Abstract
The polyene macrolide rimocidin, produced by Streptomyces rimosus M527, is highly effective against a broad range of fungal plant pathogens, but at low yields. Elicitation is an effective method of stimulating the yield of bioactive secondary metabolites. In this study, the biomass and filtrate of a culture broth of Escherichia coli JM109, Bacillus subtilis WB600, Saccharomyces cerevisiae, and Fusarium oxysporum f. sp. cucumerinum were employed as elicitors to promote rimocidin production in S. rimosus M527. Adding culture broth and biomass of S. cerevisiae (A3) and F. oxysporum f. sp. cucumerinum (B4) resulted in an increase of rimocidin production by 51.2% and 68.3% respectively compared with the production under normal conditions in 5-l fermentor. In addition, quantitative RT-PCR analysis revealed that the transcriptions of ten genes (rimA to rimK) located in the gene cluster involved in rimocidin biosynthesis in A3 or B4 elicitation experimental group were all higher than those of a control group. Using a β-glucuronidase (GUS) reporter system, GUS enzyme activity assay, and Western blot analysis, we discovered that elicitation of A3 or B4 increased protein synthesis in S. rimosus M527. These results demonstrate that the addition of elicitors is a useful approach to improve rimocidin production.
Key Points
• An effective strategy for enhancing rimocidin production in S. rimosus M527 is demonstrated.
• Overproduction of rimocidin is a result of higher expressed structural genes followed by an increase in protein synthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptomyces sp. is well known to produce several valuable natural products with structural complexity and with diverse biological activities (Kemung et al. 2018; Liu et al. 2018; Olanrewaju and Babalola 2019). Rimocidin, a 28-membered tetraene macrolide comprising a large lactone ring and a sugar moiety, belongs to the polyene class of macrolide polyketides (Sowiński et al. 1995). Like many macrolides, rimocidin is synthesized in Streptomyces sp. by so-called type I modular polyketide synthases (Seco et al. 2004). The antifungal activity of rimocidin lies in its interaction with cell membranes through ergosterol-forming channels, and can lead to a loss of ions, destruction of electrochemical gradients, and ultimately cell death (Neumann et al. 2010). Because of the broad range of its biological activities against pathogenic fungi, rimocidin is a potential fungicide that can control plant diseases, and it has become an attractive target for research. In this context, recent observations have focused on rimocidin-producing strain isolation (Jeon et al. 2016; Lu et al. 2016), genetic transfer system development (Phornphisutthimas et al. 2010; Song et al. 2019), and rimocidin biosynthetic mechanisms (Seco et al. 2004; Escudero et al. 2015). For industrial applications, microbial strains producing high titers of the compound will be required.
In nature, streptomycetes exists as cohorts along with other microbes in complex habitats. Interactions between different bacteria are discussed to be the driving force for the production of secondary metabolites such as antibiotics (Bertrand et al. 2014). In this context, elicitation is considered to be an effective strategy to increase the production of bioactive metabolites through the introduction of microbial cells or extracts to the production medium. There are some reports on enhanced production of bioactive secondary metabolites in plant (Ramirez-Estrada et al. 2016; Shakya et al. 2019; Nourozi et al. 2019), in fungi (Somjaipeng et al. 2016; Subban et al. 2019; Toghueo et al. 2018), and in bacteria (Luti and Mavituna 2011; Recio et al. 2006; Wang et al. 2013; Wang et al. 2017) through the use of elicitors. In the work described by Recio et al. (2006), glycerol, propanediol, and ethylene glycerol were used to increase pimaricin production in the PI factor-defective strain Streptomyces natalensis npi287. In the work described by Luti and Mavituna (2011), dead Bacillus subtilis and Staphylococcus aureus cells are used to increase undecylprodigiosin production in Streptomyces coelicolor and in the work of Wang et al. (2013), four fungal elicitors can increase the production of natamycin in Streptomyces natalensis HW-2. Some aspects of the action mechanism of elicitors are known in plants (Zhai et al. 2017; Zhao et al. 2005) and fungal cells (Schroeckh et al. 2009; Wu et al. 2015), but little is known about the activation of gene clusters in Streptomyces sp. (Abdelmohsen et al. 2015).
The antagonistic strain and rimocidin producer Streptomyces rimosus M527 (China Center for Type Culture Collection (CCTCC) M2013270) was originally isolated in 2011 from a soil sample (Lu et al. 2016). Unfortunately, rimocidin production is very low in this strain. The successful enhancement of rimocidin production through the addition of an elicitor has yet to be reported. This study aimed to assess the effect of biotic elicitors on rimocidin production and morphological differentiation. In addition, the effects of elicitation on transcription of rim genes involved in rimocidin biosynthesis and on protein biosynthesis were determined.
Materials and methods
Materials
Strains
Microorganisms were used as follows: the rimocidin producer Streptomyces rimosus M527 has been deposited at the CCTCC (2013270), Wuhan, China. Recombinant strain S. rimosus M527-ES was obtained in our previous study (Song et al. 2019). Escherichia coli JM109, Bacillus subtilis WB600, Saccharomyces cerevisiae, and Fusarium oxysporum f. sp. cucumerinum were used as biotic elicitors in this study.
Media and cultivation condition
S. rimosus M527 and recombinant strain M527-ES were incubated at 28 °C, and grown in a solid mannitol soya flour (MS) medium (Zhao et al. 2019) for sporulation. The CP liquid medium, which was used as a seed medium, had the same composition as that described in an earlier study (Zhao et al. 2018). The fermentation medium contained 20 g/l of soya flour, 20 g/l of d-mannitol, and 0.06 g/l of K2HPO4·3H2O (adjusted to a pH 7.0–7.2 before sterilization). S. rimosus M527 and strain M527-ES were grown on MS medium for 4–5 days at 28 °C. In all experiments, the number of spores that initiated the vegetative inoculum was kept to 1 × 106 spores/ml. The inoculum was incubated at 28 °C for 2 days in a 250-ml Erlenmeyer flask, shaken at 180 rpm, and then used as a vegetative inoculum at 4% (v/v) to produce pure and elicited cultures at 28 °C with constant shaking at 180 rpm for 96 h.
E. coli JM109 and B. subtilis WB600 were cultured using liquid or solid Luria-Bertani medium at 37 °C. S. cerevisiae was streaked on solid yeast-peptone-dextrose (YPD) medium (Rao et al. 2008) and grown in liquid YPD medium at 28 °C. F. oxysporum f. sp. cucumerinum was streaked on solid potato-dextrose-agar medium and grown in CP medium.
Preparation and addition of elicitor
A hemocytometer was used to adjust the concentration of living cells to approximately 1 × 107 cells/ml by adding sterile water, if necessary. Spore suspension of F. oxysporum f. sp. cucumerinum was obtained from the liquid medium by filtering through sterile cotton wool and was washed twice with sterile water. Thereafter, the spores were counted in a hemocytometer. To ensure the accuracy of the result, the amount of cells or spores inoculated was approximately constant in each experiment. To investigate the effects of biomass and filtrates on rimocidin biosynthesis, E. coli JM109 and B. subtilis WB600 (1 × 106/ml) were incubated at 37 °C with constant shaking at 180 rpm for 24 h. S. cerevisiae and F. oxysporum f. sp. cucumerinum (1 × 106/ml) were incubated at 28 °C with constant shaking at 180 rpm for 48 h.
To prepare the elicitor, the fermentation broth was centrifuged at 11,000 rpm for 15 min. The supernatant of the fermentation broth (part A) was sterilized through a 0.45-μm millipore filter. Correspondingly, the filtrates of E. coli JM109, B. subtilis WB600, S. cerevisiae, and F. oxysporum f. sp. cucumerinum were designated A1, A2, A3, and A4, respectively. The deposited cells (part B) were collected, washed twice, and then re-suspended in an equal volume of sterile water. Living E. coli JM109, B. subtilis WB600, S. cerevisiae, and F. oxysporum f. sp. cucumerinum cells were named B1, B2, B3, and B4. To obtain dead cells, the living cell suspensions were placed in boiling water for 30 min. Dead E. coli JM109, B. subtilis WB600, S. cerevisiae, and F. oxysporum f. sp. cucumerinum cells were named C1, C2, C3, and C4, respectively.
Each elicitor was introduced at 1–4% (v/v) to the fermentation medium at the same time as inoculation with S. rimosus M527. As a control for part A, a 1–4% (v/v) blank culture medium of four tested microorganism was added to the fermentation broth at the same time as inoculation with S. rimosus M527. As a control for part B and part C, 1–4% sterile water (v/v) was added to the fermentation broth at the same time as inoculation with S. rimosus M527.
Scanning electron microscopy analysis
Following a previously described method (Supaphon et al. 2013), mycelium of S. rimosus M527 was collected, washed with phosphate-buffered saline (PBS), and fixed with 2.5% glutaraldehyde at 4 °C overnight. The fixed mycelia were washed with PBS three times (15 min each) and then fixed with 1% OsO4 for 1 h. Subsequently, an ethanol concentration gradient (v/v) of 30%, 50%, 75%, 90%, and 100% was used to dehydrate the fixed mycelia sequentially. Morphological characteristics of the mycelia surface were examined using JSM-5410LV scanning electron microscopy (SEM, JEOL, Tokyo, Japan).
Analysis of gene transcriptional levels by qRT-PCR
Extraction of RNA and analysis of transcriptional levels of rim genes were performed as described previously (Ma et al. 2014; Zhao et al. 2019) with some modifications. Total RNA was extracted using an AxyPrep™ Multisource Total RNA Miniprep kit (Axygen) and cDNA first-strand synthesis was performed with a PrimeScript™ RT reagent kit (TaKaRa) according to the manufacturer’s protocol. Primers designed as described by Zhao et al. (2019) were used to analyze rim gene transcription in the control and experimental groups. Real-time quantitative reverse transcription PCR (qRT-PCR) reactions were performed and the results were analyzed as described by Zhao et al. (2019). The PCR experiments were performed in triplicate using RNA samples from three independent experiments.
GUS assay
Visual observation of GUS activity was performed according to a method described by Xu et al. (2017b). GUS enzymatic activity was determined as described by Siegl et al. (2013). All assays were performed in triplicate; the reported values were the average of three assays with the calculated standard deviation.
Protein assay
Protein concentration was determined with the Bradford method (Bradford 1976) with bovine serum albumin used as a standard.
Fermentation and analysis of rimocidin
Batch fermentation experiments of S. rimosus M527 were scaled up using a 5-l fermentor (BIOTECH-5BG, Baoxing Biological Equipment Co., Shanghai, China) with a working volume of 3 l. The seed cultures were grown in 500-ml Erlenmeyer flasks containing 100 ml medium at 28 °C for 48 h with shaking at 180 rpm. The agitation speed and aeration rate were 200 r/min and 1.5 m3/(m3·min), respectively. A dissolved oxygen level of 20–30% was used for rimocidin production. The pH was kept at 7.0 by automatic addition of 3 M NaOH, and all fermentation experiments were carried out at 28 °C (Zhao et al. 2019). Rimocidin was analyzed using high-performance liquid chromatography (HPLC) (Varian, USA). The production and analysis of rimocidin was measured according to a method described by Zhao et al. (2019). The dry cell weight (DCW) of S. rimosus M527 was measured according to a method described by Farid et al. (2000). Samples taken from bioreactors at different time points were analyzed for DCW.
Western immunoblot analysis of GUS
SDS-PAGE and Western blot analysis were performed using standard techniques (Sambrook and Russell 2001). Protein samples were separated by SDS-PAGE at 60 V for 20 min and then 80 V for 1–2 h and blotted onto polyvinylidene fluoride membrane at 100 V for 2 h with a mini trans-blot electrophoretic transfer system. Primary rabbit anti-GUS antibodies (Sigma) were used at a dilution of 1:1000 and treated with horseradish peroxidase coupled with secondary antibody goat anti-rabbit IgG (Thermo) at a dilution of 1:5000 for immunologic detection (SuperSignal West Dura Extended Duration Substrate, Thermo Pierce).
Statistical analysis
All experiments were performed at least three times, and results were expressed as mean ± standard deviations (SD). Statistical analysis was performed with Student’s t test. Samples with P values < 0.05 were considered statistically significant.
Results
Effect of filtrate on biosynthesis of rimocidin
To investigate the effect of the filtrate of fermentation broth on biosynthesis of rimocidin, A1–A4 were added to the fermentation medium at the beginning of S. rimosus M527 fermentation, respectively, and the production of rimocidin was measured. As shown in Fig. 1 a, all tested part A could enhance the rimocidin production to varying degrees. In these experiments, the best result, a yield of 0.37 g/l, was obtained when 2% A3 was added to the fermentation broth of S. rimosus M527. Compared with that of the control (0.21 g/l), the increase of the yield was approximately 76.2%. These results may indicate that the extracellular small molecule contained in A1, A2, A4, and especially A3 could promote the biosynthesis of rimocidin.
Production of rimocidin by S. rimosus M527 in the presence of fermentation broth (a), living cells (b), or dead cells (c) from the four different microorganisms. Bacteria were continuously shaken at 180 rpm for 24 h at 37 °C in an incubator. Fungi were shaken at 180 rpm for 48 h at 28 °C. The fermentation broth was centrifuged at 11,000 rpm for 15 min and the supernatant of the fermentation broth was sterilized through a 0.45-μm millipore filter. The deposited cells were collected, washed twice, and then re-suspended in an equal volume of sterile water. To obtain dead cells, these suspensions were placed in boiling water for 30 min. One to 4% (v/v) was introduced to the bioreactor at the beginning of the fermentation. Symbols used were as follows: S. rimosus M527 (0), E. coli JM109 (A1, B1, C1), B. subtilis WB600 (A2, B2, C2), S. cerevisiae (A3, B3, C3), and F. oxysporum f. sp. cucumerinum (A4, B4, C4). “ns” indicates no statistically significant results (P value > 0.05). “*” indicates statistically significant results (0.01 < P value < 0.05). “**” indicates highly statistically significant results (P value < 0.01)
Effect of biomass on biosynthesis of rimocidin
To investigate the effect of the biomass of microorganism on the biosynthesis of rimocidin, living cells (part B) and dead cells (part C) prepared from four types of microorganism were added to the broth at the same time as S. rimosus M527. As shown in Fig. 1 b, the production of rimocidin was significantly enhanced when 2% B3 or 2% B4 were added to the fermentation broth of S. rimosus M527. The rimocidin yield increased by approximately 42.9% (0.30 g/l) and 85.7% (0.39 g/l) compared with that of the control, respectively. However, B1 and B2 markedly inhibited the biosynthesis of rimocidin. As shown in Fig. 1 c, compared with the control, the rimocidin yield increased slightly with the addition of C1 or C3, whereas the addition of C2 or C4 showed no significant effect on rimocidin production.
Elicitors A3 and B4 at 2% (v/v) were used in subsequent investigations due to their superior stimulating effect.
Effects of adding elicitors A3 and B4 on S. rimosus M527
Cell growth
To investigate the effect of the elicitors on S. rimosus M527 cell growth, elicitor A3 was added to the fermentation medium at the same time as S. rimosus M527, and DCW was measured. As shown in Fig. 2, there was almost no difference in terms of cell growth in the first 12 h between the control group and the elicitation experimental group with the addition of A3. However, after 24 h of cultivation, the DCW of S. rimosus M527 growing in the presence of A3 was significantly higher (more than 20%) than that of the control group. Unfortunately, the DCW of S. rimosus M527 growing in the presence of B4 was difficult to measure due to the difficulty of separating the living cells of F. oxysporum f. sp. cucumerinum from the fermentation broth.
Effects of elicitor A3 on the dry cell weight (DCW) of S. rimosus M527 generated during the fermentation process. The growth curve of S. rimosus M527 grown in the presence of A3 is shown with white circles. The growth curve of S. rimosus M527 grown in the absence of A3 is shown with black circles. The elicitor (2% (v/v)) was added to the fermentation broth at the beginning of the fermentation
Mycelium morphology
The effects of elicitors (A3 and B4) on morphological characteristics of S. rimosus M527 were investigated by SEM. As shown in Fig. 3, the addition of A3 led to more slender hyphae and more membrane substances adhered to the surface of hyphae (Fig. 3b). The mycelium of S. rimosus M527 growing in the presence of B4 was formed short rods. The gathered hyphae were more mature, and plump, and their surfaces were much smoother (Fig. 3c).
Batch fermentation
To evaluate the effects of elicitors A3 and B4 on rimocidin production of S. rimosus M527, both elicitors were respectively added in a concentration of 2% (v/v) to the 5-l fermentor at the beginning of rimocidin fermentation. Samples were periodically collected and analyzed to determine the concentration of rimocidin in the fermentation broth using HPLC. As shown in Fig. 4, both elicitors strongly influenced rimocidin production. After 96 h, A3 and B4 increased rimocidin levels to 0.62 g/l and 0.69 g/l, resulting in an increase of 51.2% and 68.3% compared with S. rimosus M527 pure fermentation, respectively.
Effects of elicitors A3 and B4 on rimocidin production of S. rimosus M527. Production by S. rimosus M527 in the absence of an elicitor is shown with triangles, production by S. rimosus M527 in the presence of A3 is shown with diamonds, and production by S. rimosus M527 in the presence of B4 is shown with circles. Error bars were calculated from three different batches of fermentation
Transcriptional levels of the rim genes involved in rimocidin biosynthesis
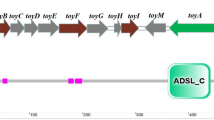
The partial sequence of the rim gene cluster involved in rimocidin biosynthesis in S. rimosus M527 had been cloned and published (GenBank accession number: MK300953). The putative functions of ten rim genes (rimA to rimK) located in cluster have been analyzed (Seco et al. 2004). To test whether the addition of elicitors stimulates the transcriptional levels of rim genes, qRT-PCR was performed after 48 h of cultivation using total RNA of S. rimosus M527 grown with and without elicitors. The qRT-PCR analysis revealed that the addition of the elicitors A3 and B4 induced the enhancement of transcriptional levels of all rim genes (Fig. 5).
qRT-PCR analysis of the transcriptional levels of the rim genes in S. rimosus M527 treated without elicitors and treated with elicitors. 16S rRNA was used as an internal control. The cells were harvested from the fermentation broth after 48 h. Error bars were produced by calculating the standard deviation of the data from three replicates of each sample. “ns” indicates no statistically significant results (P value > 0.05). “*” indicates statistically significant results (0.01 < P value < 0.05). “**” indicates highly statistically significant results (P value < 0.01). rimA: type I polyketide synthase gene; rimB: type I polyketide synthase gene; rimC: tyrosine phosphatase gene; rimD: cholesterol oxidase gene; rimE: glycosyl transferase gene; rimF: aminotransferase gene; rimG: cytochrome P450 monooxygenase gene; rimH: ferredoxin gene; rimJ: crotonyl-CoA reductase gene; rimK: acetyl-transferase gene
Protein synthesis
GUS has become a popular reporter for use in Streptomyces (Myronovskyi et al. 2011; Siegl et al. 2013; Xu et al. 2017b). We recently used the GUS reporter system in analysis of promoter activities in S. rimosus M527 (Song et al. 2019). In that study, the recombinant strain S. rimosus M527-ES, in which gusA as a reporter gene was placed under the control of permE* and integrated into the chromosome of S. rimosus M527 by intergeneric conjugation, was obtained (Song et al. 2019). In this study, our results revealed that the presence of GUS in the cells had no significant effect on the growth of S. rimosus M527 and rimocidin production (Fig. 6). And as expected, as in S. rimosus M527, rimocidin production in S. rimosus M527-ES was significantly enhanced by the addition of elicitor A3 or B4. Thus, S. rimosus M527-ES instead of S. rimosus M527 was therefore used to study the influence of A3 and B4 on protein biosynthesis.
Time courses of cell growth of the wild-type strain S. rimosus M527 (white circle) and the recombinant strain S. rimosus M527-ES containing plasmid pGUS-ermE* (black circle) (a). Effects of the elicitors on the production of rimocidin by S. rimosus M527-ES (b). Each elicitor (2% (v/v)) was added to the fermentation broth at the beginning of the fermentation. Error bars were calculated from three different batches of fermentation
As shown in Fig. 7 a, GUS activities were visible in all experimental samples harboring the gusA gene. In the presence of the elicitor A3, a dark blue color became visible, which was even darker in the presence of B4. To characterize the effect of elicitors on protein synthesis in detail, GUS enzymatic assay was performed at different time points (48 and 72 h). The data were consistent with the aforementioned intuitive observations. As shown in Fig. 7 b, GUS activity of S. rimosus M527-ES was higher than that in the presence of A3 (4.7-fold increase) and B4 (5.2-fold increase) compared with the S. rimosus M527-ES pure culture. GUS activity was slightly stronger after 72 h than after 48 h. These results were also confirmed by western blot analysis (Fig. 7c). It is worth mentioning that the Bradford method (Table 1) provided similar results, suggesting that the use of the GUS reporter is an effective method for representing total protein expression.
Influence of elicitors on total protein synthesis determined by GUS activities and Western blotting. GUS was used as reporter gene for examining protein synthesis. Each elicitor at 2% (v/v) was introduced to the fermentation medium at the beginning of the fermentation. Pure and elicited cultures were incubated at 28 °C with constant shaking at 180 rpm. “ES+A3” represents the recombinant strain S. rimosus M527-ES with the addition of A3. “ES+B4” represents the recombinant strain S. rimosus M527-ES with the addition of B4. a Visual observations of GUS activities. One milliliter liquid medium of pure and elicited cultures at different time points (48, 72 h) was taken and added with 100 μl X-Gluc (1 mmol/l), respectively. The blue color was caused by 5,5′-dibromo-4,4′-dichloro-indigo, which was formed by β-glucuronidase activity. All assays were performed in triplicate. b Detection of β-glucuronidase activity using enzyme assay. Glucuronidase activity was measured in cell lysates of the control group and the experimental group from fermentation broth at 48 and 72 h, respectively. Error bars were calculated from three different batches of fermentation. “ns” indicates no statistically significant results (P value > 0.05). “**” indicates highly statistically significant results (P value < 0.01). c Detection of expression level of GUS was performed by Western blotting in the control and experimental groups. Samples were probed with anti-GUS antibodies as described in the “Materials and methods” section
Discussion
The production of secondary metabolites by microorganisms is strongly dependent on environmental factors, such as growth conditions and biotic stresses (Bode et al. 2002; Pettit 2011; Ren et al. 2015; Tanaka et al. 2017; Wakefield et al. 2017). In this study, the culture filtrates, living cells, and dead cells from E. coli JM109, B. subtilis WB600, and S. cerevisiae, and the plant pathogenic fungi F. oxysporum f. sp. cucumerinum, were used as elicitors to investigate their effects on rimocidin production of S. rimosus M527.
In this study, culture filtrates, living and dead cell from E. coli JM109, B. subtilis WB600, and S. cerevisiae and the plant pathogenic fungi F. oxysporum f. sp. cucumerinum, were used as elicitors to investigate their effects on rimocidin production of S. rimosus M527. The fermentation broths from all four microorganisms and living cells from S. cerevisiae and F. oxysporum f. sp. cucumerinum stimulated rimocidin production even in low concentrations. Wang et al. (2013) reported that the filtrate of the broth from Penicillium chrysogenum AS 3.5163 stimulated natamycin production in Streptomyces natalensis HW-2. A low-molecular-weight substance with a polarity similar to that of butyl alcohol was identified as an elicitor. We also believe that a low-molecular-weight compound is responsible for the observed effects; however, further studies have to be performed to analyze which compounds of the strains used in our study are responsible for the observed effects. A primary HPLC analysis of the filtrate of the broth of S. cerevisiae revealed unknown elicitor components. A follow-up study is in progress, and the results will be reported in a separate paper.
Several studies confirmed that physical contact (cell-cell interaction) through signaling or defense molecules can induce the production of secondary metabolites (Pettit 2009; Marmann et al. 2014; Scherlach and Hertweck 2009; Schroeckh et al. 2009). Competition for limited nutrient sources and antagonisms are characteristics of defense mechanisms that promote biosynthesis of bioactive secondary metabolites. Rimocidin reportedly exhibits strong biological activity against F. oxysporum f. sp. cucumerinum (Lu et al. 2016; Yu et al. 2017). The high production of rimocidin, particularly in the presence of F. oxysporum f. sp. cucumerinum, indicates a kind of defense response of S. rimosus M527 against F. oxysporum f. sp. cucumerinum invasion. In contrast, living cells from bacteria (E. coli JM109, B. subtilis WB600) reduced rimocidin biosynthesis. The coexistence of two microorganisms that grow and survive in the same environment leads to their competition for limited nutrient sources, which affects microbial growth, adaptation, and development pattern (Jones and Elliot 2017; Ola et al. 2013; Slattery et al. 2001). As both E. coli and B. subtilis grow much faster than Streptomyces rimosus M527, they have an advantage in competition for limited nutrient sources, leading to the displacement of S. rimosus M527 as the dominant strain in fermentation medium. This results in a lower rimocidin production.
As model strain, S. cerevisiae was used as biotic elicitor to induce microbial secondary metabolites (Shin et al. 1998; Suh and Shin 2000). Recently, the biomass and filtrate of the broth from S. cerevisiae AS 2.2081 were used as elicitors to induce natamycin production in S. natalensis HW-2, but little stimulatory effect on natamycin was found for either the biomass or the filtrate (Wang et al. 2013). In our study, both fermentation broth and the living cells of S. cerevisiae showed a significant effect on rimocidin production. The effect was a bit higher when fermentation broth was used. Furthermore, fermentation broths and the living cells of F. oxysporum f. sp. cucumerinum showed a significant effect on rimocidin production. In this case, the stimulatory effect was higher when living cells were used. Thus, different microorganisms cause different effects. This has to be considered for the selection of the best elicitor strain.
Regardless of which dead cells of the four microorganisms were used as elicitors, including S. cerevisiae and F. oxysporum f. sp. cucumerinum, no significant stimulation effects on rimocidin biosynthesis were observed. Our results were not consistent with those of Luti and Mavituna (2011). In their study, dead cells of B. subtilis and Staphylococcus aureus increased the maximum undecylprodigiosin production by 3-fold and 5-fold, respectively, compared with S. coelicolor pure culture. It is again obvious that strains react differently on different elicitors. In this context, the formation of natural products does not follow any consistent rule.
Morphological changes and physicochemical responses usually occur with the addition of biotic elicitors (Wang et al. 2013; Wang et al. 2017). The results of our study indicate that the elicitors A3 and B4 did not result in an important change in the mycelium morphology of S. rimosus M527. Moreover, there was no significant difference in colony morphology on the plate between the experiment group and the control group (data not shown). In some cases, addition of filtrates as elicitors did not favor cell growth (Sun et al. 2011; Wang et al. 2013; Wang et al. 2017), whereas filtrates of treated mycelium from fungi used as elicitor promoted cell growth in Xanthophyllomyces dendrorhous (Wang et al. 2006). Our results indicated that both cell growth and rimocidin production were significantly promoted in the presence of A3. The maximum rimocidin production of S. rimosus M527 increased from 16.0 to 24.1 mg·g DCW−1, indicating the stimulation effect of rimocidin production caused by addition of A3 was superior to cell growth.
It has been reported that elicitors stimulate the production of secondary metabolites through activation of the expression of biosynthetic gene clusters (Zhang et al. 2019). As expected, under the A3 or B4 elicitation condition, transcriptional levels of rim genes responsible for rimocidin biosynthesis were all higher than those in the control group. This result demonstrates that increased rimocidin biosynthesis caused by elicitation of A3 or B4 can be attributed to the enhancement of transcriptional levels of all rimocidin biosynthetic genes.
Green fluorescent protein (GFP) was usually used as a reporter to evaluate protein synthesis and targeted product production (Hansen et al. 2001; Ma et al. 2014). Because of the utility of the GUS reporter system for gene expression study at the translation level in actinomycetes (Ma et al. 2016; Myronovskyi et al. 2011), we employed the GUS reporter system to investigate protein synthesis and rimocidin production between experimental groups (A3 or B4) and the control group. Our results indicate that more proteins are generated in the cell in the presence of both elicitors, suggesting that the enhanced rimocidin production may be partly attributable to an increase in the level of protein synthesis.
These results suggest that either a small molecule acts as signal molecule triggering specific genes or a general defense mechanism is activated in the presence of elicitors. However, the molecular mechanism of elicitation is not well understood and needs more research at a molecular level.
In the 5-l fermentor, the maximum rimocidin production of S. rimosus M527 was achieved by addition of B4 with 2% (v/v) at the beginning of fermentation, resulting in 0.69 g/l of production, which was 68.3% greater than that produced by S. rimosus M527 fermented without elicitor. Future study should concentrate on improving industrial rimocidin production rates. We will attempt to develop a fed-batch fermentation model working with the addition of elicitors and optimizing the whole fermentation process and conditions in a large-scale fermentor.
It has been described that elicitors can trigger the expression of silent or cryptic secondary metabolite gene clusters (Seyedsayamdost 2014; Okada and Seyedsayamdost 2017; Xu et al. 2017a). S. rimosus M527 contains 30–40 natural product biosynthetic gene clusters as predicted by antiSMASH 3.0. Therefore, it is anticipated that future studies shall concentrate on the activation of cryptic gene clusters to explore novel compounds with diverse activities using different elicitation strategies.
In conclusion, this study provides an effective strategy to improve the rimocidin production in S. rimosus M527. The use of elicitors to enhance production is reliable and can be extended to other antibiotic overproduction strains.
References
Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U (2015) Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv 33(6 Pt 1):798–811
Bertrand S, Bohni N, Schnee S, Schumpp O, Gindro K, Wolfender JL (2014) Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol Adv 32(6):1180–1204
Bode HB, Bethe B, Höfs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 3(7):619–627
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Escudero L, Al-Refai M, Nieto C, Laatsch H, Malpartida F, Seco EM (2015) New rimocidin/CE-108 derivatives obtained by a crotonyl-CoA carboxylase/reductase gene disruption in Streptomyces diastaticus var. 108: substrates for the polyene carboxamide synthase PcsA. PLoS One 10(8):e0135891
Farid MA, El-Enshasy HA, El-Diwany AI, El-Sayed ESA(2000) Optimization of the cultivation medium for natamycin production by Streptomyces natalensis. J Basic Microbiol 40:157–166
Hansen LH, Ferrari B, Sørensen AH, Veal D, Sørensen SJ (2001) Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole-cell biosensors and flow cytometry. Appl Environ Microbiol 67(1):239–244
Jeon BJ, Kim JD, Han JW, Kim BS (2016) Antifungal activity of rimocidin and a new rimocidin derivative BU16 produced by Streptomyces mauvecolor BU16 and their effects on pepper anthracnose. J Appl Microbiol 120(5):1219–1228
Jones SE, Elliot MA (2017) Streptomyces exploration: competition, volatile communication and new bacterial behaviours. Trends Microbiol 25(7):522–531
Kemung HM, Tan LT, Khan TM, Chan KG, Pusparajah P, Goh BH, Lee LH (2018) Streptomyces as a prominent resource of future anti-MRSA drugs. Front Microbiol 9:2221
Liu R, Deng Z, Liu T (2018) Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab Eng 50:74–84
Lu D, Ma Z, Xu X, Yu X (2016) Isolation and identification of biocontrol agent Streptomyces rimosus M527 against Fusarium oxysporum f. sp. cucumerinum. J Basic Microbiol 56(8):929–933
Luti KJ, Mavituna F (2011) Elicitation of Streptomyces coelicolor with dead cells of Bacillus subtilis and Staphylococcus aureus in a bioreactor increases production of undecylprodigiosin. Appl Microbiol Biotechnol. 90(2):461–466
Marmann A, Aly AH, Lin W, Wang B, Proksch P (2014) Co-cultivation--a powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar Drugs 12(2):1043–1065
Ma Z, Luo S, Xu X, Bechthold A, Yu X (2016) Characterization of representative rpoB gene mutations leading to a significant change in toyocamycin production of Streptomyces diastatochromogenes 1628. J Ind Microbiol Biotechnol 43(4):463–471
Ma Z, Tao L, Bechthold A, Shentu X, Bian Y, Yu X (2014) Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl Microbiol Biotechnol 98(11):5051–5058
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) β-Glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77(15):5370–5383
Neumann A, Baginski M, Czub J (2010) How do sterols determine the antifungal activity of amphotericin B? Free energy of binding between the drug and its membrane targets. J Am Chem Soc 132(51):18266–18272
Nourozi E, Hosseini B, Maleki R, Abdollahi Mandoulakani B (2019) Iron oxide nanoparticles: a novel elicitor to enhance anticancer flavonoid production and gene expression in Dracocephalum kotschyi hairy-root cultures. J Sci Food Agric 99(14):6418–6430
Okada BK, Seyedsayamdost MR (2017) Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev 41(1):19–33
Ola AR, Thomy D, Lai D, Brötz-Oesterhelt H, Proksch P (2013) Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J Nat Prod 76(11):2094–2099
Olanrewaju OS, Babalola OO (2019) Streptomyces: implications and interactions in plant growth promotion. Appl Microbiol Biotechnol 103(3):1179–1188
Pettit RK (2009) Mixed fermentation for natural product drug discovery. Appl Microbiol Biotechnol 83(1):19–25
Pettit RK (2011) Small-molecule elicitation of microbial secondary metabolites. Microb Biotechnol 4(4):471–478
Phornphisutthimas S, Sudtachat N, Bunyoo C, Chotewutmontri P, Panijpan B, Thamchaipenet A (2010) Development of an intergeneric conjugal transfer system for rimocidin-producing Streptomyces rimosus. Lett Appl Microbiol 50(5):530–536
Ramirez-Estrada K, Vidal-Limon H, Hidalgo D, Moyano E, Golenioswki M, Cusidó RM, Palazon J (2016) Elicitation, an effective strategy for the biotechnological production of bioactive high-added value compounds in plant cell factories. Molecules 21(2):182
Rao Z, Ma Z, Shen W, Fang H, Zhuge J, Wang X (2008) Engineered Saccharomyces cerevisiae that produces 1,3-propanediol from D-glucose. J Appl Microbiol 105(6):1768–1776
Recio E, Aparicio JF, Rumbero A, Martín JF (2006) Glycerol, ethylene glycol and propanediol elicit pimaricin biosynthesis in the PI-factor-defective strain Streptomyces natalensis npi287 and increase polyene production in several wild-type actinomycetes. Microbiology. 152(Pt 10):3147–3156
Ren XD, Chen XS, Zeng X, Wang L, Tang L, Mao ZG (2015) Acidic pH shock induced overproduction of ε-poly-L-lysine in fed-batch fermentation by Streptomyces sp. M-Z18 from agro-industrial by-products. Bioprocess Biosyst Eng 38(6):1113–1125
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Scherlach K, Hertweck C (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem 7(9):1753–1760
Schroeckh V, Scherlach K, Nützmann HW, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA (2009) Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A 106(34):14558–14563
Seco EM, Pérez-Zúñiga FJ, Rolón MS, Malpartida F (2004) Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem Biol 11(3):357–366
Seyedsayamdost MR (2014) High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A 111(20):7266–7271
Shakya P, Marslin G, Siram K, Beerhues L, Franklin G (2019) Elicitation as a tool to improve the profiles of high-value secondary metabolites and pharmacological properties of Hypericum perforatum. J Pharm Pharmacol 71(1):70–82
Shin CS, Kim HJ, Kim MJ, Ju JY (1998) Morphological change and enhanced pigment production of monascus when cocultured with Saccharomyces cerevisiae or Aspergillus oryzae. Biotechnol Bioeng 59(5):576–581
Siegl T, Tokovenko B, Myronovskyi M, Luzhetskyy A (2013) Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab Eng 19:98–106
Slattery M, Rajbhandari I, Wesson K (2001) Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb Ecol 41(2):90–96
Somjaipeng S, Medina A, Magan N (2016) Environmental stress and elicitors enhance taxol production by endophytic strains of Paraconiothyrium variabile and Epicoccum nigrum. Enzym Microb Technol 90:69–75
Song ZQ, Liao ZJ, Hu YF, Ma Z, Bechthold A, Yu XP (2019) Development and optimization of an intergeneric conjugation system and analysis of promoter activity in Streptomyces rimosus M527. J Zhejiang Univ Sci B 20(11):891–900
Sowiński P, Pawlak J, Borowski E, Gariboldi P (1995) Stereostructure of rimocidin. J Antibiot (Tokyo) 48(11):1288–1291
Subban K, Subramani R, Srinivasan VPM, Johnpaul M, Chelliah J (2019) Salicylic acid as an effective elicitor for improved taxol production in endophytic fungus Pestalotiopsis microspora. PLoS One 14(2):e0212736
Suh JH, Shin CS (2000) Analysis of the morphologic changes of Monascus sp. J101 cells cocultured with Saccharomyces cerevisiae. FEMS Microbiol Lett 193(1):143–147
Sun JL, Zou X, Liu AY, Xiao TF (2011) Elevated yield of monacolin K in Monascus purpureus by fungal elicitor and mutagenesis of UV and LiCl. Biol Res 44(4):377–382
Supaphon P, Phongpaichit S, Rukachaisirikul V, Sakayaroj J (2013) Antimicrobial potential of endophytic fungi derived from three seagrass species: Cymodocea serrulata, Halophila ovalis and Thalassia hemprichii. PLoS One 8(8):e72520
Tanaka Y, Izawa M, Hiraga Y, Misaki Y, Watanabe T, Ochi K (2017) Metabolic perturbation to enhance polyketide and nonribosomal peptide antibiotic production using triclosan and ribosome-targeting drugs. Appl Microbiol Biotechnol 101(11):4417–4431
Toghueo RMK, Sahal D, Zabalgogeazcoa Í, Baker B, Boyom FF (2018) Conditioned media and organic elicitors underpin the production of potent antiplasmodial metabolites by endophytic fungi from Cameroonian medicinal plants. Parasitol Res 117(8):2473–2485
Wakefield J, Hassan HM, Jaspars M, Ebel R, Rateb ME (2017) Dual induction of new microbial secondary metabolites by fungal bacterial co-cultivation. Front Microbiol 8:1284
Wang D, Wei L, Zhang Y, Zhang M, Gu S (2017) Physicochemical and microbial responses of Streptomyces natalensis HW-2 to fungal elicitor. Appl Microbiol Biotechnol 101(17):6705–6712
Wang D, Yuan J, Gu S, Shi Q (2013) Influence of fungal elicitors on biosynthesis of natamycin by Streptomyces natalensis HW-2. Appl Microbiol Biotechnol 97(12):5527–5534
Wang W, Yu L, Zhou P (2006) Effects of different fungal elicitors on growth, total carotenoids and astaxanthin formation by Xanthophyllomyces dendrorhous. Bioresour Technol 97(1):26–31
Wu C, Zacchetti B, Ram AF, van Wezel GP, Claessen D, Hae Choi Y (2015) Expanding the chemical space for natural products by Aspergillus-Streptomyces co-cultivation and biotransformation. Sci Rep 5:10868
Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR (2017a) Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J Am Chem Soc 139(27):9203–9212
Xu X, Wang J, Bechthold A, Ma Z, Yu X (2017b) Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J Microbiol Biotechnol 33(2):30
Yu J, Liu Q, Chen C, Qi X (2017) Antifungal activity change of Streptomyces rimosus MY02 mediated by confront culture with other microorganism. J Basic Microbiol 57(3):276–282
Zhai X, Jia M, Chen L, Zheng CJ, Rahman K, Han T, Qin LP (2017) The regulatory mechanism of fungal elicitor-induced secondary metabolite biosynthesis in medical plants. Crit Rev Microbiol 43(2):238–261
Zhao J, Davis LC, Verpoorte R (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23(4):283–333
Zhang X, Hindra, Elliot MA (2019) Unlocking the trove of metabolic treasures: activating silent biosynthetic gene clusters in bacteria and fungi. Curr Opin Microbiol 51:9–15
Zhao YF, Lu DD, Bechthold A, Ma Z, Yu XP (2018) Impact of otrA expression on morphological differentiation, actinorhodin production, and resistance to aminoglycosides in Streptomyces coelicolor M145. J Zhejiang Univ Sci B 19(9):708–717
Zhao Y, Song Z, Ma Z, Bechthold A, Yu X (2019) Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J Ind Microbiol Biotechnol. 46(5):697–708
Contributors
ZQ Song conducted the experiments and wrote this article. Z Ma and A Bechthold designed the research and revised this article. XP Yu checked the final version. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31772213, 31972320), excellent youth fund of Zhejiang province, China (LR17C140002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Song, Z., Ma, Z., Bechthold, A. et al. Effects of addition of elicitors on rimocidin biosynthesis in Streptomyces rimosus M527. Appl Microbiol Biotechnol 104, 4445–4455 (2020). https://doi.org/10.1007/s00253-020-10565-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10565-4