Abstract
Co-occurring mutations are frequently observed in acute myeloid leukemia (AML) with NPM1 mutation, and NPM1 measurable residual disease (MRD) is an effective prognostic biomarker. This retrospective study investigated the impact of gene co-mutations and NPM1 MRD on outcomes in these patients. Among 234 patients, 11.5% carried the rare type NPM1 mutation (NPM1RT). The median age was 49 years (IQR 36–58), with a median follow-up of 30.4 months (IQR 12.1–55.7). Nine genes were mutated in > 10%, with DNMT3A (53.8%) and FLT3-ITD (44.4%) being most prevalent. Univariable analysis in 137 patients showed FLT3-ITD, DNMT3A co-mutations, and MRD2 < 3 log reduction predicted poorer survival. FLT3-ITD and DNMT3A co-mutations correlated with the lowest event-free (EFS) and overall survival (OS) (3-year EFS 30.0%; 3-year OS 34.4%; both p < 0.001). FLT3-ITD alone did not worsen survival compared to patients without FLT3-ITD. Multivariable analysis identified DNMT3A co-mutation [EFS, HR = 1.9, p = 0.021; OS, HR = 2.2, p = 0.023] and MRD2 ≥ 3 log reduction (EFS, HR = 0.2; OS, HR = 0.1, both p < 0.001) as independent survival predictors. Patients with FLT3-ITD and DNMT3A co-mutations or a MRD2 < 3 log reduction were identified as high risk, but allogeneic hematopoietic stem cell transplantation (allo-HSCT) improved survival significantly compared to chemotherapy only (3-year EFS, 57.9% vs. 30.0%, p = 0.012; 3-year OS, 72.9% vs. 34.4%, p = 0.001). In AML patients with NPM1 mutation, the detrimental impact of FLT3-ITD mutation was exacerbated by DNMT3A co-mutation. Poor-risk younger patients identified by FLT3-ITD and DNMT3A co-mutations or MRD2 < 3 log reduction benefit from allo-HSCT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nucleophosmin 1 (NPM1) mutation occurs in 28–35% of acute myeloid leukemia (AML) patients [1,2,3], and AML with NPM1 mutation has been recognized as a separate entity in the World Health Organization (WHO) classification of myeloid neoplasms since 2016 [4]. AML with NPM1 mutation has been shown to be associated with a normal karyotype (NK) at a reported frequency of 48% to 53% [5,6,7]; these patients are classified into the favorable prognosis group. However, a concurrent mutation in fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD) diminishes the favorable effect of NPM1 mutation [8].
With the application of next-generation sequencing (NGS), many mutations have been observed to co-occur in AML, especially in AML with NPM1 mutation. DNMT3A mutation, which is associated with preleukemic clones, has been reported in ~ 50% of AML patients with NPM1 mutation [9], and the prognostic significance of DNMT3A mutation is conflicting. Several studies have shown that NPM1 measurable residual disease (MRD) is a favorable predictive marker for the survival of AML patients with NPM1 mutation [10, 11]. However, further investigation is required to determine the prognostic value of the interactions of FLT3-ITD and DNMT3A or other molecular mutations with NPM1 MRD in AML patients with NPM1 mutation.
This study aimed to investigate the value of concurrent mutations and NPM1 MRD for predicting survival in a retrospective cohort of AML patients with NPM1 mutation against the background of conventional chemotherapy.
Patients and methods
Patients
From January 2013 to August 2021, treatment-naïve AML patients with NPM1 mutations who received conventional induction or consolidation chemotherapy at the Peking University Institute of Hematology were included in this retrospective cohort study, and these patients were followed up until January 2024. All patients had an Eastern Cooperative Oncology Group (ECOG) performance status of 2 or less and no serious heart, lung, liver, or kidney dysfunction or severe infection. The study adhered to the principles of the Helsinki Declaration and was approved by the local institutional review boards.
Treatment
Induction therapy
Patients were treated with an anthracycline and cytarabine-based or homoharringtonine and cytarabine-based induction chemotherapy regimen following the Chinese guidelines for the diagnosis and treatment of adult AML (not acute promyelocytic leukemia) (2011) [12] and the 2010 European Leukemia Net recommendations for AML [13]. The chosen induction therapy regimens were IA (idarubicin 10 mg/m2/day, d1-3; cytarabine 100 mg/m2/day, d1-7), GAG (aclacinomycin 20 mg/day, d1-4; cytarabine 10 mg/m2 every 12 h for 7 to 14 days; granulocyte colony stimulating factor [G-CSF], 300 µg/day for 7 to 14 days) and HAA (homoharringtonine 2 mg/m2/day, d1-7; aclacinomycin 20 mg/day, d1-7; cytarabine 100 mg/m2/day, d1-7).
Consolidation therapy
Patients who achieved complete remission (CR) or CR with incomplete hematologic recovery (CRi) received four cycles of high-dose cytarabine (2 g/m2 every 12 h for 3 days). Refractory or relapsed patients received high-dose cytarabine-based regimens, such as revised CLAG (cladribine 5 mg/m2/day, d1–5; cytarabine 1 g/m2/day, d1–5; G-CSF 300 μg/day, d1–5), FLAG (fludarabine 25 mg/m2/day, d1–5; cytarabine 1 g/m2/day, d1–5; G-CSF 300 μg/day, d1–5), or venetoclax combined with demethylating agents such as azacitidine or decitabine.
Since 2014, the FLT3 inhibitor (FLT3i) sorafenib has been available in our center, and some patients with FLT3-ITD mutations received FLT3i therapy during induction, consolidation or relapse.
Patients who had FLT3-ITD mutation, who underwent morphological/molecular relapse, or who had a log reduction in NPM1 MRD after consolidation 2 (MRD2) < 3 log were indicated for allogeneic hematopoietic stem cell transplantation (allo-HSCT); the allo-HSCT protocol was performed as previously described [14, 15].
Molecular and cytogenetic analysis
Blood from the bone marrow was analyzed according to the 2009 International System for Human Cytogenetic Nomenclature (ISCN) using the G-banding technique. Types A, B and D of NPM1 mutation, as well as the rare type, and NPM1 MRD were detected by real-time quantitative polymerase chain reaction (RT-PCR) [16] at diagnosis and each end of the chemotherapy cycle.
Targeted next-generation sequencing
The DNA was extracted from bone marrow samples obtained at the time of diagnosis. Following extraction and purification of genomic DNA from patient samples, specific regions are captured using hybridization probes or PCR primers designed to target these genes. Utilizing Next-Generation Sequencing (NGS) with targeted capture, we sequenced mutation hotspots or the complete coding regions of 139 genes that are frequently mutated in myeloid neoplasms (refer to Supplementary Table 1). The raw variant results were filtered based on the following criteria: an average effective sequencing depth on target per sample of at least 2,000x, a mapping quality score of at least 30, a base quality score of at least 30, and a variant allele frequency (VAF) of at least 1% for both single-nucleotide variants (SNVs) and small insertions-deletions (InDels). Variants are then annotated based on their potential impact on protein function, using databases like the 1000 Genomes Project, COSMIC, and clinical databases.
Definitions
The criteria for CR were as follows: bone marrow blasts < 5%; absence of circulating blasts and blasts with Auer rods; absence of extramedullary disease; absolute neutrophil count (ANC) ≥ 1.0 × 109/L; and platelet (PLT) count ≥ 100 × 109/L. CRi was defined as meeting all CR criteria except for ANC or PLT count. Relapse was defined as bone marrow blasts ≥ 5%, the reappearance of blasts in the blood, or the development of extramedullary disease [9]. Molecular relapse was defined according to the European LeukemiaNet (ELN) MRD Working Party [17]. Overall survival (OS) was calculated from the date of diagnosis to the date of death due to any cause. Event-free survival (EFS) was calculated from the date of CR/CRi until the date of morphological relapse, molecular relapse, or death from any cause. MRD2 was defined as the log reduction in the transcript level of NPM1 at the end of the second cycle of consolidation compared with the baseline.
Statistical analysis
Continuous variables are reported as medians and ranges, and categorical variables are expressed as percentages. Competing risk analysis was used to calculate the cumulative incidence of relapse (CIR), and Gray’s test was used to test for differences between groups. Maximally selected rank statistics were calculated to determine the cutoff value of NPM1 MRD for predicting survival. OS and EFS were calculated by the Kaplan–Meier method, and comparisons were made with the log-rank test. A Cox regression model was used for the analysis of prognosis. P values < 0.05 were considered to indicate statistical significance. Hazard ratios (HR) were calculated with their 95% confidence intervals (CI). Data analyses were primarily conducted with the Statistical Package for the Social Sciences (SPSS), version 25.0 (SPSS Inc., IBM Corp., Armonk, NY, USA), and R software, version 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria), was used for competing risk analysis.
Results
Initial patient characteristics
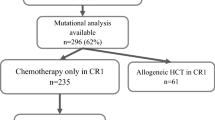
A total of 302 patients were diagnosed with AML with NPM1 mutation, and 234 patients for whom NGS data were available were included in this study (Fig. 1). The median age was 49 years (interquartile range [IQR] 36–58, and the median follow-up was 30.4 months (IQR 12.1–55.7). Forty genes were mutated > 1% of patients, and 9 genes were mutated > 10% of patients (Fig. 2A). DNMT3A was the most common co-mutation (53.8%), followed by FLT3-ITD (44.4%). The mutation interactions are shown in Fig. 2B. The pretreatment characteristics of patients are detailed in Table 1.
Overview of patients included in this study. NGS: next-generation sequencing; Chemoonly: chemotherapy without allogeneic hematopoietic stem cell transplantation; Allo-HSCT: allogeneic hematopoietic stem cell transplantation; NPM1RT: rare type of NPM1 mutation. In patients with NPM1RT mutation, 16 patients received chemotherapy only, and 11 patients received allo-HSCT. However, MRD of NPM1RT were not available, these patients were not included except the analysis of baseline
Overview of gene mutations identified by targeted sequencing in 234 AML patients with NPM1 mutation. a. Histogram showing the frequency of gene mutations detected in > 1% of patients. Bars are colored according to the functional category assigned to each gene. The FLT3 mutation included 25(49.0%) FLT3-TKD. b. Association of gene co-mutations in AML patients with NPM1 mutation
Treatment
Sorafenib
A total of 44.9% (93/207) of patients had FLT3-ITD mutation, and 32.3% (30/93) of those patients received FLT3i treatment. A total of 26.7% (8/30) of these patients received sorafenib only during induction chemotherapy, 10.0% (3/30) of patients received sorafenib from induction to consolidation therapy, and 6.7% (2/30) of patients received sorafenib from induction to maintenance therapy. A total of 40.0% (12/30) of patients used sorafenib from induction to allo-HSCT, and 6.7% (2/30) of patients used it from consolidation to allo-HSCT. A total of 10.0% (3/30) of patients received sorafenib when they relapsed.
Allo-HSCT
Among the 207 patients who received induction chemotherapy followed by consolidation chemotherapy, 31.9% (66/207) underwent allo-HSCT. The reasons for allo-HSCT in these 66 patients were FLT3-ITD mutation (54.5%, 36/66), molecular or morphological relapse (21.2%, 14/66), continuous positive MRD (16.7%, 11/66), MRD from negativity to positivity (4.5%, 3/66), and CR after 2 courses of induction (3.0%, 2/66). The median time from diagnosis to HSCT was 7.1 (IQR 5.8–10.8) months. The differences in baseline characteristics between the chemoonly group (not including 4 patients who died early) and the allo-HSCT group are shown in Table 2.
Response, relapse and death
Overall, 207 patients received induction therapy, during which 1.9% (4/207) died of infection or hemorrhage; therefore, the CR/CRi rate was available for 203 patients in the cohort. The CR/CRi rate after one induction course was 76.8% (156/203), the cumulative CR/CRi rate after two induction courses was 91.6% (186/203), and the total CR/CRi rate was 91.6% (186/203), with a median age of 50.0 (IQR 35–58) years. The median age of the 17 patients who did not achieve CR/CRi was 55 (IQR 50–61) years.
A total of 44.6% (83/186) and 4.3% (8/186) of patients experienced morphological relapse and molecular relapse, respectively, and the median relapse times were 9.8 (IQR 4.1–18.9) months and 15.7 (IQR 12.3–25.5) months, respectively. Among the 83 patients with morphological relapse, 10.8% (9/83) relapsed after allo-HSCT. Overall, 9.6% (8/83) and 25% (2/8) of patients received a venetoclax-based regimen after morphological relapse and molecular relapse, respectively. The cumulative incidence of morphological relapse (CIRm) at 12 months, 24 months and 36 months was 25.8% (95% CI 19.8–32.3%), 40.5% (95% CI 33.3–47.6%) and 43.8% (95% CI 36.4–51.0%), respectively.
A total of 42.0% (87/207) of patients died, and 1.9% (4/207) of patients died during induction therapy. The survival rates at 12 months, 24 months and 36 months were 79.0% (95% CI 73.5–84.5%), 67.0% (95% CI 60.5–73.5%) and 60.3% (95% CI 53.2–67.3%), respectively.
In the chemotherapy group, 46.0% (63/137) of the patients died due to relapse (63.5%, 40/63), no response to chemotherapy (25.4%, 16/63), infection (7.9%, 5/63), or hemorrhage (3.2%, 2/63). In the allo-HSCT group, 30.3% (20/66) of patients died, 35.0% (7/20) of whom died due to morphological relapse, 10.0% (2/20) due to no response to chemotherapy, and 55% (11/20) due to treatment-related mobility (TRM).
Among the 5 patients with complex karyotypes, 80% (4/5) achieved CR, and 75% (3/4) experienced morphological relapse, with relapse-free survival (RFS) of 20.6, 39.6, and 6.5 months and OS of 81.2, 45.1, and 8.5 months, respectively. One patient who achieved a consistent CR status was still alive at the end of the study, with an OS of 93.9 months. One patient who did not achieve CR died after 6.7 months.
Survival analysis
As NPM1 MRD data for rare type mutations were not available, survival analysis was conducted for 137 patients who received chemotherapy only, excluding NPM1RT patients.
Optimal cutoff value of MRD
MRD was analyzed in 137 chemoonly patients, and the optimal cutoff value for MRD2 reduction for both EFS (p < 0.001) and OS (p < 0.001) was a reduction ≥ 3 log. A reduction in MRD2 ≥ 3 log was significantly associated with increased EFS and OS compared with a reduction in MRD2 < 3 log (p < 0.001, p < 0.001) (Fig. 3).
Conversion of MRD status from negative to positive
NPM1 MRD in 60% (85/137) of patients converted to a negative status at a median time of 4.1 (IQR 2.8–5.9) months. MRD in 40.0% (34/85) of these patients converted from negative to positive (MRDneg to pos) thereafter, with a median time of 12.5 (IQR 4.6–25.9) months, and the median transcription level of NPM1 was 0.06% (IQR 0.01–0.57). Patients whose MRD was consistently negative (MRDconsist neg) had greater EFS (Fig. 4A) and OS (Fig. 4B) than patients with MRDneg to pos (both p < 0.001).
Co-occurring mutations
Genes with a prevalence ≥ 10% and those predicted to be at high risk by the 2022 ELN recommendation in the 137 chemoonly patients with NGS data are listed in Table 3. The most prevalent genes were DNMT3A (57.7%) and FLT3-ITD (40.1%).
Univariable and multivariable analyses of survival were performed for 137 chemoonly patients with NGS data. Univariate analysis revealed that age, FLT3-ITD co-mutation, DNMT3A co-mutation and a decrease in MRD2 < 3 log were associated with poorer EFS and OS, and the white blood cell (WBC) count only was a prognostic factor for poor OS. However, in multivariate analysis, DNMT3A co-mutation (EFS, HR = 1.9, p = 0.021; OS, HR = 2.2, p = 0.023) and MRD2 reduction ≥ 3 log (EFS, HR = 0.1; OS, HR = 0.1, both p < 0.001) were found to be independent prognostic factors for survival, and age was associated with lower EFS (HR = 1.3, p = 0.013) (Table 4).
The patients were divided into the following 4 groups according to the status of DNMT3A co-mutation and FLT3-ITD co-mutation: both FLT3-ITD and DNMT3A negative, FLT3-ITD-positive and DNMT3A-negative (12.5% [2/16] of patients received sorafenib), FLT3-ITD-negative and DNMT3A-positive, and both FLT3-ITD- and DNMT3A-positive (35.0% [14/40] of patients received sorafenib). EFS was calculated in 89.8% (123/137) of patients who achieved CR/CRi. The co-occurrence of FLT3-ITD and DNMT3A mutations was associated with the lowest EFS and OS (3-year EFS rate 30.0%, p < 0.001; 3-year OS rate 34.4%, p < 0.001). However, patients with only the FLT3-ITD co-mutation did not have poorer survival than patients without the FLT3-ITD co-mutation (3-year EFS rate 71.4% vs. 72.8%, p = 0.998; 3-year OS rate 75.0% vs. 74.6%, p = 0.789) (Fig. 5).
Survival of co-occurrent mutation status of FLT3-ITD and DNMT3A. (a) EFS, (b) OS. neg negative, pos positive. 7.1% (1/14) patient and 43.3% (13/30) patients received Sorafenib in G2 and G4 respectively in (a). 12.5% (2/16) patients and 35% (14/40) patients received Sorafenib in G2 and G4 respectively in (b)
Re-stratifying the risk groups
The favorable, intermediate, and poor-risk groups were re-stratified according to the status of FLT3-ITD and DNMT3A co-mutations and NPM1 MRD2 reduction. The poor-risk group comprised patients with a < 3 log reduction in MRD2 or with both FLT3-ITD and DNMT3A co-mutations achieved the lowest EFS (3-year EFS rate 30.0%) and OS (3-year OS rate 34.4%). The favorable-risk group comprised patients with FLT3-ITDneg + DNMT3Aneg and an MRD2 ≥ 3 log reduction, with a 3-year EFS rate of 80.8% and a 3-year OS rate of 90.3% (Fig. 6).
Survival of patients re-stratified by the status of FLT3-ITD and DNMT3A mutation and NPM1 reduction of MRD2. (a) EFS, (b) OS. Favorable risk group, patients with FLT3-ITDneg + DNMT3Aneg and MRD2 ≥ 3log reduction. Poor risk group, FLT3-ITDpos + DNMT3Apos or MRD2 < 3log reduction. Intermediate risk group, neither favorable nor poor risk group. In (a), the percentage of patients received Sorafenib were 2.3% (1/44), 0(0/47), 43.3% (13/30) in three different risk groups, while 2.3% (1/44), 0(0/48), 35.0% (14/40) in (b)
HSCT improved the survival of poor-risk patients
A total of 29.6% (60/203) of the 203 patients who had NGS data, excluding those with NPM1RT or who died early, were classified into the poor-risk group. A total of 66.7% (40/60) of patients received chemotherapy only, and 33.3% (20/60) of patients underwent allo-HSCT. HSCT improved EFS (3-year EFS rate 57.9% vs. 30.0%, p = 0.012) compared with patients who received chemotherapy only in 81.7% (49/60) of patients who achieved CR/CRi (Fig. 7a). Additionally, the OS in the allo-HSCT group was greater than that in the chemotherapy group (3-year OS rate 72.0% vs. 34.4%, p = 0.001) (Fig. 7b).
Discussion
AML with NPM1 mutation is the most common subtype of adult AML and has relatively strong heterogeneity [9]. Risk stratification of AML patients with NPM1 mutation according to international guidelines was mainly based on whether an FLT3-ITD co-mutation was present, namely, the favorable-risk group included patients without an FLT3-ITD co-mutation, and the intermediate-risk group included patients with an FLT3-ITD co-mutation [8]. Only approximately 3.4% of patients have chromosomal abnormalities that cause adverse risk [18] and are categorized into the adverse-risk group [8].
NPM1 mutation is an ideal target for MRD monitoring to predict relapse. However, different centers have defined different prognostic thresholds and time points of NPM1 MRD. In the study by Max et al. of 158 patients from the AMLCG 1999, 2004 and 2008 trials, a reduction of 3 log in NPM1 MRD after induction or consolidation therapy was the cutoff for predicting relapse (p = 0.001, p = 0.001) [19]. In the ALFA-0701 trial, patients with NPM1 MRD positivity (defined as > 0.1% in the bone marrow) after induction and at the end of treatment also had a greater risk of relapse, but OS did not differ [20]. Balsat et al. also studied the relationship between peripheral blood (PB) NPM1 MRD and relapse or OS, and a > 4 log reduction in PB MRD after induction therapy was significantly associated with a lower rate of CIR and shorter OS [10]. Our center’s results are consistent with our previously published data, in which a reduction in MRD2 ≥ 3 log was associated with increased disease-free survival and OS [21]. Moreover, Ivey et al. confirmed that a positive NPM1 MRD in the PB after the second chemotherapy course was associated with a greater risk of relapse in patients in the AML17 trial (3-year CIR rate 82% vs. 30%, p < 0.001) [22]. Fabio Guolo et al. reported that in 19% (8/42) of AML patients with NPM1 mutation who experienced morphological relapse, the recurrence of NPM1 MRD contributed to relapse, with a median time of 4.5 months [23]. However, our study aimed to explore the significance of the shift from negative to positive MRD, rather than simply comparing negative and positive MRD. Univariable analysis in our study demonstrated that patients with continuously negative NPM1 MRD had higher EFS and OS than those patients who shifted from negative to positive MRD, but this finding still needs to be confirmed via multivariable analysis.
However, the prognosis of AML patients with NPM1 mutation could be affected by multiple factors, since the relapse rate was not very low. More co-mutations have been identified in AML patients with NPM1 mutation than in any other subtype of AML. Accumulating evidence shows that the outcome of AML patietns with NPM1 mutation may vary because of co-mutations other than the FLT3-ITD mutation. AML patients with NPM1 and FLT3-ITD co-mutations were classified into the intermediate-risk group [8]; however, the survival of AML patients with NPM1 and FLT3-ITD co-mutations was similar to that of AML patients with only NPM1 mutations in our study. In our study, DNMT3A co-mutation were found in 53.8% of AML patients with NPM1 mutation, while FLT3-ITD co-mutation were found in 44.4%. In the FLT3-ITD co-mutation group, 61.5% of patients were FLT3-ITDpos + DNMT3Apos; therefore, it was hypothesized that these patients contributed to the poor survival rates rather than the patients with FLT3-ITD mutation alone. Specifically, only patients with both FLT3-ITD and DNMT3A co-mutations had a poor prognosis. Elli Papaemmanuil et al. also reported that the deleterious effect of FLT3-ITD mutation was most clinically relevant in patients with concomitant NPM1 and DNMT3A co-mutations [24], which were shown to be associated with a high leukemia stem cell frequency and synergistic upregulation of specific leukemia stem cell regulators [25]. The co-occurrence of NPM1/FLT3-ITD/DNMT3A mutations was associated with decreased OS and disease-free survival [25]. Furthermore, a link between the co-occurrence of NPM1/FLT3-ITD/DNMT3A mutations and AML resistance to anthracycline has been identified in functional studies [26]. The effect of FLT3-ITD mutation on survival was less obvious when it co-occurred with either an NPM1 or DNMT3A mutation alone [24].
Other co-mutation, such as TET2, PTPN11, IDH1, IDH2 and NRAS had no prognostic on OS, which were coincidence with the latest report including 1357 patients [27]. However, FLT3-TKD was controversial. In the 1357 patients’ study and a recent study from China, FLT3-TKD did not influence OS [27, 28], other study verified it improved OS [29, 30], and our study found it decreased OS. Maybe different treatment background such as FLT3i may change the outcome.
In addition, we combined MRD and NPM1 co-mutations to achieve a more accurate risk re-stratification of AML patients with NPM1 mutation. Maël Heiblig et al. reported that in patients aged older than 60 years (median age 66.1 years), patients with co-occurring NPM1/FLT3-ITD/DNMT3A mutations, or with co-occurring NPM1/DNMT3A mutations and MRD reduction < 4 log after the first cycle of induction were classified as poor risk, with a median of 7.7 months of leukemia-free survival [11]. Our study verified that the co-occurrence of NPM1/FLT3-ITD/DNMT3A mutations or an MRD2 reduction < 3 log could be used to identify the poor-risk group among younger elderly patients with a median age of 54 years. Moreover, the conversion from negative to positive MRD status was also associated with poor outcomes, but multivariate analysis needs to be performed for verification.
More than half of the patients were diagnosed before FLT3i were widely used; furthermore, owing to the financial limitations of using FLT3i at that time, only a small proportion of patients received sorafenib during treatment—11.7% (16/137) of patients in the chemoonly group and 21.2% (14/66) of patients in the HSCT group. However, in our survival analysis, more patients received sorafenib in the FLT3-ITDpos + DNMT3Apos group than in the FLT3-ITDpos + DNMT3Aneg group (14 patients vs. 2 patients), and the prognosis of patients in the FLT3-ITDpos + DNMT3Apos group was still poor. In the current era of targeted therapy, the Bcl-2 inhibitor venetoclax is effective for treating AML with NPM1 mutation [31], and our previous data also suggested that the rate of CR/CRi after one induction cycle was 77.8% (14/18) [32]. Venetoclax–based low-intensity chemotherapy results in 84% of molecular failure.
AML patients with NPM1 mutation achieved an MRD response, and 71% of these patients became MRD negative [33]. In refractory/relapsed (R/R) AML patients with NPM1 mutation, venetoclax combination therapy has a greater response rate of CR/CRi (46%) [34]. In our study, a total of 10 patients received venetoclax-based therapy after molecular or morphological relapse, which prolonged survival. Sorafenib plus triple therapy with venetoclax, azacitidine, and homoharringtonine (VAH) was well tolerated and highly effective against R/R AML with FLT3-ITD mutation [35], and venetoclax- and FLT3i-based therapy may be more suitable and more effective for treating AML patients with NPM1 mutation who have FLT3-ITD and DNMT3A co-mutations.
In conclusion, the deleterious effect of FLT3-ITD mutation is more pronounced when concomitant with DNMT3A mutation in AML patients with NPM1 mutation. A < 3 log reduction in MRD2 was also an independent prognostic factor for poor survival, which could be improved by allo-HSCT. In addition, the shift from negative to positive MRD status was also associated with poor EFS and OS according to the univariable analysis.
Data availability
No datasets were generated or analysed during the current study.
References
Thiede C et al (2006) Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood 107(10):4011–4020
Falini B et al (2005) Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med 352(3):254–266
Patel JP et al (2012) Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 366(12):1079–1089
Polyatskin IL, Artemyeva AS, Krivolapov YA (2019) Revised WHO classification of tumors of hematopoietic and lymphoid tissues, 2017 (4th edition):lymphoid tumors. Arkh Patol 81(3):59–65
Döhner K et al (2005) Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 106(12):3740–3746
Schnittger S et al (2005) Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 106(12):3733–3739
Schlenk RF et al (2008) Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 358(18):1909–1918
Döhner H et al (2022) Diagnosis and management of AML in adults: 2022 ELN recommendations from an international expert panel on behalf of the ELN. Blood 140:1345–1377
Döhner H et al (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129(4):424–447
Balsat M et al (2017) Postinduction minimal residual disease predicts outcome and benefit from allogeneic stem cell transplantation in acute myeloid leukemia with NPM1 mutation: a study by the Acute Leukemia French Association Group. J Clin Oncol 35(2):185–193
Heiblig M et al (2021) The impact of DNMT3A Status on NPM1 MRD predictive value and survival in elderly AML patients treated intensively. Cancers (Basel) 13(9):2156
中华医学会血液学分会, 成人急性髓系白血病(非急性早幼粒细胞白血病)中国诊疗指南(2011年版). Chinese J Hematol 2011. 32: 804–807
Döhner H et al (2010) Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 115(3):453–474
Xu L et al (2018) The consensus on indications, conditioning regimen, and donor selection of allogeneic hematopoietic cell transplantation for hematological diseases in China-recommendations from the Chinese Society of Hematology. J Hematol Oncol 11(1):33
Lu DP et al (2006) Conditioning including antithymocyte globulin followed by unmanipulated HLA-mismatched/haploidentical blood and marrow transplantation can achieve comparable outcomes with HLA-identical sibling transplantation. Blood 107(8):3065–3073
Ruan GR et al (2009) Nucleophosmin mutations in Chinese adults with acute myelogenous leukemia. Ann Hematol 88(2):159–166
Schuurhuis GJ et al (2018) Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood 131(12):1275–1291
Angenendt L et al (2019) Chromosomal abnormalities and prognosis in NPM1-Mutated Acute Myeloid Leukemia: a pooled analysis of individual patient data from nine international cohorts. J Clin Oncol 37(29):2632–2642
Hubmann M et al (2014) Molecular response assessment by quantitative real-time polymerase chain reaction after induction therapy in NPM1-mutated patients identifies those at high risk of relapse. Haematologica 99(8):1317–1325
Lambert J et al (2014) MRD assessed by WT1 and NPM1 transcript levels identifies distinct outcomes in AML patients and is influenced by gemtuzumab ozogamicin. Oncotarget 5(15):6280–6288
Zhao T et al (2017) Prognostic significance of early assessment of minimal residual disease in acute myeloid leukemia with mutated NPM1 patients. Zhonghua Xue Ye Xue Za Zhi 38(1):10–16
Ivey A et al (2016) Assessment of minimal residual disease in standard-risk AML. N Engl J Med 374(5):422–433
Guolo F et al (2019) Longitudinal minimal residual disease (MRD) evaluation in acute myeloid leukaemia with NPM1 mutation: from definition of molecular relapse to MRD-driven salvage approach. Br J Haematol 186(6):e223–e225
Papaemmanuil E et al (2016) Genomic classification and prognosis in Acute Myeloid Leukemia. N Engl J Med 374(23):2209–2221
Bezerra MF et al (2020) Co-occurrence of DNMT3A, NPM1, FLT3 mutations identifies a subset of acute myeloid leukemia with adverse prognosis. Blood 135(11):870–875
Guryanova OA et al (2016) DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med 22(12):1488–1495
Othman J et al (2024) Molecular, clinical and therapeutic determinants of outcome in NPM1 mutated AML. Blood 144:714–728
Yao Y et al (2024) Co-mutation landscape and its prognostic impact on newly diagnosed adult patients with NPM1-mutated de novo acute myeloid leukemia. Blood Cancer J 14(1):118
Perry M et al (2018) FLT3-TKD Mutations Associated With NPM1 Mutations Define a Favorable-risk Group in Patients With Acute Myeloid Leukemia. Clin Lymphoma Myeloma Leuk 18(12):e545–e550
Boddu P et al (2017) Co-occurrence of FLT3-TKD and NPM1 mutations defines a highly favorable prognostic AML group. Blood Adv 1(19):1546–1550
DiNardo CD et al (2020) Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. N Engl J Med 383(7):617–629
Yu WJ et al (2022) Short-term efficacy of venetoclax combined with azacitidine in acute myeloid leukemia: a single-institution experience. Zhonghua Xue Ye Xue Za Zhi 43(2):134–140
Jimenez-Chillon C et al (2024) Venetoclax-based low intensity therapy in molecular failure of NPM1-mutated AML. Blood Adv 8(2):343–352
Stahl M et al (2021) Clinical and molecular predictors of response and survival following venetoclax therapy in relapsed/refractory AML. Blood Adv 5(5):1552–1564
Yu S et al (2024) Sorafenib plus triplet therapy with venetoclax, azacitidine and homoharringtonine for refractory/relapsed acute myeloid leukemia with FLT3-ITD: A multicenter phase 2 study. J Intern Med 295(2):216–228
Acknowledgements
We thank medical staff and patients’ participants.
Funding
This work was funded by The Grants from the Beijing Municipal Science and Technology Commission (Z221100007422008), Peking University People’s Hospital Research and Development Funds (RDL2022-13), Peking University Medicine Fund for world’s leading discipline or discipline cluster development (No.71003Y3035).
Author information
Authors and Affiliations
Contributions
Hao Jiang and Xiaojun Huang designed the study. Wenbing Duan collected and analyzed the data. Hao Jiang, Wenbing Duan prepared the typescript. All authors approved the final typescript, take responsibility for the content, and agreed to submit for publication.
Corresponding authors
Ethics declarations
This study was approved by the Ethics Committee of Peking University People’s Hospital (approval number 2022PHD019-001, 2023PHB164-001) and was conducted according to the principles of the Helsinki Declaration. Informed consent was obtained from all individual participants included in the study. The authors declare no known no potential conflicts of interest.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Duan, W., Jia, J., Wang, J. et al. Only FLT3-ITD co-mutation did not have a deleterious effect on acute myeloid leukemia patients with NPM1 mutation, but concomitant with DNMT3A co-mutation or a < 3log reduction of MRD2 predicted poor survival. Ann Hematol (2024). https://doi.org/10.1007/s00277-024-06001-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00277-024-06001-6