Abstract
FK520 (ascomycin), a 23-membered macrolide with immunosuppressive activity, is produced by Streptomyces hygroscopicus. The problem of low yield and high impurities (mainly FK523) limits the industrialized production of FK520. In this study, the FK520 yield was significantly improved by strain mutagenesis and genetic engineering. First, a FK520 high-producing strain SFK-6-33 (2432.2 mg/L) was obtained from SFK-36 (1588.4 mg/L) through ultraviolet radiation mutation coupled with streptomycin resistance screening. The endogenous crotonyl-CoA carboxylase/reductase (FkbS) was found to play an important role in FK520 biosynthesis, identified with CRISPR/dCas9 inhibition system. FkbS was overexpressed in SFK-6-33 to obtain the engineered strain SFK-OfkbS, which produced 2817.0 mg/L of FK520 resulting from an increase in intracellular ethylmalonyl-CoA levels. In addition, the FK520 levels could be further increased with supplementation of crotonic acid in SFK-OfkbS. Overexpression of acetyl-CoA carboxylase (ACCase), used for the synthesis of malonyl-CoA, was also investigated in SFK-6-33, which improved the FK520 yield to 3320.1 mg/L but showed no significant inhibition in FK523 production. To further enhance FK520 production, FkbS and ACCase combinatorial overexpression strain SFK-OASN was constructed; the FK520 production increased by 44.4% to 3511.4 mg/L, and the FK523/FK520 ratio was reduced from 9.6 to 5.6% compared with that in SFK-6-33. Finally, a fed-batch culture was carried out in a 5-L fermenter, and the FK520 yield reached 3913.9 mg/L at 168 h by feeding glycerol, representing the highest FK520 yield reported thus far. These results demonstrated that traditional mutagenesis combined with metabolic engineering was an effective strategy to improve FK520 production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
FK520 (ascomycin) is a 23-membered macrocyclic antibiotic produced by Streptomyces hygroscopicus var. ascomyceticus ATCC 14891 (Bérdy 2005). FK520 has the same macrolide structure as tacrolimus (Kosec et al. 2012); therefore, it exhibits satisfactory immunosuppressive activity and has been used for the chemical synthesis of pimecrolimus, which is clinically applied for the treatment of atopic dermatitis (Eichenfield et al. 2002). Although FK520 and its derivatives are highly valued due to their pharmacological importance and clinical applications, the low yield of FK520 produced by the S. hygroscopicus ATCC 14891 strain substantially limits its industrial production. Intense efforts have been made to improve the production of FK520 in S. hygroscopicus in recent decades, including traditional random mutagenesis (Qi et al. 2012), medium optimization (Yu et al. 2019), exogenous feeding strategies (Qi et al. 2014b), fermentation process control (Qi et al. 2014a), and genetic engineering (Qi et al. 2017; Song et al. 2017). Among these approaches, traditional mutagenesis is a powerful and easily operated method for strain improvement in Streptomyces sp., especially for microbes with less understanding of genomic information and metabolic mechanisms. Although some FK520 high-producing strains have been developed with titanium sapphire laser (Qi et al. 2012), atmospheric and room temperature plasma (Yu et al. 2019), and chemical mutagens (Qi et al. 2014b), it is definitely laborious and time-consuming to improve FK520 production by simply relying on random mutagenesis. Thus, traditional mutagenesis has been necessarily coupled with rational metabolic engineering strategies to shorten the process of strain breeding.
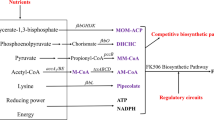
Metabolic engineering strategies have been widely used in the improvement of natural products, especially for the production of secondary metabolites in microbes. Currently, the enhanced production of secondary metabolites through metabolic engineering mainly focuses on adjustment of the biosynthetic pathway (Wang et al. 2017a), investigation of regulatory genes (Wei et al. 2018), improvement of precursor supply (Li et al. 2019), and comparative metabolomics profiling analysis (Xia et al. 2013). Among those strategies, enhanced precursor supply has been shown to be a direct and precise way to increase the accumulation of desired natural products. For example, an increased supply of isobutyryl-CoA, a precursor for the biosynthesis of ansamitocin P-3 (AP-3), led to about 4-fold improvement of AP-3 production in Actinosynnema pretiosum with isobutanol supplementation (Lin et al. 2011). Additionally, an increased supply of propionyl-CoA carboxylase (PCC) along with the addition of propionate was found to be an effective way to increase the concentration of intracellular methylmalonyl-CoA for rapamycin biosynthesis in S. hygroscopicus (Jung et al. 2011). As illustrated in Fig. 1, three important precursors are involved in the biosynthesis of FK520, namely, ethylmalonyl-CoA, methylmalonyl-CoA, and malonyl-CoA. A structural analogue FK523 is produced during the biosynthesis of FK520 in S. hygroscopicus (Dunn and Khosla 2013). This analogue has a methyl group in the macrolide skeleton C21 position rather than an ethyl group (Fig. 1) and is difficult to separate from FK520 by conventional separation and purification methods. It is necessary to apply preparative high-performance liquid chromatography (prep-HPLC) to separate FK523 from FK520 in industrial production, leading to a high manufacturing cost. The synthesis of FK523 results from the nonspecific substrate recognition of acyltransferase in the polyketide synthase (PKS) module for ethylmalonyl-CoA or methylmalonyl-CoA. Therefore, it is possible to decrease the production of FK523 by increasing the levels of intracellular ethylmalonyl-CoA, which is synthesized by crotonyl-CoA carboxylase/reductase (CCR) in S. hygroscopicus. Although Wang et al. integrated an exogenous CCR derived from S. coelicolor into a low-yield strain FS35 (Wang et al. 2017b), the effect of FkbS on FK523 accumulation and its role in the biosynthesis of FK520 in higher production strains are still not clearly understood. In addition, ACCase plays an important role in converting acetyl-CoA to malonyl-CoA. Overexpression of ACCase led to an increase in precursor supply for actinorhodin production in S. coelicolor (Ryu et al. 2006). However, there have been no study reporting the enhancement of FK520 production by improving the supply of malonyl-CoA in Streptomyces hygroscopicus. Therefore, ethylmalonyl-CoA and malonyl-CoA may be recognized as promising targets for metabolic modification to improve FK520 production and inhibit the accumulation of FK523 in S. hygroscopicus.
Proposed biosynthetic pathway of CoA-esters involved in FK520 biosynthesis. Overexpression targets (fkbS and accA2BE) in this study are marked with a deep gray box. In the structure of FK520, the part consisting of malonyl-CoA was marked in red and the C21 part composed of ethylmalonyl-CoA was marked in blue. Compared with FK520, the C21 part of FK523 was synthesized by methylmalonyl-CoA and labeled in yellow
In this study, we aimed to enhance FK520 production and decrease the biosynthesis of FK523. A high-yield strain (SFK-6-33) was obtained through traditional mutagenesis using ultraviolet radiation along with the streptomycin resistance screening method. The enhanced supplementation of ethylmalonyl-CoA and malonyl-CoA led to an increase in FK520 production and decrease in FK523 production through the overexpression of FkbS and ACCase together in SFK-6-33. Finally, the scale-up fermentation showed significantly enhanced FK520 production by the genetically engineered strain supplemented with glycerol. This example of combining classical mutagenesis and metabolic engineering for increasing desirable product yields and lowering impurity levels presents a cost-effective method for industrialized production of FK520.
Materials and methods
Strains, plasmids, primers, and culture conditions
All strains and plasmids used in this study are listed in Table 1. The primers used are listed in Table S1. The original strain S. hygroscopicus SFK-36 was generated from S. hygroscopicus var. ascomyceticus ATCC 14891. The slant/plate medium, seed medium, and culture conditions for S. hygroscopicus were prepared according to previously described methods (Yu et al. 2019). The seed culture was transferred into fermentation medium containing glycerol (80.0 g/L), soybean meal (40.0 g/L), KH2PO4 (0.2 g/L), FeSO4·7H2O (0.01 g/L), and ZnSO4·7H2O (0.01 g/L) at pH 6.5 and incubated at 28 °C and 200 rpm for 8 days in shaking flasks. For the fed-batch culture, aliquots of glycerol stock (500 g/L) were prepared, and 10 g/L of glycerol was added into the fermentation medium in flasks once a day. Scale-up fermentation of S. hygroscopicus in a 5-L fermenter was conducted as reported previously (Yu et al. 2019).
Traditional mutagenesis of S. hygroscopicus
Spores of S. hygroscopicus suspended in phosphate-buffered saline (PBS, 0.05 mol/L, pH 7.0) were prepared for the ultraviolet (UV) or N-methyl-N′-nitro-N-nitrosoguanidine (NTG) mutation treatments. For UV mutation, 2 mL of spore suspension (107 spores/mL) was placed into a sterilized plate and then subjected to UV radiation (254 nm, 15 W power) for 80 s at a distance of 30 cm. After mutation, spores were placed in a dark room for 30 min for dark repair. The UV-irradiated spores were diluted 10-fold and then plated onto a plate medium containing streptomycin at 0.4, 0.8, or 1.6 mg/L. For NTG mutation, NTG (final concentration at 1 mg/mL) was added into the spore suspension (107 spores/mL) at 28 °C and 200 rpm for 2 h. The NTG-treated spores were diluted and spread onto a plate medium containing streptomycin at 0.4 or 0.8 mg/L. After mutagenesis, 60 mutant strains were randomly selected and inoculated into the fermentation medium in shaking flasks at 200 rpm for 8 days. The mutant with the highest FK520 yield was obtained for the next round of mutagenesis. Additionally, the stability of these high-producing mutants obtained in each round of mutagenesis was detected by natural selection.
Genetic manipulation
All plasmids constructed in this study were introduced into S. hygroscopicus SFK-6-33 according to the Escherichia coli–Streptomyces conjugation method reported previously (Bierman et al. 1992; Kieser et al. 2000). The genomic DNA of S. hygroscopicus and S. coelicolor were obtained with a DNeasy Blood & Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions. All primers used are listed in Table S1 in Supplementary Materials. To investigate the importance of CCR encoded by fkbS for FK520 biosynthesis in S. hygroscopicus, plasmid pSET-dCas9-fkbS was constructed to inhibit the expression of fkbS. Since the sequence of fkbS was reported incompletely (Wu et al. 2000), sgRNA was designed based on the partial known sequence of fkbS and amplified with primers fkbS-D-F/fkbS-D-R. The fragment was then inserted into the SpeI/EcoRI sites of pSET-dCas9 using the ClonExpress II One Step Cloning Kit (Vazyme Biotech, China) according to the instructions provided by the manufacturer. Two genes, fkbS and ccr, both encoding CCR were amplified using primers fkbS-F/fkbS-R and ccr-F/ccr-R from the genomic DNA of S. hygroscopicus and S. coelicolor, respectively. The overexpression plasmids pSET-fkbS and pSET-ccr were obtained by inserting these two genes into the NdeI/AscI sites of pSET152. Plasmid pSET-2fkbS with two copies of fkbS was constructed based on pSET-fkbS. The fragment containing promoters PermE* and fkbS was cloned from pSET-fkbS with primers fkbS-F-2/fkbS-R-2 and then inserted into the AscI/NotI sites of pSET-fkbS. To construct pSET-3fkbS with three fkbS copies, another PermE*-fkbS fragment was amplified by fkbS-F-3/fkbS-R-3 and then inserted into the NotI/EcoRV sites of pSET-2fkbS. The DNA fragment accA2BE, composed of three genes encoding acetyl-CoA carboxylase (ACCase), namely, accA2, accB, and accE, was obtained by amplification from plasmid PLCB1-A2BE using primers A2BE-F/A2BE-R. Then, accA2BE was cloned into the NdeI/AscI sites of pSET152 to construct the overexpression plasmid pSET-A2BE. To realize the integration of multiple copies of ACCase in S. hygroscopicus, plasmid PLCB1-A2BE with two integrases (ΦC31, ΦBT1) and PLCBR1-A2BE with three integrases (ΦC31, ΦBT1, R4) were applied in this study. To overexpress the genes fkbS, accA2, accB, and accE together, accA2BE was amplified with primers fkbS-A2BE-F/fkbS-A2BE-R and then inserted into the AscI/EcoRV sites of pSET-fkbS to generate the combinatorial overexpression plasmid pSET-fkbS-A2BE.
The complete gene fkbS was amplified from genomic DNA of S. hygroscopicus SFK-6-33 using the degenerate primers fkbS-J-F/fkbS-J-R (Table S1) and then sequenced. PCR amplification was performed using PrimeSTAR GXL DNA Polymerase (Takara, China) with the following program: 98 °C for 5 min; 30 cycles of 98 °C for 30 s and 68 °C for 1 min; and 68 °C for 10 min.
Analysis of transcriptional levels by quantitative real-time PCR
The comparison of transcriptional levels of the FK520 biosynthetic gene cluster between S. hygroscopicus SFK-36 and SFK-6-33 by quantitative real-time PCR (qPCR) was performed according to the method reported previously. To determine the transcriptional levels of the fkbS gene in SFK-6-33, SFK-OfkbS, and SFK-DfkbS, the total RNA of these strains was isolated from culture broth at 3, 5 and 7 d using an Ultrapure RNA Kit (CWBIO, China). The residual DNA was removed with RNase-free DNase I (Takara, China). The quality of RNA samples was determined using a Nano-100 system (Hangzhou Allsheng Instruments Co., Ltd., China). In addition, the elimination of DNA was confirmed by PCR amplification with primers 16S-RT-F/16S-RT-R (Table S1). The fold changes of the fkbS gene in different samples were quantified using the 2-ΔΔCt method as described previously (Livak and Schmittgen 2001). The 16S rRNA gene was used as the internal control to normalize the transcriptional levels in qPCR analysis (Yu et al. 2019). The primers used in qPCR analysis are listed in Table S1.
Extraction and analysis of intracellular CoA-esters
Fermentation culture broth (1 mL) of S. hygroscopicus strains SFK-6-33, SFK-OfkbS, SFK-OA2BE, and SFK-OASN was collected at 6 days. Each sample was centrifuged at 12,000×g for 5 min to obtain mycelium and then washed twice with PBS buffer. The extraction of intracellular CoA-esters was carried out using a modified method reported previously (Armando et al. 2012; Xie et al. 2019). The mycelium was resuspended in 1 mL of PBS buffer and lysed by adding 2 mL of formic acid with intermittent vortexing for 30 min. The extraction was placed on ice for 30 min and then centrifuged at 12000×g for 5 min at 4 °C to collect the supernatant. The supernatant was instantly quenched and extracted by acetonitrile/methanol/0.1% glacial acetic acid (45:45:10, v/v). These samples were analyzed using ultra-high-performance liquid chromatography (UPLC, Waters, USA)-electrospray ionization (ESI)-tandem mass spectrometry (MS, AB SCIEX QTRAP 6500) as described previously with some modifications (Xia et al. 2013). The column ACQUITY UPLC HSS T3 1.8 μm, 2.1 × 100 MVK, was used in this study. The mobile phase composed of solvent A (0.1% formic acid in water) and solvent B (35% acetonitrile/65% water) was programmed as follows: 0–1.00 min, 98% A/2% B; 1.00–1.50 min, a linear gradient from 98% A/2% B to 40% A/60% B; 1.50–1.70 min, 40% A/60% B; 1.70–1.71 min, a linear gradient from 40% A/60% B to 98% A/2% B; and 1.71–2.00 min, 98% A/2% B. The concentration of CoA-esters was quantified using multiple reaction monitoring (MRM) mode in MS/MS with two mass ions: m/z parent > m/z daughter (malonyl-CoA, 854 > 347; methylmalonyl-CoA, 868 > 361; ethylmalonyl-CoA, 882 > 375) in positive-mode UPLC-ESI-MS/MS (Fig. S1). All authentic CoA-ester standards were purchased from Cayman Chemical (USA).
Analysis methods
To better separate FK523 from FK520, an analytical gradient elution method for HPLC was applied using an Agilent C18 column (4.6 × 150 mm, 3.5 μm), following the program with solvent A (0.01% acetic acid/10% acetonitrile/90% water) and solvent B (0.01% acetic acid in acetonitrile): 0–25 min, 60% A/40% B; 25–35 min, a linear gradient from 60% A/40% B to 55% A/45% B; 35–45 min, a linear gradient from 55% A/45% B to 30% A/70% B; 45–47 min, a linear gradient from 30% A/70% B to 10% A/90% B; 47–50 min, a linear gradient from 10% A/90% B to 60% A/40% B. The UV detection was set at 205 nm with a flow rate of 2 mL/min at 60 °C.
The biomass of each fermentation sample was determined as packed mycelium volume (PMV) carried out using a method presented previously (Yu et al. 2019). Dry cell weight (DCW) of different samples was measured according to the method reported previously (Wang et al. 2017b). The residual glycerol was quantified using HPLC with a refractive index detector (RID-10A, Shimadzu, Japan) in a Welch Hilic-NH2 column (5 μm, 4.6 × 250 mm). The mobile phase was composed of deionized water with acetonitrile (20:80, v/v) at a flow rate of 1.0 mL/min.
Statistics
Each experiment was replicated three times, with the error bars showing the standard deviations (SDs). To compare the difference between the test and control data, P values were calculated by Student’s t test (P < 0.001).
The complete sequence of fkbS cloned from S. hygroscopicus SFK-6-33 has been deposited in GenBank with the Accession No. MN372206.
Results
Mutation and screening for FK520 high-producing strains
To obtain FK520 high-producing strains, S. hygroscopicus was treated with UV radiation or NTG mutagen and screened by the streptomycin resistance screening model. The genealogical tree of S. hygroscopicus SFK-6-33 is presented in Fig. 2a. Initially, spores of S. hygroscopicus SFK-36 stored in 20% (v/v) glycerol were diluted and then spread on the plate medium to restore its genetic characteristics. After multi-round of natural selection, a stable high-producing strain SFK-1-15 (1630.3 mg/L) was chosen for traditional mutagenesis. Under the selective pressure of 0.4 mg/L of streptomycin, SFK-2-3 was obtained with UV mutagenesis, and the FK520 production of this mutant reached 1905.1 mg/L, which is 16.9% higher than that of SFK-1-15. However, NTG mutagenesis combined with 0.4 mg/L streptomycin had no obvious effect on improving the yield of FK520 in S. hygroscopicus. The FK520 highest yield of the mutants generated by NTG was 1725.5 mg/L, showing no significant difference compared with that of SFK-1-15. To further enhance FK520 production, a higher streptomycin addition (0.8 mg/L) combined with UV radiation was applied for mutant breeding. The obtained mutant SFK-3-48 presented an FK520 yield of 2214.3 mg/L, attaining a 16.2% increase compared with SFK-2-3. However, in subsequent rounds of mutagenesis, the improvement of FK520 production in each round of mutagenesis decreased to less than 5%, even when streptomycin addition was increased to 1.6 mg/L. Finally, a stable high-producing mutant SFK-6-33 was obtained by UV radiation and the streptomycin resistance screening model. The FK520 production by SFK-6-33 reached 2432.2 mg/L, which is 53.2% higher than that of the original strain SFK-36.
Traditional mutagenesis breeding for S. hygroscopicus. a Genealogical tree of S. hygroscopicus SFK-6-33 with UV radiation and streptomycin resistance screening model. SMr*, 0.4 mg/L streptomycin; SMr**, 0.8 mg/L streptomycin; SMr***, 1.2 mg/L streptomycin. b The time course of FK520 production and growth of SFK-36 and SFK-6-33 in shaking flasks. c Relative expression levels of FK520 biosynthetic gene cluster in S. hygroscopicus SFK-6-33, compared with those of SFK-36. In total, seven genes fkbW, fkbU, fkbR1, fkbE, fkbB, fkbO, and fkbS were selected to indicate the expression level of each co-transcription units. RNA samples were collected from the culture broth of SFK-6-33 and SFK-36 grown in a fermentation medium at 4 and 6 days, respectively. The gene 16S rRNA was used as an internal control to normalize the transcriptional levels. Error bars show standard deviations; ***P < 0.001
The time course of FK520 production clearly revealed that the mutant SFK-6-33 could produce more FK520 than SFK-36, but with no significant difference in biomass during the fermentation process in flasks (Fig. 2b). The relative expression levels of the FK520 biosynthetic gene cluster between SFK-6-33 and SFK-36 were also investigated by qPCR analysis. The biosynthetic gene cluster of FK520 consisting of 22 genes was divided into seven co-transcription units (Wu et al. 2000; Yu et al. 2019). As depicted in Fig. 2c, the results indicated that transcriptional levels of most of the co-transcription units were significantly increased in mutant SFK-6-33 (P < 0.001), including fkbW, fkbU, fkbR1/fkbR2, fkbB/C/L/K/J/I/H, fkbO/P/A/D/M, and fkbS/Q/N, which may closely lead to the improvement of FK520 accumulation in S. hygroscopicus SFK-6-33.
Identification of FkbS using the CRISPR/dCas9 inhibition system
Since the complete genomic information of S. hygroscopicus was unknown and the sequence of the gene fkbS encoding CCR was reported to be incomplete (only 460 bp has been identified), the investigation and application of endogenous FkbS in S. hygroscopicus were limited. To determine the vital role of FkbS in the biosynthesis of FK520, the CRISPR/dCas9 system was applied to repress the expression of fkbS. As shown in Fig. 3a, the biomass of SFK-DfkbS showed no significant differences after introduction of the plasmid pSET-dCas9-fkbS, indicating that the CRISPR/dCas9 system constructed in S. hygroscopicus has no adverse effect on the growth of mycelium. However, the production of FK520 by SFK-OfkbS decreased to 841.4 mg/L due to the sharply decreased expression of fkbS compared with that in SFK-6-33. These results firmly confirmed that FkbS plays an important role in the biosynthesis of FK520. Additionally, the FK523/FK520 impurity proportion increased to 21.8%, suggesting that the inhibited activity of FkbS exerts no influence on the accumulation of FK523.
The functional identification of FkbS and enhancement of FK520 production with overexpression of fkbS. a Changes in FK520 production and impurity ratio of FK523/FK520 between parent strain SFK-6-33 and engineering strain SFK-DfkbS. CRISPR/dCas9 system was used to repress the expression of fkbS in SFK-DfkbS. The culture broth of SFK-6-33, SFK-DfkbS, and SFK-dCas9(−) were collected at 8 days to determine the biomass and production of FK520 and FK523. Error bars represent the standard deviations. b The transcriptional levels of fkbS in SFK-DfkbS and SFK-OfkbS compared with parent strain SFK-6-33 by qPCR. RNA samples were collected from the culture broth of these three strains at 3, 5, and 7 days in a fermentation medium. The error bars show the standard deviations of three independent replicates. ***P < 0.001. c Amino acid alignments of partial sequence of crotonyl-CoA carboxylase/reductases (CCR) of different Streptomyces spp. The comparison of first seven amino acids in amino acid sequence from nine CCRs was marked with a red box. In total, CCRs listed here were from nine different strains including S. coelicoflavus (NCBI Accession No. WP_007387963), S. coelicolor (NCBI Accession No. NP_630556), S. mirabilis (NCBI Accession No. WP_037706421), S. griseofuscus (NCBI Accession No. WP_037654730), S. Venezuela (NCBI Accession No. AGX26728), S. tsukubaensis (NCBI Accession No. EIF93549), S. lasaliensis (NCBI Accession No. WP_137304740), S. NBRC 110611 (NCBI Accession No. WP_066930901), and S. rapamycinicus (NCBI Accession No. WP_020866310). d The amplification of gene fkbS from genome of S. hygroscopicus SFK-6-33. 1, PCR amplification product of fkbS with primers fkbS-J-F/ fkbS-J-R; M, 1-kb gene ruler. e Comparison of FK520 production and FK523/FK520 impurity ratio between SFK-6-33 and its engineering strains, including SFK-OfkbS, SFK-OccrC, and SFK-pSET(−). Crotonic acid (Cr, 1 g/L) was added into the fermentation medium for SFK-6-33 and SFK-OfkbS. Each culture broth was collected for HPLC analysis of FK520 and FK523 production at 8 days in shaking flasks. The error bars represent the standard deviations of three biological replicates. f Concentrations of intracellular CoA-esters of SFK-6-33, SFK-OfkbS, and SFK-OfkbS supplemented with 1 g/L Cr. The mycelium samples for LC-MS/MS analysis were isolated from culture broth at 6 days. The error bars show the standard deviations of three independent experiments
Cloning fkbS with degenerate oligonucleotide primers
Because the complete sequence of FkbS was unknown, degenerate primers were used to amplify the fkbS fragment from the genome of S. hygroscopicus. Amino acid alignments of nine CCRs from different Streptomyces spp. are listed in Fig. 3c. It is found that the first seven amino acids in the sequences are relatively conserved. Valine or methionine is listed in the first position; thus, the corresponding codon is RTG. While there are multiple possibilities in the second position, the corresponding codon is set as NNN. Codon GAN is used to represent aspartic acid or glutamic acid in the third position. Isoleucine is located in the fourth position among the nine genes; therefore, its corresponding codon is ATH. The fifth amino acid is isoleucine or leucine, so the codon is set as HTN. In the sixth position, codon NNN is set to include various possibilities. Alanine is the only amino acid in the seventh position according to the sequence alignments; thus, its corresponding codon is GCN. In general, the degenerate primer fkbS-J-F was set as 5′-RTGNNNGANATHHTNNNNGCN-3′ (R=A, G; H= A, T, C; N= A, T, C, G). The DNA fragment of fkbS (1362 bp) was amplified from the genome of S. hygroscopicus with primers fkbS-J-F/fkbS-J-R (Fig. 3d). Finally, the complete sequence of fkbS was determined with sequencing, and it is consistent with the reported partial sequence (Wu et al. 2000).
Overexpression of FkbS in S. hygroscopicus
FkbS, an endogenous CCR of S. hygroscopicus, can convert crotonyl-CoA to ethylmalonyl-CoA, which is the major precursor for FK520 biosynthesis (Wu et al. 2000). To enhance CCR activity, fkbS under the control of PermE* was integrated into SFK-6-33 through conjugal transfer to obtain the two-copy CCR strain SFK-OfkbS. A significant increase in the transcriptional level of fkbS was detected during fermentation in SFK-OfkbS (Fig. 3b). For comparison, an exogenous CCR derived from S. coelicolor was also integrated into the ΦC31 integration site of S. hygroscopicus SFK-6-33, generating the engineered strain SFK-OccrC. The amino acid alignment of FkbS and CCR is listed in Fig. S2, showing 70.8% sequence identity. Overexpression of FkbS increased the production of FK520 from 2432.2 to 2817.0 mg/L, representing a 15.8% increase in FK520 yield, and decreased the FK523/FK520 impurity proportion to 6.3% as well (Fig. 3e). However, overexpression of the exogenous CCR showed no significant effect on FK520 production and FK523 biosynthesis. The results indicated that endogenous CCR may have a better effect than exogenous CCR for improving FK520 production in the high-yield strain SFK-6-33. Compared with SFK-6-33, a nearly 40% increase in intracellular ethylmalonyl-CoA concentration was detected in SFK-OfkbS (Fig. 3f), which strongly implied that a sufficient supply of ethylmalonyl-CoA (147.9 pmol/g DCW) could lead to enhanced FK520 biosynthesis in S. hygroscopicus as well as reduced production of the impurity FK523. Afterwards, two or three additional copies of fkbS were introduced into SFK-6-33, respectively (Fig. S3). When two copies of fkbS were introduced into SFK-6-33, FK520 production reached 2649.5 mg/L without further increase compared with SFK-OfkbS. For the three-copy strain SFK-OfkbS-3, the biomass decreased to 19.6% of PMV, indicating the inhibited growth of mycelium, which led to a marked decrease in FK520 production (1943.3 mg/L).
Increased ethylmalonyl-CoA supply in SFK-OfkbS supplemented with crotonic acid
According to the proposed synthesis pathway of ethylmalonyl-CoA (Fig. 1), crotonyl-CoA could be catalytically converted into ethylmalonyl-CoA by FkbS for the biosynthesis of FK520. Therefore, we tried to increase the concentration of intracellular crotonyl-CoA by adding exogenous crotonic acid into the fermentation medium to further increase the intracellular pool of ethylmalonyl-CoA in SFK-OfkbS. As depicted in Fig. 3e, when SFK-OfkbS was cultured in fermentation medium supplemented with 1.0 g/L crotonic acid, a 1.3-fold (3063.6 mg/L) enhanced FK520 yield was attained compared with that in SFK-6-33. Additionally, the FK523/FK520 impurity proportion further decreased to 5.3%. In addition, an increased concentration of intracellular ethylmalonyl-CoA (174.1 pmol/g DCW) was observed in the SFK-OfkbS strain supplemented with crotonic acid. In contrast, the concentrations of malonyl-CoA and methylmalonyl-CoA showed no significant difference in crotonic acid-supplemented SFK-OfkbS compared with SFK-6-33. These results suggested that crotonic acid can be converted into intracellular crotonyl-CoA and then transformed into ethylmalonyl-CoA by FkbS. The increased supply of intracellular ethylmalonyl-CoA in SFK-OfkbS with crotonic acid addition could significantly increase the biosynthesis of FK520 and reduce the production of the impurity FK523.
Subsequently, the effects of different crotonic acid additions on mycelium growth and FK520 production in SFK-OfkbS were also investigated. As shown in Fig. S4, the highest FK520 production of 3063.6 mg/L was obtained in SFK-OfkbS with 1.0 g/L crotonic acid. A decrease in biomass and FK520 yield was observed when the crotonic acid addition was more than 1.0 g/L, indicating that excessive crotonic acid addition could inhibit the growth of mycelium.
Overexpression of ACCase in S. hygroscopicus
It has been reported that acetyl-CoA carboxylase can convert acetyl-CoA into malonyl-CoA (Ryu et al. 2006), and the latter is an important precursor of the FK520 macrocyclic structure (Wu et al. 2000). Therefore, ACCase was cloned from S. coelicolor and then integrated into SFK-6-33, generating the recombinant strain SFK-OA2BE. As presented in Fig. 4a, compared with that in SFK-6-33, a 2.1-fold higher (478.3 pmol/g DCW) malonyl-CoA concentration was observed in SFK-OA2BE during fermentation in shaking flasks. The significantly increased malonyl-CoA supply led to a 36.1% increase in FK520 production (3320.1 mg/L) in SFK-OA2BE (Fig. 4b). Interestingly, an increased intracellular methylmalonyl-CoA concentration (99.2 pmol/g DCW) was detected in SFK-OA2BE, which was 41.7% higher than that in SFK-6-33. However, the concentration of ethylmalonyl-CoA between SFK-OA2BE and SFK-6-33 showed no significant difference. It is likely that the increased malonyl-CoA levels could increase the concentration of methylmalonyl-CoA, but no significant increase was observed in the ethylmalonyl-CoA supply, which may lead to a failure in reducing the FK523/FK520 impurity ratio in SFK-OA2BE. To further enhance the activity of ACCase, two or three copies of A2BE were then integrated into SFK-6-33. However, the growth of mycelium was inhibited, leading to decreased FK520 production in S. hygroscopicus (Fig. S5).
The concentrations of intracellular CoA-esters and production of FK520 and FK523 in engineering strains. a Changes of concentrations of intracellular CoA-esters in SFK-6-33 and its engineering strains. Four strains, including parent strain SFK-6-33, SFK-OfkbS, SFK-OA2BE, and SFK-OASN, were cultured in the fermentation medium supplemented with 1 g/L crotonic acid and the samples prepared for CoA-esters analysis by LC-MS/MS were collected at 6 days. Error bars indicate the standard deviations. b Comparison of FK520 production and ratio of FK523/FK520 between SFK-6-33, SFK-OfkbS, SFK-OA2BE, and SFK-OASN. These four strains were cultured in the fermentation medium supplemented with 1 g/L crotonic acid for 8 days. The error bars represent the standard deviations of three biological replicates. c HPLC analysis of FK520 and FK523 production of SFK-6-33, SFK-OfkbS, SFK-OA2BE, and SFK-OASN in fermentation broths cultured in shaking flasks for 8 days
Combinatorial overexpression of FkbS and ACCase in S. hygroscopicus
When ACCase was overexpressed in SFK-6-33, enhanced malonyl-CoA biosynthesis led to the increased production of methylmalonyl-CoA; however, no obvious increase in ethylmalonyl-CoA supply was detected. This phenomenon indicated that some limiting factors in the ethylmalonyl-CoA biosynthesis pathway may hinder the conversion from malonyl-CoA to ethylmalonyl-CoA. The above results confirmed that overexpression of FkbS could enhance the biosynthesis of ethylmalonyl-CoA. Therefore, we attempted to further increase the intracellular pool of ethylmalonyl-CoA through combinatorial overexpression of FkbS and ACCase in S. hygroscopicus SFK-6-33. As illustrated in Fig. 4a, the malonyl-CoA concentration decreased to 399.6 pmol/g DCW in SFK-OASN, which was 16.5% lower than that in SFK-OA2BE. The concentration of methylmalonyl-CoA also slightly decreased to 80.7 pmol/g DCW. Nevertheless, the supply of ethylmalonyl-CoA significantly increased to 219.9 pmol/g DCW, which was 77.8% higher than that in SFK-OA2BE. Figure 4b shows that co-overexpression of FkbS and ACCase could simultaneously increase the supply of ethylmalonyl-CoA and malonyl-CoA to further enhance the production of FK520 (3511.4 mg/L), also reducing the FK523/FK520 impurity ratio to 5.6%.
Effect of glycerol addition on FK520 production by SFK-OASN in shake flask culture
It has been reported that microbial secondary metabolite production can be promoted by adding appropriate nutrients (Xia et al. 2013). A fed-batch culture strategy for SFK-OASN was then developed to increase FK520 production in shake flask culture. In the fermentation medium, the initial glycerol concentration was 80 g/L. To determine the optimal glycerol feeding strategy, 10 g/L of glycerol was added to the fermentation medium once a day, totalling three, four, or five times (Fig. 5). The glycerol addition started on the third day, when the residual glycerol in the culture was approximately 30 g/L. The time course of SFK-OASN cultured in fermentation medium without batch addition of glycerol is shown in Fig. 5a. Glycerol was rapidly consumed from 80 to 32 g/L in the period of 0 to 3 days, and the consumption rate of glycerol reached 0.67 g/(L h). During this period, the biomass increased to 19.0%, and FK520 production reached 1038.7 mg/L. The consumption rate of glycerol decreased to 0.38 g/(L h) in the period of 3 to 5 days, and the biomass reached the highest value of 31.5%. Subsequently, biomass decreased gradually with decreasing residual glycerol from the fifth day, indicating that glycerol was inadequate for the growth of SFK-OASN in the late stage of fermentation. The FK520 yield reached 3511.4 mg/L at 8 days. Similar results were observed when the fermentation medium was supplemented with glycerol three times (Fig. 5b) compared with no addition. However, when the culture broth was fed for four times at a total glycerol supplement of 40 g/L, the emergence of the highest biomass was postponed to the sixth day, reaching 34.5% (Fig. 5c). These results indicated that this feeding strategy provided sufficient glycerol supply for mycelium growth and delayed the ageing time of mycelium as well. The FK520 yield was 3753.4 mg/L, attaining a 6.8% increase compared with no addition. When glycerol was added to the culture for five times (Fig. 5d), the biomass also reached the highest value of 35.5% on the sixth day. However, the final glycerol concentration was still higher than 20 g/L owing to the decreased metabolic capacity of SFK-OASN in the late fermentation stage (7–8 days). Excessive glycerol concentration in the late stage could inhibit the growth and metabolism of mycelium, leading to decreased FK520 production (3343.2 mg/L).
Effect of glycerol supplement on FK520 production of S. hygroscopicus SFK-OASN in shaking flasks. a Time course of FK520 production and growth of SFK-OASN in a flask containing fermentation medium, with no supplemental glycerol. In the fed-batch culture, 10 g/L of glycerol was added to the fermentation medium once a day from 3 to 5 days (b); from 3 to 6 days (c); from 3 to 7 days (d). Fk520 production, glycerol, and PMV are the mean values of triplicate detections
Scale-up fermentation in a 5-L bioreactor
The fed-batch culture of S. hygroscopicus SFK-OASN was carried out in a 5-L fermenter. As depicted in Fig. 6, the residual glycerol decreased from 80.0 to 39.3 g/L during the early stage of fermentation (0–72 h), reflecting a fast consumption of glycerol in the fermenter, which is similar to that in shaking flasks. Based on the results of the glycerol feeding strategy in flasks, at 72 h, 10 g/L of glycerol was added into the fermenter once a day, totalling four times. A total of 40 g/L of glycerol was added into the fermenter to maintain a residual glycerol level higher than 20 g/L in the period from 72 to 168 h. With the consumption of glycerol, the PMV remained higher than 30%, but the pH decreased continuously. To avoid the inhibition of cell growth caused by low pH conditions, the pH was maintained at approximately 5.6 by 1 mol/L ammonia according to our previous report. Finally, 3913.9 mg/L FK520 with a 5.1% FK523/FK520 ratio was obtained at 168 h. The scale-up fermentation performed in a 5-L fermenter confirmed that the rational strategy of feeding glycerol in the fermentation could greatly enhance the biosynthesis of FK520 and reduce the proportion of the impurity FK523 in S. hygroscopicus SFK-OASN.
Scale-up fermentation of S. hygroscopicus SFK-OASN cultured in a 5-L fermenter. The aeration was kept at 1.0 vvm (air/culture volume/min). The agitation rate was set automatically to maintain dissolved oxygen levels higher than 20% during the culture process, and pH was maintained higher than 5.6 using 1 mol/L ammonia. For the fed-batch culture, 10 g/L of glycerol was added into the fermenter every 24 h from 72 to 144 h. Error bars show the standard deviations of three samples collected from the fermenter
Discussion
To date, some efforts have been made to improve FK520 production in S. hygroscopicus through traditional mutagenesis. For example, a titanium sapphire laser and shikimic acid resistance screening were applied to S. hygroscopicus ATCC 14891 to generate the two high-yield strains FS35 and SA68, with FK520 yields of 305.6 mg/L and 450.0 mg/L, respectively (Qi et al. 2012; Qi et al. 2014b). In our previous work, a stable high-producing strain SFK-36 yielding 1476.9 mg/L FK520 was obtained though atmospheric and room temperature plasma mutagenesis combined with medium optimization (Yu et al. 2019). In this study, classical ultraviolet radiation coupled with streptomycin resistance mutagenesis was used with SFK-36 to further improve FK520 production. Ribosome engineering can introduce random mutations on ribosomes in microbes, therefore generating drug-resistant mutants with the enhanced production of secondary metabolites (Ochi and Hosaka 2013). Our work is the first reported application of ribosome engineering in S. hygroscopicus to improve FK520 accumulation. The stable high-producing strain SFK-6-33 (2432.2 mg/L) was successfully obtained from original strain SFK-36 through traditional mutagenesis, suggesting that this combination mutagenesis was an effective method for improving FK520 production in S. hygroscopicus and providing a parent strain with good characteristics for further genetic modification.
Since the genomic information of S. hygroscopicus was unknown and the biosynthesis gene cluster of FK520 was reported to be incomplete, functional investigation of the fkbS gene encoding CCR was restricted (Wu et al. 2000). The CRISPR/dCas9 inhibition system contains a nuclease-deficient Cas9, which can bind to the target gene location to block transcription without gene cleavage. This approach is a convenient way to repress gene expression compared with traditional gene knockout or knockdown and has recently been used for functional gene identification in microbes (Wang et al. 2016; Zhao et al. 2018). In this study, the CRISPR/dCas9 system targeting a partially reported sequence of the fkbS gene was established to repress the expression of fkbS in S. hygroscopicus. The significantly decreased FK520 production verified the important role of FkbS in the biosynthesis of FK520. Therefore, the complete sequence of fkbS was cloned from S. hygroscopicus using degenerate primers and then integrated into SFK-6-33 to enhance the activity of FkbS. A significant increase in FK520 production and decrease in FK523 accumulation were observed in FkbS overexpression strain SFK-OfkbS. It has been reported that introduction of an exogenous CCR cloned from S. coelicolor into the low-yield strain FS35 increased FK520 production from 305.6 to 361.7 mg/L (Wang et al. 2017b). However, when we integrated this exogenous CCR into the high-producing strain SFK-6-33, it exerted no significant influence on FK520 and FK523 accumulation, suggesting that FkbS played a more important role than exogenous CCR for increasing FK520 production. Differences in the amino acid sequences between FkbS and CCR may lead to differences in catalytic activity (Fig. S2).
Acyl-CoA precursors can be catalyzed by polyketide synthase and directly participate in the synthesis of the macrolide skeleton of antibiotics (Chen et al. 2012; Lu et al. 2016). An increased supply of CoA-esters has been reported to promote the accumulation of antibiotics in microbes, such as S. coelicolor (Ryu et al. 2006), S. albus (Lu et al. 2016), and Actinosynnema pretiosum (Lin et al. 2011). Three important CoA esters (ethylmalonyl-CoA, methylmalonyl-CoA, and malonyl-CoA) are involved in the biosynthesis of FK520 in S. hygroscopicus (Fig. 1). Ethylmalonyl-CoA can be recognized by acyltransferase, but the substrate specificity of acyltransferase is not strict, usually leading to the production of some analogues in Streptomyces sp., such as FK523 in FK520 production (Dunn and Khosla 2013), FK523 and FK520 in tacrolimus production (Kosec et al. 2012), and monensin B in monensin A production (Liu and Reynolds 1999). It is difficult to separate these structural analogues due to their similar structures and chemical properties. In this work, the ethylmalonyl-CoA supply was increased by overexpressing endogenous FkbS, leading to an enhanced FK520 yield (2817.0 mg/L) in S. hygroscopicus. The FK523/FK520 impurity proportion was also reduced from 9.6 to 6.3% due to adjustment of the CoA-ester supply (Fig. 3). According to the metabolic pathways of ethylmalonyl-CoA reported previously (Chan et al. 2009; Jung et al. 2014), there are still some other potential targets to enhance ethylmalonyl-CoA accumulation, including PCC overexpression or deletion of ethylmalonyl-CoA mutase encoded by MeaA. However, enhanced PCC can also increase methylmalonyl-CoA concentration by converting propionyl-CoA to methylmalonyl-CoA (Jung et al. 2011), and may lead to the increase of impurity FK523. In addition, the deletion of MeaA is difficult to carry out because the sequence of MeaA in S. hygroscopicus is unknown. It is noted that the exogenous addition of crotonic acid could further increase the intracellular concentration of ethylmalonyl-CoA, promoting FK520 accumulation in SFK-OfkbS. It has been reported that the addition of precursors is an efficient way to increase secondary metabolite production in microbes. For example, increased rapamycin production was obtained by adding 1 mM of propionate to a propionyl-CoA carboxylase overexpression strain UV2-2/PCC (Jung et al. 2011). Additionally, the malonyl-CoA supply was increased in SFK-OA2BE with the integration of ACCase, leading to increased FK520 production in SFK-OA2BE. Interestingly, the intracellular concentration of ethylmalonyl-CoA exhibited no increase compared with methylmalonyl-CoA and malonyl-CoA, suggesting that an inadequate ethylmalonyl-CoA supply may be the limiting factor for further increasing FK520 accumulation in SFK-OA2BE. Therefore, FkbS and ACCase were overexpressed in SFK-6-33 to increase the supply of ethylmalonyl-CoA and malonyl-CoA at the same time, achieving an increased FK520 production and decreased impurity ratio simultaneously.
Above all, the stable high-producing strain, S. hygroscopicus SFK-6-33, was obtained through multiple rounds of traditional mutagenesis by combining ultraviolet radiation with streptomycin resistance screening. Afterwards, the intracellular pools of ethylmalonyl-CoA and malonyl-CoA increased with the overexpression of FkbS and ACCase in SFK-6-33, and the increased supply of these CoA-esters significantly enhanced FK520 biosynthesis and reduced the proportion of the impurity FK523. This study is the first to report a strategy to inhibit production of the impurity FK523 by adjusting the CoA-ester supply, showing a cost-effective method for FK520 production. A fed-batch culture strategy for SFK-OASN was also developed in a 5-L fermenter, attaining the highest FK520 yield reported thus far. These findings suggest that traditional mutagenesis coupled with metabolic engineering is an effective method for improving FK520 production in S. hygroscopicus.
References
Armando JW, Boghigian BA, Pfeifer BA (2012) LC-MS/MS quantification of short-chain acyl-CoA’s in Escherichia coli demonstrates versatile propionyl-CoA synthetase substrate specificity. Lett Appl Microbiol 54(2):140–148
Bérdy J (2005) Bioactive Microbial Metabolites. J Antibiot 58(1):1–26
Bierman M, Logan R, Obrien K, Seno ET, Rao RN, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116(1):43–49
Chan YA, Podevels AM, Kevany BM, Thomas MG (2009) Biosynthesis of polyketide synthase extender units. Nat Prod Rep 26(1):90–114
Chen D, Zhang Q, Zhang Q, Cen P, Xu Z, Liu W (2012) Improvement of FK506 production in Streptomyces tsukubaensis by genetic enhancement of the supply of unusual polyketide extender units via utilization of two distinct site-specific recombination systems. Appl Environ Microbiol 78(15):5093–5103
Dunn BJ, Khosla C (2013) Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J R Soc Interface 10(85):20130297–20130297
Eichenfield LF, Lucky AW, Boguniewicz M, Langley RG, Cherill R, Marshall K, Bush C, Graeber M (2002) Safety and efficacy of pimecrolimus (ASM 981) cream 1% in the treatment of mild and moderate atopic dermatitis in children and adolescents. J Am Acad Dermatol 46(4):495–504
Jung WS, Yoo YJ, Park JW, Park SR, Han AR, Ban YH, Kim EJ, Kim E, Yoon YJ (2011) A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl Microbiol Biotechnol 91(5):1389–1397
Jung WS, Kim E, Yoo YJ, Ban YH, Kim EJ, Yoon YJ (2014) Characterization and engineering of the ethylmalonyl-CoA pathway towards the improved heterologous production of polyketides in Streptomyces venezuelae. Appl Microbiol Biotechnol 98(8):3701–3713
Kieser T, Bibb MJ, Butter MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics: a laboratory manual. The John Innes Foundation, Norwich
Kosec G, Goranovic D, Mrak P, Fujs S, Kuscer E, Horvat J, Kopitar G, Petkovic H (2012) Novel chemobiosynthetic approach for exclusive production of FK506. Metab Eng 14(1):39–46
Li L, Wei K, Liu X, Wu Y, Zheng G, Chen S, Jiang W, Lu Y (2019) aMSGE: advanced multiplex site-specific genome engineering with orthogonal modular recombinases in Actinomycetes. Metab Eng 52:153–167
Lin J, Bai L, Deng Z, Zhong J (2011) Enhanced production of ansamitocin P-3 by addition of isobutanol in fermentation of Actinosynnema pretiosum. Bioresour Technol 102(2):1863–1868
Liu H, Reynolds KA (1999) Role of crotonyl coenzyme A reductase in determining the ratio of polyketides monensin A and monensin B produced by Streptomyces cinnamonensis. J Bacteriol 181(21):6806–6813
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25(4):402–408
Lu C, Zhang X, Jiang M, Bai L (2016) Enhanced salinomycin production by adjusting the supply of polyketide extender units in Streptomyces albus. Metab Eng 35:129–137
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97(1):87–98
Qi H, Xin X, Li S, Wen J, Chen Y, Jia X (2012) Higher-level production of ascomycin (FK520) by Streptomyces hygroscopicus var. ascomyceticus irradiated by femtosecond laser. Biotechnol Bioprocess Eng 17(4):770–779
Qi H, Zhao S, Fu H, Wen J, Jia X (2014a) Enhancement of ascomycin production in Streptomyces hygroscopicus var. ascomyceticus by combining resin HP20 addition and metabolic profiling analysis. J Ind Microbiol Biotechnol 41(9):1365–1374
Qi H, Zhao S, Wen J, Chen Y, Jia X (2014b) Analysis of ascomycin production enhanced by shikimic acid resistance and addition in Streptomyces hygroscopicus var. ascomyceticus. Biochem Eng J 82:124–133
Qi H, Lv M, Song K, Wen J (2017) Integration of parallel 13C-labeling experiments and in silico pathway analysis for enhanced production of ascomycin. Biotechnol Bioeng 114(5):1036–1044
Ryu Y, Butler MJ, Chater KF, Lee KJ (2006) Engineering of primary carbohydrate metabolism for increased production of actinorhodin in Streptomyces coelicolor. Appl Environ Microbiol 72(11):7132–7139
Song K, Wei L, Liu J, Wang J, Qi H, Wen J (2017) Engineering of the LysR family transcriptional regulator FkbR1 and its target gene to improve ascomycin production. Appl Microbiol Biotechnol 101(11):4581–4592
Wang Y, Zhang ZT, Seo SO, Lynn P, Lu T, Jin Y, Blaschek HM (2016) Gene transcription repression in Clostridium beijerinckii using CRISPR-dCas9. Biotechnol Bioeng 113(12):2739–2743
Wang C, Liu J, Liu H, Wang J, Wen J (2017a) A genome-scale dynamic flux balance analysis model of Streptomyces tsukubaensis NRRL18488 to predict the targets for increasing FK506 production. Biochem Eng J 123:45–56
Wang J, Wang C, Song K, Wen J (2017b) Metabolic network model guided engineering ethylmalonyl-CoA pathway to improve ascomycin production in Streptomyces hygroscopicus var. ascomyceticus. Microb Cell Fact 16(1):169
Wei K, Wu Y, Li L, Jiang W, Hu J, Lu Y, Chen S (2018) MilR2, a novel TetR family regulator involved in 5-oxomilbemycin A3/A4 biosynthesis in Streptomyces hygroscopicus. Appl Microbiol Biotechnol 102(20):8841–8853
Wu K, Chung L, Revill WP, Katz L, Reeves CD (2000) The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC 14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251(1):81–90
Xia M, Huang D, Li S, Wen J, Jia X, Chen Y (2013) Enhanced FK506 production in Streptomyces tsukubaensis by rational feeding strategies based on comparative metabolic profiling analysis. Biotechnol Bioeng 110(10):2717–2730
Xie H, Zhao Q, Zhang X, Kang Q, Bai L (2019) Comparative functional genomics of the acarbose producers reveals potential targets for metabolic engineering. Synth Syst Biotechnol 4(1):49–56
Yu Z, Shen X, Wu Y, Yang S, Ju D, Chen S (2019) Enhancement of ascomycin production via a combination of atmospheric and room temperature plasma mutagenesis in Streptomyces hygroscopicus and medium optimization. AMB Express 9(1):25
Zhao Y, Li L, Zheng G, Jiang W, Deng Z, Wang Z, Lu Y (2018) CRISPR/dCas9-Mediated multiplex gene repression in Streptomyces. Biotechnol J 13(9):1800121
Acknowledgments
We thank the support from Professor Yinhua Lu (College of Life and Environmental Sciences, Shanghai Normal University), and Professor Weihong Jiang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for kindly providing the plasmid pSET-dCas9, PLCB1-A2BE and PLCBR1-A2BE.
Funding
This study was sponsored by Program of Shanghai Technology Research Leader (19XD1433200).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 565 kb)
Rights and permissions
About this article
Cite this article
Yu, Z., Lv, H., Wu, Y. et al. Enhancement of FK520 production in Streptomyces hygroscopicus by combining traditional mutagenesis with metabolic engineering. Appl Microbiol Biotechnol 103, 9593–9606 (2019). https://doi.org/10.1007/s00253-019-10192-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10192-8