Abstract
The chitinases are gaining much attention based on their role in the defense against pathogen attacks and harmful insects. The partially chitinase produced by Bacillus licheniformis strain J24 exhibited a large antifungal spectrum, and the highest activity was obtained toward Fusarium species in vitro on PDA and in vivo on corn seeds. The chitinase was inducible by the presence of autoclaved Fusarium conidia in the medium culture and it was active at 70 °C and pH 7 and not affected by the tested chemical agents EDTA and SDS. The nucleotide and amino acid sequences encoding chitinase showed the close phylogenetic relation with chitinase from Bacillus paralicheniformis species. Based on the analysis of the putative domain active, the described chitinase from strain J24 was belonging to the GH family-18 and the novelty of its structure was revealed. Here the combination of functional and structural antifungal extremely chitinase proves its importance in biotechnology area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The chitinases (EC 3.2.1.14) catalyze the hydrolysis of chitin, a linear homopolymer of N-acetyl-d-glucosamine residues. Chitin is a fundamental compound of the cell wall of the phytopathogens fungi, the exoskeletons of arthropods, crustaceans, and nematodes. Enzymes implicated in the lysis of fungal cell walls have been common in the mechanism of bio-control by various bacterial agents [1, 2] The microbial chitinase production and characterization attracted research attention, not only because of its various industrial applications or its antifungal properties but, also due to the absence of an effective performance enzyme production method [3, 4]. Bacterial chitinases were largely studied and several bacterial producing species were identified, the best known genera include Aeromonas, Serratia, Vibrio, Streptomyces and Bacillus. Chitinases were produced and identified from a variety of Bacillus spp. species such as B. cereus, B. licheniforrmis B. thuringiensis, Virgibacillus marismortui [5,6,7]. Nowadays, the research highlights toward the isolation and identification of the gene encoding for the chitinase to be used to control fungal plant pathogens Founded on the amino acid sequences similarity in the catalytic domains, the chitinases are classified into GH family-18 and GH family-19 [8]. In the literature, the family 18 of glycosyl hydrolase has been found widely in bacteria, and the presence of the rectangle area (IDGIDIDYE) in the amino acid sequences indicates the affiliation to the GH family 18. Finally, chitinases can be divided into the endochitinases which cleave chitin at internal sites to generate multimers of N-acetyl-d-glucosamine and the exochitinases catalyze the hydrolysis of the chitin into chitobiose and chitotriose and N-acetyl-d-glucosamine [9].
During post-harvest storage and transport of horticultural products, molds can cause economic losses and threaten food security. Among these fungi, Fusarium species are responsible for considerable losses on cereal products. Fusarium are polyphagous pathogens causing wilting of plants, rotting of fruits and production of mycotoxins on cereal grains [10, 11]. To control these toxigenic pathogens, the biological control arises as an effective and safe alternative, and the most used biocontrol agents were Bacillus species [12]. In this study, a new halotolerant chitinolytic biocontrol bacterium Bacillus licheniformis strain J24 previously isolated from Tunisian saline soil was investigated for its prominent chitinolytic production [13]. In particular, this paper highlights their use to: (i) optimize the chitinase production, (ii) determine the antifungal spectrum of the partial purified enzyme, (iii) test the ability of the isolate to control disease on corn seeds, (vi) extract, purify and gene sequence characterize of the chitinase produced by B. licheniformis isolate J24.
Materials and Methods
Bacterial Strain and Culture Conditions
A moderately halophilic bacterium B. licheniformis strain J24 was previously isolated from a shallow salt lake in Tunisia and its corresponding nucleotide sequence of 16S RNA has been deposited in the GenBank database under the accession number EF471920 [13]. The strain J24 was carried out in various media M1, M2, M3 and M4: M1 medium containing 5 g L−1 Tryptone: 1 g L−1 glucose: 5 g L−1 YE, 1 g L−1 K2HPO4 and NaCl: 5 g L−1, pH 7.2. To test induction on enzyme production, the same medium M1 was added with 106 spores ml−1 (M2) and M1 with simple modification (0.1 g L−1 glucose and 106 spores ml−1 (M3) and M1 added with 0.5% (w/v) of colloidal chitin (M4). Before use, the fungal conidia suspension was adjusted to 106 spores into sterile water ml−1, then the solution was emulsified and homogenized by the use of T25 digital ultra-turrax, then this obtained solution has undergone three autoclave cycles to avoid the germination of fungal conidia. All culture was incubated at 37 °C for 3 days on a rotary shaker (120 rpm). After centrifugation at 12,000 rpm for 10 min, the cell-free supernatants were collected from each medium for Chitinase assay with three independent replications. The salinity effect was evaluated by growing the bacterium in the optimum medium M2 supplemented with a gradient of salinity (0–5–10-15-20-25 and 30% NaCl, w/v), at 37 °C for 3 days.
Chitinase Assay
Chitinase was determined according to the method of Gomez Ramirez et al. [14]. The mixture volume per volume (v/v) of Chitinase solution and colloidal chitin suspension (10%) was incubated for 1 h at 50 °C. The reaction was stopped by adding 1 ml of 1% NaOH and shaking. The product was determined by 3,5-dinitrosalicylic acid assay (DNS) and the absorbance was measured at 535 nm. The chitinase activity was defined as the amount of enzyme required to produce 1 μmol of N-acetylglucosamine (NAG, Sigma) per h per mg of protein.
Antifungal Activity Assay
A well diffusion method was performed to assess the potential antibiosis of cell free supernatant from B licheniformis J24. Antifungal activity was determined against Fusarium pseudograminearum, Fusarium avenaceum, Sclerotinia sp., Penicillium digitatum and Botrytis cinerea. Fungal strains were maintained on potato dextrose agar (PDA) slants, grown at 28 °C for 72 h. The fungal growth was examined daily for the formation of inhibition zone.
Chitinase Enzyme Partial Purification and Characterization
Strain J24 was grown in 100 ml of M2 medium in 250 ml Erlenmeyer flask at 37 °C for 3 days. The culture broth was centrifuged for 20 min at 12,000 rpm at 4 °C. The supernatant used for enzyme purification, was brought to 80% precipitation with ammonium sulfate and left standing overnight at 4 °C. The precipitate was collected by centrifugation at 12,000 rpm for 20 min and redissolved in 50 mmol Tris HCl buffer pH 7.
The optimum temperature was determined by monitoring the enzyme activity at various temperatures ranging from 40 to 100 °C. The heat stability was determined by measuring the residual activity after pre-incubation step for 30 min at each tested temperatures (40–100 °C) [15].
The pH optimum was resolute by applying a substrate solution at different pH from 4 to 12 and was measured at optimum temperature (70 °C). The pH stability was examined by incubating enzymes in the various pH value for 1 h at 4 °C before adding the substrate. The remaining activities (%) were subsequently determined. The experiments were repeated three times and mean values were taken [15].
Effect of EDTA, SDS and Mercaptoethanol Reagents
In order to explore the stability of the enzyme to the reducing agents (2-mercaptoethanol), the chelating agent EDTA, detergents (SDS), it was incubated with various reagents (direct exposure) for 1 h at 70 °C, pH 7 and then the residual activity of the exposed enzyme was determined. In each case the residual activity was compared to untreated tube (enzyme without any reagent) set as 100% [16].
In Vivo Effect’s of the Partial Purified Chitinase
Fusarium species were grown at 25 °C on PDA for 10 days. Sterile water (20 ml) was added to each plate and the surface was scraped gently with a sterile loop to release the spores. The resulting spore suspension was filtered through a sterile 30 μm filter to remove any mycelia fragments and adjusted to the concentration of 106 spores ml−1.
A number of corn seeds were disinfected and artificially infested with Fusarium pseudograminearum (50 μl of 106 spores ml−1), after 1 h, the same volume of the partially purified chitinase was added (50 μl) to each well infested corn seeds. Each assay was repeated four times. The diameter of fungal growth on seeds was recorded and compared to untreated one. The disease control value was calculated using the following formula: disease control (%) = [(A − B)/A] × 100, where A is the diameter of mycelia growth on untreated seeds and B was the diameter of fungal growth on treated seeds by a volume of the partially purified chitinase.
Cloning and Purification of Chitinase Gene Sequence
A pair of degenerated oligonucleotide primers designed on the basis of chitinase gene sequences from bacterial species deposited in the GenBank database NCBI forward primer, KbF1: 5′-GCCAGGATGAAAACGAGTG-3′; reverse primer, KbR1: 5′-CCACCATTTCGCTTCATACA-3′ were used to amplify the chitinase encoding gene by polymerase chain reaction. The PCR cycling conditions were 3 min at 94 °C for pre-denaturation step, 34 cycles of 30 s at 95 °C, 45 s at 50 °C and 30 s at 72 °C for amplification and 10 min at 72 °C for extension. The PCR product (1400 pb) was purified with QIAEXgel elution kit (Qiagen) and cloned using the Gen jet Kit Cloning (Fermentas) and transformed into INVαF′ competent cells (Invitrogen, La Jolla, CA). Cells carrying the recombinant plasmids were screened and transferred individually in LB medium supplemented with 50 μg ml−1 ampicillin. After growth on a rotary shaker at 37 °C for 20 h, cells were harvested by centrifugation (8000 rpm, 10 min). The pellet obtained was retained for the extraction and purification of recombinants plasmids using the Gene jet TM plasmid miniprep Kit (Fermentas), plasmid DNA was eluted in distilled water [17].
Sequencing and Phylogenetic Analysis of Chitinase Gene Sequence
DNA sequencing was performed on an automated system (GATC Biotech, Germany).Gene sequence was identified by comparison with sequences available at the NCBI database (http://www.ncbi.nlm.nih.gov) based on Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nig.gov/blast) [18]. The nucleotide sequence of the chitinase gene determined here has been deposited with the NCBI Gen Bank database under the accession number MF76595. For phylogenetic assessment, chitinase gene sequences were first aligned with Clustal W software [19]. Then the evolutionary history was inferred by using the Neighbor joining method and evolutionary distances were computed using Jukes and Cantor method. Tree topology was evaluated by performing bootstrap analysis of 1000 data sets using MEGA 6 (Molecular Evolutionary Genetics Analysis). Others NCBI tools were also used as Conserved Domain Search, Conserved Domain Architecture Retrieval Tool, MView.
Results
In the presence of different carbon sources, B. licheniformis J24 can produce extracellular, chitin degrading enzymes in all tested media. The maximum production was observed in M2 medium with 253UA ml−1 (Fig. 1). The high chitinase production occurred in the medium M2 containing glucose 1 g L−1 and 106 spores ml−1, show that the descried enzyme was induced by the presence of pathogen in growth medium however the same result did not appear in the presence of chitin medium M2. So the described enzyme can be suggested as an inducible antifungal chitinase. In addition, the strain J24 produced a halotolerant chitinase with an optimum at 5% NaCl (w/v).
Antifungal Properties of Chitinase
To evaluate the antifungal activities of strain J24, the effect of its cell-free supernatant on mycelium growth and spore germination of a list of phytopathogenic fungi was undertaken. The strain J24 was able to inhibit six fungal species among seven used with a high percentage of inhibition exceeding 94% (Table 1).
During the process of purification of the enzyme produced by the strain J24, we observed a decrease of chitinase activity expressed in U. The ammonium sulfate fraction contained 84.98%, and the dialysate contained only 73.7% with specific activity 6.61 U mg−1. These decreases of total activity (U) were probably due to the loss of chitinase after the purification steps (Table 2). The partial purified chitinase was used to the characterization and the in vivo essays.
The efficiency of the chitinase produced by strain J24 was also screened in vivo on corn seeds and the results make that the presence of the partial purified enzyme could reduce the disease of about 71.08% compared to untreated seeds (Fig. 2a). The in vitro anti-Fusarium activity of the partial purified chitinase observed in Fig. 2b, showed a clear inhibition zone compared to untreated well.
(a) In vivo antifungal effect of the partially purified chitinase produced by the strain J24 of Bacillus licheniformis on disease reduction on corn seeds and (b) antifungal activity in vitro on PDA medium against Fusarium pseudograminearum. C: untreated, B bacterial Chitinase treated and T: sterile distilled water
Characterization of Chitinase Enzyme
Moreover, the chitinase described here was thermotolerant, with optimum temperature at 70 °C, and thermostable since it can retain more than 87.2% of its activity at temperatures from 40 to 80 °C and 71.8% at 90 °C (Fig. 3a). The enzyme revealed optima activity at pH 7 but less than 37% was retained from pH value 7 to 12. Yet the enzyme was more stable at pH 5 and 6, which retained more than 90% of its residual activity (Fig. 3b). Moreover, we demonstrate that the enzyme produced by strain J24 was not affected by the presence of 2-mercaptoethanol, EDTA or SDS (Fig. 3c).
Molecular Structure of Gene Coding to Chitinase
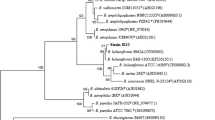
The nucleotide sequence of the chitinase gene is available in the NCBI EMBL/GenBank database under the accession number MF765958.The phylogeny analysis showed its high homology to the chitinase from Bacillus paralicheniformis strain B1–09 (accession CP10524) with 99% similarity (Fig. 4). The phylogeny evolution of the chitinase sequences from B. licheniformis species showed that: on one hand, the chitinase sequences are separated into three groups: group I containing chitinase from numerous B. licheniformis species related to chitinase A. A group III containing the chitinase produced from major Bacillus paralicheniformis species. On the other hand, the chitinase sequence produced by the strain J24 are related to a subgroup II containing those produced by B. paralicheniformis species and B. licheniformis gh18A (Fig. 4). The BLAST analysis of the deduced amino acid chitinase sequence of strain J24 (Accession Number AXG21593), shows 100% identity to the chitinase from B. paralicheniformis (WP_075754039). The identity of the putative domain active in the chitinase from strain J24 is established with the domain active of superfamily c110447 (PSSMID 3245582). The localization of the functional consensus pattern for the chitinase family, by multiple alignment sequences was reported here. And the results of the research of the best pattern for functional protein analysis by logicial EBITools /Protein Functional Analysis /Patt, show that the best functional pattern was A containing the listed amino acid “YDPKAKEAGVPNAETYNI” from position 195 to 212 (Fig. 5).
Phylogenetic tree showing clustering of bacterial chitinases genes constructed by the neighbor-joining method. The tree was constructed with chitinase sequences taken from GenBank . The evolutionary history was inferred using the neighbor-joining method. The scale represents 0.05 substitutions per site. Numbering at each branch indicates bootstrap supports. Tree was constructed using the MEGA software package, version 6.0. Paenibacillus sp. FSL (CP009282.1) was taken as an outgroup
Sequence alignment of the catalytic domain in the Chitinase of strain J24, with those in other GH18 bacterial chitinases. EMBL EBI Alignment by multiple sequence alignment tools MView version 1.63 were used and the amino acid residues were colored by identity. The aligned sequences (Accession Number) are the following: 1: amino acid sequence of Chitinase produced by the strain J24 (2259625), 2 (AAF23368.1), 3(ABI15082.1), 4(ACF40833.1), 5(KYDO1898.1), 6(WP-025810802.1), 7 (WP-031305218.1), 8 (WP-039073260), 9(WP-075212995.1) and 10 (WP-07574039).The hash symbol (#) denotes the putative catalytic residue
Discussion
From a global viewpoint, here, we report a novel antifungal chitinase useful for biotechnological applications as an attractive alternative to control plant disease without the negative impact of chemical fungicides. Since microbial production of chitinase attracts attention of many researchers and industrials, in this work an attempt was made to optimize the enzyme production for that four media composition, temperature, and salinity effect’s on chitinase production were undertaken. Here we firstly reported a strongest chitinase inducible by the presence of Fusarium. Previously, similar chitinase induction was observed from Planococcus rifitoensis in the presence of the pathogen Botrytis cinerea [17]. Besides, the result of the inducible chitinase, correlate with those reported from Stachybotrys elegans [20] and Bacillus thuringiensis pakistani [21]. In the present work, we have used a partial purified enzyme from the supernatant cell free of the strain J24 grown in the optimum medium M2. It is worth noting that in the case of its total purification, the obtained enzyme will show the highest antifungal activity as previously demonstrated by [6], compared to other chitinases reported anti-Fusarium activities; such as Streptomyces sporovirgulis [22], Bacillus cereus YQQ308 [23], B. pumilus SG2 [24], Enterobacter spNRG4 [25], and Stenotrohomonas maltophilia [26]. Evidence is provided that our chitinase was thermostable, similar optima temperature 70 °C was also observed by the thermostable chitinase synthesized by B. licheniformis isolated from Red Palm Weavil [27], and 60 °C from Bacillus sp. [28]. Contradictory the pH optima of others chitinases from B. licheniformis was between 5 and 6 [24], or between 6 and 8 from B. licheniformis S-K1 [29], but similar alkaline chitinases (pH 7 and 8) were reported from Bacillus sp [28]. In fact, not only the chitinase produced by B. licheniformis appear to be stable and resistant to ß-mercaptoethanol, chitinases from Serratia marcesens and Chromobacterium spp also harbor the similar characteristics [28]. The stability of the enzyme to EDTA and the resistance to SDS, suggest its promising potential use in detergent industries. Markedly, the data of the sequence analysis demonstrate the difference in the phylogeny evolution of the nucleotide sequence coding to the chitinase from the strain J24 which were certainly related to its isolation origin and environmental adaptation. It is well known that, in the carbohydrate-active enzyme (CAZY) database, based on the amino acid sequence similarity analysis of the catalytic domain, chitinases are classified into two families GH18 and GH19 (http://www.cazy.org/, [30]. Moreover, the similarity of the N-terminal region of enzyme with the catalytic domain CD of the chitinases belonging to family GH18 proved its closely relation to the family GH18 by involving the active site motif typical of this family [9]. For that it was classified as a member of the gh18 (glycosylhydrolase, family 18) type II chitinase which hydrolyses chitin an abundant polymer of beta 1.4 linked N-acetyl glucosamine. The analysis shows the detection of six conserved motifs corresponding to a putative catalytic residue in the sequence [7]. In the present study, first, we described the biological function of the enzyme as antifungal, thermoresistant and halotolerant enzyme. Second, we developed the structural properties of the enzyme. In addition to the phylogeny analysis of chitinase produced by strain J24, here we analyzed the most discriminative patterns and their distribution. Previously Waldeck et al. [7], reported that the chitinase gene is almost identical to 99–100% among all B. licheniformis strains. In the present finding based on the molecular analysis as well as the biochemical and antifungal properties of the described enzyme, the novelty of the chitinase from strain J24 is confirmed. Other work detailing the separation of the chitinase gene sequence (nucleotide and amino acid) into different clusters revealing the dissimilarity in the sequences [28]. Rare information was reported on the characterization of chitinase gene from halophilic origin such marine source or saline soil [13, 17, 31]. Compared to literature, this study represents the first occasion in which halotolerant chitinase gene from B. licheniformis isolated from saline soil located in Tunisia has been cloned and characterized. In addition, the zymography analysis has proved the presence of a single chitinase in the cell free supernatant (data not published). Furthermore, on the one hand, we develop the correlation between the structure and function of bacterial chitinases and we highlight the influence of the extreme environment on the adaptation, the function and the structure of enzyme. On the other hand, our results could be useful to achieve a mutagenesis site in order to produce a modified enzyme for biotechnological purposes for example.
Finally the results indicated here associated with our previous work [5, 32] emphasize a potential use of the strain J24 as biological control agents. Fortunately the described chitinase was found to be slightly different from other reported chitinases from bacteria regarding to its induction by the presence of the pathogen and its resistance to acidic medium, temperature, SDS and EDTA, these characteristics stimulate the use of such chitinase as an efficient environmentally safe solutions in plant protection.
References
Liu D, Caia J, Xie C, Liu C, Chen Y (2010) Purification and partial characterization of a 36 kDa chitinase from Bacillus thuringiensis subsp colmeri and its biocontrol potential. Enzyme Microbial Technol 46:252–256
Swiontek BM, Jankiewicz U, Burkowska A, Walczak M (2014) Chitinolytic microorganisms ans their possible application in environmental protection. Curr Microbiol 68:71–81
Nampoothiri MK, Baiju TV, Sandhya C, Sabu A, Szakacs G et al (2004) Process optimization for antifungal chitinase production by Trichoderma harzianum. Process Biochem 39:1583–1590
Matroudi S, Zamani MR, Motallebi M (2008) Molecualr cloning of chitinase 33 (chit 33) gene from Trichoderma atroviride. Braz J Microbiol 39:433–437
Essghaier B, Hedi A, Bejji M, Jijakli H, Boudabous et al (2011) Characterization of a novel chitinase from a moderately halophilic bacterium Virgibacillus marismortui strain M3-23. Ann Microbiol 62(2): 835–841
Ghorbel-Bellaaj O, Manni L, Jellouli K, Hmidet N, Nasri M (2012) Optimization of protease and chitinase production by Bacillus cereus SV1 on shrimp shell waste using statistical experimental design. Biochemical and molecular characterization of the chitinase. Ann Microbiol 62:1255–1268
Waldeck J, Daum G, Bisping B, Meinhardt F (2006) Isolation and molecular characterization of chitinase deficient Bacillus licheniformis strains capable of deproteinization of shrimp shell waste to obtain highly viscous chitin. Appl Environ Microbiol 72:7879–7885
Ohnuma T, Sorlie M, Fukuda T, Kawamato N, Taira T, Fukamizo T (2011) Chitin oligosaccharide binding to a family GH19 chitinase from the moss Bryum coronatum. FEBS J 278:3991–4001
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 293(3):781–788
Duan C, Qin Z, Yang Z, Li W, Sun S et al (2016) Identification of pathogenic Fusarium spp causing maize ear rot and potential mycotoxin production in China. Toxins 8(6):186
McGovern RJ (2015) Management of tomato diseases caused by Fusarium oxysporum. Crop Protect 73:78–92
Khan N, Maymon M, Hirsch AN (2017) Combating fusarium infection using bacillus-based antimicrobials. Microorganisms 5:75
Essghaier B, Fardeau ML, Cayol JL, Hajlaoui MR, Boudabous A et al (2009a) Biological control of grey mold in strawberry fruits by halophilic bacteria. J App Microbiol 106(3):833–846
Gomez Ramirez M, Rojas Avelizapa LI, Rojas Avelizapa NG, Cruz Camarillo R (2004) Colloidal chitin stained with Remazol Brillant blue R, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J Microbiol Methods 56(2):213–219
Issam SM, Mohamed G, Farid L, Sami F, Thierry M, et al (2003) Production, purification and biochemical characterization of two beta-glucosidases from Sclerotinia sclerotiorum. Appl Biochem Biotechnol 111 (1): 29–40
Ellouze O, Mejri M, Smaali I, Limam F, Marzouki MN (2007) Induction, properties and application of xylmanase activity from Sclerotinia sclerotiorum S2 Fungus. J Food Biochem 31:96–107
Essghaier B, Bejji M, Jijakli H, Boudabous A, Sadfi-Zouaoui N (2009b) High salt-tolerant protease from a potential biocontrol agent Bacillus pumilus M3-16. Ann Microbiol 59(3):553–558
Altschul SF, Madden TL, Schaffer AA, Zhang Z, Miller W et al (1997) Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Taylor G, Jabaji-hare S, Charest PM, Khan W (2002) Purification and characterization of an extracellular exochitinase, β-N-acdetylhexosaminidase, from the fungal mycoparasitte Stachybotrys elegans. Can J Microbiol 48:311–311
Thamthiankul S, Suan-Ngay S, Tantimavanich S, Panbangred W (2001) Chitinase from Bacillus thuringiensis subsp. Pakistani. Appl Microbiol Biotechnol 56:396–401
Swiontek BM, Jankiewicz U, Lisiecki K (2013) Optimization of cultural conditions for the production of antifungal chitinase by Streptomyces sporovirgulis. Appl Biochem Microbiol 49(2):154–159
Chang WT, Chen YC, Jao CL (2007) Antifungal activity and enhancement of plant growth by Bacillus cereus grown on shellfish chitin wastes. Bioresour Technol 98:1224–1230
Ghasemi S, Ahmadian G, Jelodar NB, Rahimian H, Ghandili S et al (2010) Antifungal chitinases from Bacillus pumilus SG-2: preliminary report. World J Microbiol Biotechnol 26:1437–1443
Dahiya N, Tewari RP, Hoondal GS (2005) Production of an antifungal chitianse from Enterobacter sp NRG4 and its application in protoplast production. World J Microbiol Biotechnol 21:1611–1616
Jankiewicz U, Swiontek BM, Saks E (2012) Identification and characterization of a chitinase of Stenotrophomonas maltophilia, a bacterium that is antagonistic towards fungal phytopathogens. J Biosci Bioeng 113(1):30–35
Khiyami M, Masmali I (2008) Characteristics of thermostable chitinase enzymes of Bacillus licheniformis isolated from Red Palm Weavil Gut. Aus J Basic Appl Sci 2(4):943–948
Yuli PE, Suhartono MT, Rukayadi Y, Hwang JK, Pyun YR (2004) Characteristics of thermostable chitinase enzymes from the Indonesian Bacillus sp 13.26. Enzyme Microbial Technol 35:147–153
Kudan S, Rath P (2009) Purification and characterization of termostable chitinase from Bacillus licheniformis SK-1. Appl Biochem Biotechnol 157:23–35
Lombard V, Ramulu HG, Drula E, Corytinho PM, Henrissat B (2014) The carbohydrate active enzymes CAZY in 2013. Nucleic Acids Res 42:490–495 (http://www.cazy.org/)
Arguelles Arias A, Ongena M, Halimi B, Lara Y, Brans A et al (2009) Bacillus amyloquefasciens GA 1as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb Cell Factories 8(63)
Essghaier B, Rouaissi M, Boudabous A, Jijakli H, Sadfi-Zouaoui N (2010) Production and partial characterization of chitinase from a halotolerant Planococcus rifitoensis strain M2-26. World J Microbiol Biotechnol 26:977–984
Acknowledgements
This work was supported by funds from the Ministry of Higher Education and Scientific Research of Tunisia (LR16ES05).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Essghaier, B., Zouaoui, M., Najjari, A. et al. Potentialities and Characterization of an Antifungal Chitinase Produced by a Halotolerant Bacillus licheniformis. Curr Microbiol 78, 513–521 (2021). https://doi.org/10.1007/s00284-020-02329-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02329-0