Abstract
Precursor engineering is an effective strategy for the overproduction of secondary metabolites. The polyene macrolide rimocidin, which is produced by Streptomyces rimosus M527, exhibits a potent activity against a broad range of phytopathogenic fungi. It has been predicted that malonyl-CoA is used as extender units for rimocidin biosynthesis. Based on a systematic analysis of three sets of time-series transcriptome microarray data of S. rimosus M527 fermented in different conditions, the differentially expressed accsr gene that encodes acetyl-CoA carboxylase (ACC) was found. To understand how the formation of rimocidin is being influenced by the expression of the accsr gene and by the concentration of malonyl-CoA, the accsr gene was cloned and over-expressed in the wild-type strain S. rimosus M527 in this study. The recombinant strain S. rimosus M527-ACC harboring the over-expressed accsr gene exhibited better performances based on the enzymatic activity of ACC, intracellular malonyl-CoA concentrations, and rimocidin production compared to S. rimosus M527 throughout the fermentation process. The enzymatic activity of ACC and intracellular concentration of malonyl-CoA of S. rimosus M527-ACC were 1.0- and 1.5-fold higher than those of S. rimosus M527, respectively. Finally, the yield of rimocidin produced by S. rimosus M527-ACC reached 320.7 mg/L, which was 34.0% higher than that of S. rimosus M527. These results confirmed that malonyl-CoA is an important precursor for rimocidin biosynthesis and suggested that an adequate supply of malonyl-CoA caused by accsr gene over-expression led to the improvement in rimocidin production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces are soil-dwelling, gram-positive bacteria that produce several kinds of secondary metabolites [1,2,3]. Among them, polyketides are an outstanding group of secondary metabolites because of their structural diversity and the range of their applications, such as antibiotics (e.g., tetracycline, erythromycin), antifungals (e.g., amphotericin B), antivirals (e.g., balticolid), anticancer agents (e.g., bleomycin, doxorubicin), immunosuppressants (e.g., rapamycin, FK506), and antiparasitics (e.g., avermectins, milbemycins) [4]. However, their yields are too low to meet the requirements of industrial fermentation. Therefore, the genetic modification of producing strains is normally required to improve the production of these compounds [5,6,7].
Glycolysis, tricarboxylic acid cycle, and pentose phosphate pathway are the primary metabolic pathways involved in the production of precursors that are utilized for the biosynthesis of secondary metabolites [8, 9]. The precursors of polyketides come directly or indirectly from primary metabolism, so polyketide production is often limited by the availability of these essential precursors [10]. Therefore, precursor engineering has been applied to redesign and/or optimize the metabolic pathways to increase the intracellular pool of precursors and to redirect the flux toward the targeted polyketide biosynthesis [11,12,13]. Malonyl-CoA is usually involved in numerous essential primary metabolic pathways, as well as a starter and extender units for polyketide biosynthesis [14]. Malonyl-CoA is mainly synthesized through the carboxylation of acetyl-CoA by acetyl-CoA carboxylase (ACC) [15]. It was reported that the introduction and heterologous expression/over-expression of genes encoding ACC are practicable approaches for the improvement of polyketides [16]. For example, increasing the copy number of the acc gene encoding ACC remarkably improved the FK520 titers in Streptomyces hygroscopicus [17]. Similarly, Ryu et al. [18] over-expressed the acc gene, which led to an increase in precursor supply for actinorhodin production in S. coelicolor.
Rimocidin is a 28-membered tetraene macrolide comprising a large lactone ring with a sugar moiety [19], and it is produced by type I modular polyketide synthases (PKS) in S. rimosus M527 [20]. Rimocidin is a promising fungicide utilized in controlling the occurrence of plant fungal diseases in agricultural field [21]. Constant efforts were always made to enhance the yield of rimocidin in S. rimosus M527 to meet the commercial requirements. Various strategies, such as medium optimization [22], addition of elicitors [23], genetic engineering [24], and ribosomal engineering [25], have been adopted to achieve this goal.
A rimocidin synthetic pathway was predicted [20]. Butyryl-CoA was used as a starter unit by the loading module; macrolactone rimocidin was formed through elongation steps in which acetate was mainly incorporated by the decarboxylative condensation of malonyl-CoA [20]. However, it is still unclear whether malonyl-CoA is a real and important precursor involved in rimocidin biosynthesis in S. rimosus M527. In a previous study, based on the analysis of comparative transcriptome data for rimocidin production under normal and optimized conditions, a differential expression gene that encodes ACC was found in S. rimosus M527.
Therefore, the aim of this study was to investigate the relationships between the differential expression gene, malonyl-CoA, and rimocidin in S. rimosus M527.
Materials and Methods
Materials
Q5 High-Fidelity Master Mix with GC-buffer was purchased from NEB. PCR reagents, restriction endonucleases, Miniprep kits, and gel extraction kits were purchased from TaKaRa Biotechnology Co., Ltd. Malonyl-CoA ELISA kit and ACC assay kit were purchased from Shanghai Jianglai Biotechnology Co., Ltd. Oligonucleotide primer synthesis and DNA sequencing of PCR products were performed by Beijing TSINGKE Biotechnology Co., Ltd., China.
Bacterial Strains and Plasmids
The strains and plasmids used in this study are listed in Table S1. Streptomyces rimosus M527, a rimocidin producer, was used as a host for gene expression and rimocidin production; it has been deposited at the CCTCC (M2013270), Wuhan, China. Escherichia coli JM109 was used as a general host for gene cloning and plasmid construction. The methylation-deficient strain E. coli ET12567/pUZ8002 was used as a donor for intergeneric conjugation. Plasmid pIB139 harboring a permE* promoter is a shuttle vector that replicates in E. coli and integrates into Streptomyces chromosomes site specifically.
Media and Culture Conditions
E. coli strains were cultured in liquid or solid Luria–Bertani (LB) medium, containing appropriate antibiotics at 37 °C. Apramycin (50 µg/mL), chloramphenicol (25 µg/mL), ampicillin (100 µg/mL), kanamycin (50 µg/mL), and nalidixic acid (50 µg/mL) were added as needed.
S. rimosus M527 and its derivatives were incubated according to the method described by Zhao et al. [25]. S. rimosus M527 was incubated at 28 °C and grown in a solid mannitol soya flour (MS) medium (20 g/L soya flour, 20 g/L mannitol, 20 g/L agar, and tap water) for sporulation. Moreover, 2CMC solid media containing 10 g/L starch, 2 g/L tryptone, 1 g/L NaCl, 2 g/L (NH4)2SO4, 2 g/L MgSO4·7H2O, 2 g/L CaCO3, 2 g/L casamino acids, 1 g/L K2HPO4·3H2O, 1 g/L FeSO4·7H2O, 1 g/L MgCl2·6H2O, 1 g/L ZnSO4·7H2O, and 20 g/L agar (adjusted to pH 7.2 by NaOH) were used for conjugation [22].
S. rimosus M527 spores (1 × 106/mL) were inoculated into a 250-mL Erlenmeyer flask containing 50 mL of seed medium and was shaken at 28 °C and 180 r/min. The CP liquid medium used as the seed medium had the same composition as that described in an earlier study [26].
DNA Manipulations
The general procedures for DNA manipulation were conducted according to the method described by Sambrook and Russel [27]. The intergeneric conjugation of Streptomyces and E. coli was performed as described by Kieser et al. [28] with minor modifications. S. rimosus M527 spores were collected from solid medium, filtrated by sterile absorbent cotton in order to remove mycelia and then incubated at 50 °C for 10 min to induce germination. The donor strain, E. coli ET12567/pUZ8002 harboring constructed plasmid, was grown in LB with the appropriate antibiotics until an OD600 of 0.4–0.6. The cells were washed twice with LB and resuspended in a final volume of 500 µL of LB. E. coli (108/mL) as donor cells and the S. rimosus M527 spores (108/mL) as recipients were mixed and spread on 2CMC agar plate containing 10 mmol/L MgCl2. The plates were incubated at 28 °C for 10–20 h and overlaid with 500 µL fresh LB medium containing 100 µg/mL nalidixic acid and 300 µg/mL apramycin. The plates were further incubated at 28 °C for 3–4 days, and the exconjugants were counted [22].
Clone of acc sr Gene and Construction of Recombinant Strains
Using the genomic DNA of S. rimosus M527 as a template, a 1524 bp accsr gene open reading frame (ORF) was amplified by PCR using primers P1 and P2 (Table S1). High-fidelity PCR was performed using Q5 DNA polymerase (NEB) to obtain DNA fragments to be used for plasmid construction. PCR amplification was started at 95 °C for 5 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 60 °C for 20 s, and extension at 72 °C for 45 s. After 30 cycles, a 5-min extension at 72 °C was done. Then, the PCR product was digested with NdeI and NotI and inserted into the corresponding site of plasmid pIB139, yielding a plasmid pIB139-accsr. Sequencing results confirmed that the gene did not contain any mutation.
Subsequently, the introduction of the constructed pIB139-accsr into S. rimosus M527 was performed by intergeneric conjugation [22] to the recombinant strain S. rimosus M527-ACC. Recombinant strains were confirmed using apramycin resistance and PCR.
Determination of Intracellular Malonyl-CoA and Enzyme Assay
Fermentation culture broth (1 mL) of S. rimosus M527 and S. rimosus M527-ACC was collected at 12 h, 24 h, 36 h, 48 h, 72 h, and 96 h. Each sample was centrifuged at 11,000×g for 5 min to obtain mycelium; then, each was washed twice with PBS buffer. For extraction and quantification of intracellular malonyl-CoA, the instructions described in the ELISA kits were followed [29]. The standard sample was diluted to different concentrations with assay buffer to prepare a standard curve. The samples for testing were diluted 1:5 by adding 10 µL of sample into 40 µL of Sample Diluent. The solutions were incubated at 37 °C for 60 min and the TMB chromogenic substrate was added. We measured the light absorption value at 450 nm and then measured intracellular concentration of malonyl-CoA in each sample according to the standard curve.
ACC activity was assayed using the ACC assay kit as described by the kit instructions. ACC activity was measured by determining the increased inorganic phosphorus content using the ammonium molybdate phosphate method; one unit of ACC activity is defined as the amount of inorganic phosphorus per hour per mg of tissue protein produced by 1 mol of inorganic phosphorus [30].
Analysis of Gene Transcriptional Level by qRT-PCR
We extracted RNA and analyzed the transcriptional level of accsr and some structural genes (rimA, rimE, rimJ, rimK) located in the rimocidin biosynthetic gene cluster between the recombinant strain S. rimosus M527-ACC and WT strain, as described by Zhao et al. [25]. The thermal profiles consisted of initial denaturation at 94 °C for 3 min, followed by previously indicated cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and a final step at 72 °C for 10 min. The 16S rRNA gene in S. rimosus M527 was used as a positive internal control for the quantitative RT-PCR (qRT-PCR) assay. The qRT-PCR experiments were carried out in triplicate using RNA samples from three independent experiments. The primers used to analyze transcriptional levels of accsr and some rim genes in WT strain M527 and recombinant strains S. rimosus M527-ACC are listed in Table S1.
Production and HPLC Analysis of Rimocidin
Rimocidin production and its analysis were performed according to the method described by Zhao et al. [25]. Rimocidin presence was analyzed and confirmed using high-performance liquid chromatography (HPLC) with a column of Supersil ODS2 (4.6 × 150 mm, 5 μm) maintained at 30 °C. The percentage volume of methanol was varied as follows: linearly increased from 5 to 83% (0–20 min), held at 83% (20–30 min), linearly increased to 100% (30–35 min), and then linearly decreased to 5% (35–40 min). The UV detection of rimocidin was conducted at 304 nm and the solvent flow rate was 1.0 mL/min.
Construction of the Phylogenetic Tree
Phylogenetic analysis of acc gene using NCBI BLAST. The nucleotide sequences of accsr from different prokaryotic species were collected from the NCBI database, and phylogenetic analysis of accsr was performed with MEGA 7.0, using the neighbor-joining method in the Jukes–Cantor model [31].
Statistical Analysis
All experiments were conducted at least three times, and the results are expressed as means ± standard deviations. Statistical analysis was performed using Student’s t test.
Data Availability
The nucleotide sequence of accsr gene was submitted to the GenBank database under the accession number MT176432.
Results
Cloning of acc sr from S. rimosus M527
It is predicted that malonyl-CoA is an important extender unit for rimocidin biosynthesis in Streptomyces diastaticus var. 108 [20, 32]. The acc gene encodes ACC which can catalyze the carboxylation of acetyl-coenzyme A (acetyl-CoA) to produce malonyl-CoA. In this context, two obvious questions are as follows: Which gene is responsible for malonyl-CoA formation in S. rimosus M527? Whether increase of malonyl-CoA is beneficial to overproduction of rimocidin in S. rimosus M527?
According to the analysis of comparative transcriptome data of S. rimosus M527 (GenBank accession No. GCA_004196335.1) in different conditions, a differential expression gene from S. rimosus M527 was cloned. The 1524 bp gene encodes a protein with 508 amino acids. As shown in Fig. 1, among these, it was identical to the acc from S. rimosus strain WT5260 and S. rimosus strain ATCC 10970. This gene was named accsr, and its nucleotide sequence was submitted to the GenBank database under the accession number MT176432.
Phylogenetic analysis of acc gene using NCBI BLAST. The nucleotide sequences of accsr from different prokaryotic species were collected from the NCBI database, and phylogenetic analysis of accsr was performed with MEGA 7.0, using the neighbor-joining method in the Jukes–Cantor model [31]. Bootstrap values (> 50%) based on 1000 replicates were shown at the branch nodes. Bar, 0.05 substitutions per nucleotide positions
Construction of S. rimosus M527-ACC
To verify the correlation among the differential expression gene accsr, malonyl-CoA synthesis, and rimocidin production, a DNA fragment containing the accsr gene was cloned as described in the Materials and methods section. The gene accsr was placed under the control of promoter ermE* in plasmid pIB139 to create pIB139-accsr (Fig. S1). Then, the plasmid pIB139-accsr was introduced into S. rimosus M527 by intergeneric conjugative transfer between E. coli/Streptomyces to generate the recombinant strain S. rimosus M527-ACC, which was resistant to 300 µg/mL apramycin (Fig. S2). The integration of plasmid pIB139-accsr into the chromosome of S. rimosus M527 was verified by PCR (Fig. S3).
Over-expression of acc sr Gene Improved the Rimocidin Production in S. rimosus M527
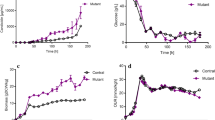
To further investigate the effect of accsr gene on rimocidin production, a batch fermentation experiment in a 250-mL Erlenmeyer flask was performed using S. rimosus M527-ACC and the wild-type (WT) strain S. rimosus M527, as described in the Materials and methods section. As shown in Fig. 2a, a significant increase in the rimocidin production of S. rimosus M527-ACC was observed compared to that of S. rimosus M527 throughout the fermentation process. After 96 h, the amount of rimocidin produced by S. rimosus M527-ACC reached the highest level of 320.7 mg/L, which was 34.0% higher than the rimocidin produced by S. rimosus M527 (239.4 mg/L). There was no significant difference between S. rimosus M527-ACC and S. rimosus M527 in terms of dry cell weight (DCW) (Fig. 2b). In addition, the expression of empty plasmid pIB139 in S. rimosus M527 had no effect on rimocidin production (data not shown).
Analysis and comparison of rimocidin concentration (a) and cell growth (b) of control strain M527 and recombinant strain M527-ACC. The error bars were calculated from three different batches of fermentation. **Indicates highly statistically significant results (P value < 0.01). ns indicates no statistically significant results (P value > 0.05)
The transcriptional level of the accsr gene was quantified by RT-qPCR analysis using the total RNA isolated from the recombinant strain S. rimosus M527-ACC and WT strain S. rimosus M527 at three time points of fermentation (36 h, 48 h, and 72 h). RT-qPCR results indicated that the transcriptional level of the accsr gene was obviously increased in S. rimosus M527-ACC compared to that in S. rimosus M527 due to the introduction of an extra copy of accsr gene (Fig. 3a).
Comparison of the transcription levels of accsr gene (a) and rim genes (rimA, rimE, rimJ, rimK) (b) in different strains obtained by quantitative reverse transcription-PCR. M527: S. rimosus M527; M527-ACC: S. rimosus M527-ACC. The cells were harvested from the fermentation 36 h, 48 h, and 72 h. Error bars were calculated by measuring the standard deviations of the data from three replicates of each sample. **Indicates highly statistically significant results (P value < 0.01). ns indicates no statistically significant results (P value > 0.05)
The effect of over-expression of gene accsr on transcriptional level of rim structural genes involved in rimocidin biosynthesis was also determined. As compared to those of the WT strain S. rimosus M527, recombinant strain S. rimosus M527-ACC exhibited higher transcriptional level of rimA and had no significant difference in transcriptional levels of rimE, rimJ, and rimK (Fig. 3b).
Enhancement of the Enzymatic Activity of ACC and Intracellular Malonyl-CoA in S. rimosus M527
We speculated that the increased transcription of accsr gene could improve the specific activity of ACC and intracellular concentration of malonyl-CoA in S. rimosus M527. To confirm this hypothesis, the specific activity of ACC and intracellular concentration of malonyl-CoA in S. rimosus M527-ACC and S. rimosus M527 was determined. The results confirmed that accsr over-expression promoted the enzymatic activity of ACC (Fig. 4a). The enzymatic activity of ACC in S. rimosus M527-ACC harboring the over-expression of accsr reached the highest level of 2.20 U/mg/DCW, which was onefold higher than that in WT strain S. rimosus M527 (1.08 U/mg/DCW).
Comparison of the acetyl-CoA carboxylase activity (a) and intracellular malonyl-CoA (b) in S. rimosus M527 and S. rimosus M527-ACC. Error bars were calculated by measuring the standard deviations of the data from three replicates of each sample. **Indicates highly statistically significant results (P value < 0.01)
In addition, this also indicated that higher intracellular concentration of malonyl-CoA was caused by the higher enzymatic activity of ACC (Fig. 4b). In S. rimosus M527-ACC, the intracellular concentration of malonyl-CoA was 547 ng/g/DCW, which was 1.5 times higher than that in S. rimosus M527 (364 ng/g/DCW).
Discussion
Rimocidin is a member of polyene macrolides that are commercially important because of their antifungal properties. Rimocidin, like all polyenes, is produced through the action of type I modular PKS. Based on its chemical structure, a model for the biosynthetic pathway of rimocidin was proposed. During its elongation process, acetate was incorporated as an elongation unit in all modules through the decarboxylative condensation of malonyl-CoA, except in modules 7 and 13. Thus, malonyl-CoA is an essential precursor for rimocidin biosynthesis. Increasing the supply of acyl-CoA, which is used as a starter unit or extender unit, has been an effective strategy for the enhancement of polyene macrolides [33, 34]. However, precursor engineering in S. rimosus M527 has not been performed so far. Malonyl-CoA is mainly synthesized by ACC carboxylation. In addition, the transcriptome microarray data of S. rimosus M527 fermented in different conditions were obtained; here, the differentially expressed gene called accsr that encodes ACC exhibited a higher transcriptional level in an optimized condition than in a normal condition. This provides a good opportunity to verify the relationships between accsr gene over-expression, intracellular malonyl-CoA concentrations, and rimocidin biosynthesis.
In this study, the accsr gene from S. rimosus M527 was cloned and over-expressed in its own host. The results showed that the over-expression of the accsr gene could enhance the specific enzymatic activity of ACC, increase the intracellular concentration of malonyl-CoA, and further promote the production of rimocidin. The results also confirmed that increasing the supply of malonyl-CoA is beneficial for the enhancement of rimocidin production. It is worth noting that we also attempted to knock out the accsr gene and confirm the correlation between the accsr gene, malonyl-CoA concentration, and rimocidin synthesis. However, these attempts remained unsuccessful after many trials. Because of poor growth, only the fermentation performance and cell characteristics of few accsr deletion mutants can be analyzed. A possible explanation for this phenomenon is the lack of adequate malonyl-CoA, which is involved in essential cell growth and primary metabolism, leading to difficulty in isolating mutant strains that harbor accsr gene deletion.
It is important to note that another tetraene, CE-108, which is a structural analog of rimocidin, was also found in the fermentation broth of S. rimosus M527 [35]. The two tetraenes differed in aglycone moiety, with a methyl side chain in CE-108 instead of the propyl group in rimocidin. In the proposed model for rimocidin and CE-108 biosynthesis, the PKS loading module seems to play a crucial role in recognizing acetyl-CoA or butyryl-CoA as starter units, giving rise to methyl or propyl side chains and determining the biosynthesis of CE-108 and rimocidin [20]. There is a common elongation module that is responsible for polyketide chain formation in CE-108 and rimocidin biosynthesis [35], malonyl-CoA is also an important precursor for CE-108 formation during the elongation step. As a result, the over-expression of the accsr gene also resulted in the improvement of CE-108 (Fig. S4). On the contrary, CE-108 exhibited a weaker antifungal activity than rimocidin [32, 36]. Therefore, an individual-targeted increase of rimocidin instead of CE-108 is worth considering.
The significance of this study is that it provides a new method to promote rimocidin production by precursor engineering via accsr gene over-expression. To further specifically increase the rimocidin production, the specificity of the loading module to recognize butyryl-CoA as a starter unit can be improved by modifying the site-specific acyltransferase domain in the loading module. For further improvement of rimocidin production in S. rimosus M527, the supply of butyryl-CoA as a starter unit and malonyl-CoA as an extender can also be increased at the same time by metabolic engineering technology. Further investigations will be continued in our studies.
Conclusion
In this study, the gene called the accsr encoding ACC, which exhibits differential expression from comparative transcriptome data of S. rimosus M527 in different conditions, was cloned and over-expressed in original strain S. rimosus M527. The recombinant strain S. rimosus M527-ACC harboring the over-expressed accsr gene exhibited higher transcriptional level of accsr gene, enzymatic activity of ACC, malonyl-CoA concentration, and rimocidin production, compared to S. rimosus M527. These results not only verified the accuracy of transcriptome data in which a higher expression level of accsr gene was found in S. rimosus M527 under optimization condition but also illustrated the relationships between the accsr gene, malonyl-CoA, and rimocidin biosynthesis.
References
Liu R, Deng Z, Liu T (2018) Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab Eng 50:74–84
Kemung HM, Tan LT, Khan TM et al (2018) Streptomyces as a prominent resource of future anti-MRSA drugs. Front Microbiol 9:2221
Quinn GA, Banat AM, Abdelhameed AM et al (2020) Streptomyces from traditional medicine: sources of new innovations in antibiotic discovery. J Med Microbiol 69:1040–1048
Zhang LX, Demain AL (2005) Natural products: drug discovery and therapeutic medicine. The Humana Press, Totowa
Jung W, Kim E, Yoo YJ et al (2014) Characterization and engineering of the ethylmalonyl-CoA pathway towards the improved heterologous production of polyketides in Streptomyces venezuelae. Appl Microbiol Biotechnol 98(8):3701–3713
Li L, Liu X, Jiang W et al (2019) Recent advances in synthetic biology approaches to optimize production of bioactive natural products in Actinobacteria. Front Microbiol 10:2467
Palazzotto E, Tong Y, Lee SY et al (2019) Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol Adv 37(6):107366
Blažič M, Kosec G, Baebler Š et al (2015) Roles of the crotonyl-CoA carboxylase/reductase homologues in acetate assimilation and biosynthesis of immunosuppressant FK506 in Streptomyces tsukubaensis. Microb Cell Fact 14:164
Jin XM, Chang YK, Lee JH et al (2017) Effects of increased NADPH concentration by metabolic engineering of the pentose phosphate pathway on antibiotic production and sporulation in Streptomyces lividans TK24. J Microbiol Biotechnol 27(10):1867–1876
Li S, Li Z, Pang S et al (2021) Coordinating precursor supply for pharmaceutical polyketide production in Streptomyces. Curr Opin Biotechnol 69:26–34
Liu H, Reynolds KA (2001) Precursor supply for polyketide biosynthesis: the role of crotonyl-CoA reductase. Metab Eng 3(1):40–48
Olano C, Lombó F, Méndez C et al (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10(5):281–292
Wilson MC, Moore BS (2012) Beyond ethylmalonyl-CoA: the functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep 29(1):72–86
Hertweck C (2009) The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl 48(26):4688–4716
Wei J, Zhang Y, Yu T et al (2016) A unified molecular mechanism for the regulation of acetyl-CoA carboxylase by phosphorylation. Cell Discov 2:16044
Milke L, Marienhagen J (2020) Engineering intracellular malonyl-CoA availability in microbial hosts and its impact on polyketide and fatty acid synthesis. Appl Microbiol Biotechnol 104(14):6057–6065
Yu Z, Lv H, Wu Y et al (2019) Enhancement of FK520 production in Streptomyces hygroscopicus by combining traditional mutagenesis with metabolic engineering. Appl Microbiol Biotechnol 103(23–24):9593–9606
Ryu YG, Butler MJ, Chater KF et al (2006) Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl Environ Microbiol 72(11):7132–7139
Sowiński P, Pawlak J, Borowski E et al (1995) Stereostructure of rimocidin. J Antibiot (Tokyo) 48(11):1288–1291
Seco EM, Pérez-Zúñiga FJ, Rolón MS et al (2004) Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem Biol 11(3):357–366
Jeon BJ, Kim JD, Han JW et al (2016) Antifungal activity of rimocidin and a new rimocidin derivative BU16 produced by Streptomyces mauvecolor BU16 and their effects on pepper anthracnose. J Appl Microbiol 120(5):1219–1228
Song Z, Liao Z, Hu Y et al (2019) Development and optimization of an intergeneric conjugation system and analysis of promoter activity in Streptomyces rimosus M527. J Zhejiang Univ Sci B 20(11):891–900
Song Z, Ma Z, Bechthold A et al (2020) Effects of addition of elicitors on rimocidin biosynthesis in Streptomyces rimosus M527. Appl Microbiol Biotechnol 104(10):4445–4455
Liao Z, Song Z, Xu J et al (2020) Identification of a gene from Streptomyces rimosus M527 negatively affecting rimocidin biosynthesis and morphological differentiation. Appl Microbiol Biotechnol 104(23):10191–10202
Zhao Y, Song Z, Ma Z et al (2019) Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J Ind Microbiol Biotechnol 46(5):697–708
Zhao Y, Lu D, Bechthold A et al (2018) Impact of otrA expression on morphological differentiation, actinorhodin production, and resistance to aminoglycosides in Streptomyces coelicolor M145. J Zhejiang Univ Sci B 19(9):708–717
Sambrook J, Russel DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Kieser T, Bibb MJ, Buttner MJ et al (2000) Practical Streptomyces genetics: a laboratory manual. John Innes Foundation, Norwich
Pierangeli SS, Harris EN (2018) A protocol for determination of anticardiolipin antibodies by ELISA. Nat Protoc 3(5):840–848
Arabolaza A, Shillito ME, Lin TW et al (2010) Crystal structures and mutational analyses of acyl-CoA carboxylase beta subunit of Streptomyces coelicolor. Biochemistry 49(34):7367–7376
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Escudero L, Al-Refai M, Nieto C et al (2015) New rimocidin/CE-108 derivatives obtained by a crotonyl-CoA carboxylase/reductase gene disruption in Streptomyces diastaticus var. 108: substrates for the polyene carboxamide synthase PcsA. PLoS ONE 10(8):e0135891
Tippelt A, Nett M (2021) Saccharomyces cerevisiae as host for the recombinant production of polyketides and nonribosomal peptides. Microb Cell Fact 20(1):161
Ren J, Cui Y, Zhang F et al (2014) Enhancement of nystatin production by redirecting precursor fluxes after disruption of the tetramycin gene from Streptomyces ahygroscopicus. Microbiol Res 169(7–8):602–608
Pérez-Zúñiga FJ, Seco EM, Cuesta T et al (2004) CE-108, a new macrolide tetraene antibiotic. J Antibiot (Tokyo) 57(3):197–204
Caffrey P, Aparicio JF, Malpartida F et al (2008) Biosynthetic engineering of polyene macrolides towards generation of improved antifungal and antiparasitic agents. Curr Top Med Chem 8(8):639–653
Funding
This work was supported by the National Natural Science Foundation of China (31772213, 31972320), the Zhejiang Province Natural Science Foundation (LY20C140006), and the Key Program of Zhejiang Province Natural Science Foundation (LZ22C140002).
Author information
Authors and Affiliations
Contributions
ZJL, JYZ, and YS conducted experiments. ZM and YYZ designed the experiments and wrote this article. AB and XPY checked the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liao, Z., Zhang, J., Shi, Y. et al. Improvement of Rimocidin Biosynthesis by Increasing Supply of Precursor Malonyl-CoA via Over-expression of Acetyl-CoA Carboxylase in Streptomyces rimosus M527. Curr Microbiol 79, 174 (2022). https://doi.org/10.1007/s00284-022-02867-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02867-9