the cell wall. Int J Syst Bacteriol 46:234–239
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM, Schrempf H (eds) (1985) Genetic manipulation of streptomyces. A laboratory manual. John Innes Foundation, Norwich
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, vol 3. Academic Press, New York, pp 21–132
Kämpfer P (1994) Limits and possibilities of total fatty acid analysis for classification and identification of Bacillus species. Syst Appl Microbiol 17:86–98
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kosowski K, Schmidt M, Pukall R, Hause G, Kämpfer P, Lechner U (2014) Bacillus pervagus sp. nov. and Bacillus andreesenii sp. nov., isolated from a composting reactor. Int J Syst Evol Microbiol 64:88–94
Lee JS, Shin YK, Yoon JH, Takeuchi M, Pyun YR, Park YH (2001) Sphingomonas aquatilis sp. nov., Sphingomonas koreensis sp. nov. and Sphingomonas taejonensis sp. nov., yellow-pigmented bacteria isolated from natural mineral water. Int J Syst Evol Microbiol 51:1491–1498
Logan NA, Lebbe L, Verhelst A, Goris J, Forsyth G, Rodríguez-Díaz M, Heyndrickx M, De Vos P (2004) Bacillus shackletonii sp. nov., from volcanic soil on Candlemas Island, South Sandwich archipelago. Int J Syst Evol Microbiol 54:373–376
Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P (2013) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by highperformance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nielsen P, Fritze D, Priest FG (1995) Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 141:1745–1761
Obataa O, Akunnab JC, Walkera G (2015) Hydrolytic effects of acid and enzymatic pre-treatment on the anaerobic biodegradability of Ascophyllum nodosum and Laminaria digitata species of brown seaweed. Biomass Biogenerg 80:140–146
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC News Lett 20:1–6
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Methods Microbiol 18:123–156
Schlesner H, Bartels C, Sittig M, Dorsch M, Stackebrandt E (1990) Taxonomic and phylogenetic studies on a new taxon of budding, hyphal Proteobacteria, Hirschia baltica gen. nov., sp. nov. Int J Syst Bacteriol 40:443–451
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, pp 607–655
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA 6: molecular evolutionary genetics analysis 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P (2010) Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:249–266
Yang SJ, Cho JC (2008) Gaetbulibacter marinus sp. nov., isolated from coastal seawater, and emended description of the genus Gaetbulibacter. Int J Syst Evol Microbiol 58:315–318
Acknowledgments
This research was supported by the International Science and Technology Cooperation Project of the Chinese Ministry of Science and Technology (2012DFA31120), the Natural Science Foundation of China (NSFC) (31370059), National Science and Technology Major Project of China (2016ZX10004217) and the Doctoral Research Fund of Fujian Academy of Agricultural Sciences (2014BS-3).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, YX., Liu, GH., Liu, B. et al. Bacillus mesophilus sp. nov., an alginate-degrading bacterium isolated from a soil sample collected from an abandoned marine solar saltern. Antonie van Leeuwenhoek 109, 937–943 (2016). https://doi.org/10.1007/s10482-016-0692-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0692-7