Abstract
Rhodosporidium toruloides is an oleaginous yeast capable of producing a variety of biofuels and bioproducts from diverse carbon sources. Despite numerous studies showing its promise as a platform microorganism, little is known about its metabolism and physiology. In this work, we investigated the central carbon metabolism in R. toruloides IFO0880 using transcriptomics and metabolomics during growth on glucose, xylose, acetate, or soybean oil. These substrates were chosen because they can be derived from plants. Significant changes in gene expression and metabolite concentrations were observed during growth on these four substrates. We mapped these changes onto the governing metabolic pathways to better understand how R. toruloides reprograms its metabolism to enable growth on these substrates. One notable finding concerns xylose metabolism, where poor expression of xylulokinase induces a bypass leading to arabitol production. Collectively, these results further our understanding of central carbon metabolism in R. toruloides during growth on different substrates. They may also help guide the metabolic engineering and development of better models of metabolism for R. toruloides.
Key points
• Gene expression and metabolite concentrations were significantly changed.
• Reduced expression of xylulokinase induces a bypass leading to arabitol production.
• R. toruloides reprograms its metabolism to allow growth on different substrates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhodosporidium toruloides is a red basidiomycete yeast that can produce a variety of biofuels and bioproducts from diverse carbon sources, including lignocellulosic hydrolysates and lignin-derived aromatics (Fei et al. 2016; Hu et al. 2009; Huang et al. 2016; Kitahara et al. 2014; Singh et al. 2016; Wiebe et al. 2012; Yaegashi et al. 2017; Zhang et al. 2016b). It is an oleaginous yeast that accumulates triglycerides during growth under nitrogen, phosphorous, or sulfur limitation (Wang et al. 2018; Wiebe et al. 2012; Wu et al. 2011; Zhang et al. 2016b, b). In addition, R. toruloides can natively produce the sugar alcohols arabitol and galactitol from xylose and galactose, respectively (Jagtap et al. 2019; Jagtap and Rao 2018a,b). Recently, R. toruloides has been engineered for enhanced lipid production and production of fatty alcohols, terpenes, and methyl ketones (Dinh et al. 2019; Fillet et al. 2015, 2017; Geiselman et al. 2020; Lee et al. 2014, 2016; Liu et al. 2020; Wehrs et al. 2019; Wen et al. 2020; Yaegashi et al. 2017; Zhang et al. 2016a,b). Finally, R. toruloides can provide a natural source for carotenoids and industrially relevant enzymes such as L-phenylalanine ammonia lyase and D-amino acid oxidase (Cui et al. 2014; Fernández-Lafuente et al. 1998; Gilbert et al. 1985; Hoskins et al. 1980; Pilone and Pollegioni 2002).
Despite its ability to produce a wide range of value-added products, little is known about the physiology of this organism aside from a few key studies. In one seminal study, researchers measured changes in gene and protein expression in R. toruloides NP11 during growth on glucose in minimal medium under nitrogen limitation (Zhu et al. 2012). Based on these data, they were able to propose a model for lipid accumulation arising from nitrogen starvation. The same group also profiled changes in the lipid-droplet proteins in R. toruloides NP11 grown on glucose during nitrogen and phosphorous starvation (Zhu et al. 2015). In another study of note, phosphate limitation was found to increase the expression of genes involved in phosphate metabolism, RNA degradation, and lipid biosynthesis, while those involved in the tricarboxylic acid (TCA) cycle and ribosome biosynthesis were decreased (Wang et al. 2018). In another seminal study, researchers constructed a barcoded library of single-gene deletion mutants for R. toruloides IFO0880. Using this library, they were able to identify over a thousand essential genes and 150 genes affecting lipid production (Coradetti et al. 2018). Recently, RNAseq analysis was performed on R. toruloides IFO0880 cultivated on synthetic defined medium on multiple carbon sources to identify the promoters that can constitutively express native genes at high and medium levels. In the process, the authors identified 12 mono-directional and 8 bi-directional promoters for R. toruloides (Nora et al. 2019). Lastly, a recent study investigated the metabolic pathways and genes involved in carbon consumption in R. toruloides IFO0880 using multi-omics data. In addition, they have developed a genome scale metabolic network model by validating against growth phenotyping and gene fitness data (Kim et al. 2021).
In this work, we investigated the metabolism of glucose, xylose, acetate, and soybean oil by R. toruloides IFO0880. These substrates were chosen because they represent potential substrates for bioconversion by R. toruloides. To understand the metabolism of these substrates, we measured changes in gene expression using RNAseq and intracellular metabolites using GC–MS. We then mapped these data onto the central carbon metabolic pathways of R. toruloides in order to understand how they are regulated during growth on different carbon sources.
Materials and methods
Strains, media, and growth conditions
R. toruloides IFO0880, mating type A2, was obtained from the NITE Biological Resource Center in Japan (NBRC 0880). YPG medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) was used for growth of R. toruloides. A single colony from a YPG agar plate was inoculated into 2 mL of YPG liquid medium to obtain R. toruloides seed cultures. Seed cultures were then used to inoculate 25 mL of YPG, YPX medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L xylose), YPA medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L sodium acetate, pH 7.0), YPS medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L soybean oil), and YP medium (10 g/L yeast extract and 20 g/L peptone) in a 125-mL baffled shake flask with a starting OD600 of 1. The optical density at 600 nm or OD600 was used to monitor the cell density in liquid cultures. OD600 of 1.0 corresponds to roughly 107 cells per mL. The cells were then grown at 30 °C and 250 rpm.
Sample extraction for RNA-seq and metabolomics
Seed cultures at exponential phase were collected and centrifuged at 6000 × g for 3 min at 4 °C. Supernatant was discarded and the pellets were resuspended in 1 mL of ddH2O. Seed cultures then used to inoculate 25 mL of YPG, YPX, YPA, YPS, and YP medium in a 125-mL baffled shake flask with a starting OD600 of 1 and incubated at 30 °C and 250 rpm. Growth experiments are performed with three biological replicates. Samples from YPG, YPX, YPA, and YP media were collected after 24 h incubation, and sample from YPS media was collected after 16 h incubation.
Experimental procedure for RNA-Seq
The cell cultures containing a total OD of 30 were collected in centrifuge tubes and centrifuged at 6000 × g for 3 min at 4 °C. Supernatant was discarded and pellet was used for RNA extraction. Total RNA was extracted using the RNeasy mini kit (Qiagen, Hilden, Germany) as previously described, with a slight modification (Jagtap et al. 2019; Zhang et al. 2019). R. toruloides cell pellet was resuspended in 350 ul of Buffer RLT from the RNeasy mini kit (Qiagen, Hilden, Germany). Approximately 500 μl of acid-washed glass beads (acid washed, 425–600 μm; Sigma, St. Louis, MO, USA) was added and homogenized using a FastPrep-24 homogenizer (MP Biomedicals, Irvine, CA, USA), beaten at a speed of 5 m/s for 30 s six times with cooling on ice between beatings. The cell lysates were purified according to the kit’s protocol “purification of total RNA from yeast.” Extracted RNA was then treated with Turbo RNase-free DNase kit (ThermoFisher, Waltham, MA, USA) according to the manual and purified again with the RNeasy mini kit protocol “RNA clean up.” The stranded RNAseq libraries were prepared with Illumina’s TruSeq Stranded mRNA Sample Prep kit. The libraries were quantitated by qPCR and sequenced on one lane for 101 cycles from one end of the fragments on a HiSeq 4000 (Illumina, San Diego, CA, USA). Fastq files with 100 bp reads were generated and demultiplexed with the bcl2fastq v2.17.1.14 Conversion Software (Illumina, San Diego, CA, USA). Raw sequencing files were uploaded to NCBI (NCBI Bioproject Accession: PRJNA660884, NCBI SRA Accession: SRR12567876 to SRR12567890).
RNA-Seq data analysis
To obtain gene expression profiles during growth of R. toruloides IFO0880 on different substrates, total RNA was extracted, and a mRNA focused library was sequenced. Adaptor sequences and low-quality reads were trimmed using Trimmomatic (Bolger et al. 2014). Trimmed reads were analyzed for quality scores using FastQC (Andrews 2010). Reads were mapped to the R. toruloides IFO0880 v4.0 reference genome (NCBI Accession GCA_000988875.2) with STAR version 2.5.4a (Coradetti et al. 2018; Dobin et al. 2012). Between 95 and 98% of the reads were successfully mapped to the genome for each sample. Read counts were calculated using featureCounts from the Subread package, v1.5.2 (Liao et al. 2014). Differential expression analysis was performed on the reads counts in R v4.0.5 (Cite R core team) using edgeR v3.32.1 and limma v3.46.0 (Ritchie et al. 2015; Robinson et al. 2010). Graphical representation of expression data was constructed using R packages: PCAtools v2.2.0, gplots v3.1.1, and Glimma v2.0.0 (Blighe K 2021; Su et al. 2017; Warnes 2011). Before plotting heatmaps, the data was normalized row-wise (using the scale function in R), first by centering (subtracting the row mean from each value) and then scaling (dividing each data point by row’s standard deviation). Heatmaps were plotted using heatmap.2 function from gplots. Genome sequence, gene models, and functional annotation of R. toruloides was downloaded from the DOE Joint Genome Institute’s Mycocosm portal (Coradetti et al. 2018; Grigoriev et al. 2014). Data analysis scripts, along with the results, were uploaded at https://github.com/raogroupuiuc/rt88_growth.
Sample preparation for metabolome analysis
A fast filtration sampling method was used for intracellular metabolite analysis, as previously described, with a slight modification (Kim et al. 2013; Yun et al. 2018). Briefly, 0.5 mL of cells was collected from all medias and vacuum-filtered using a vacuum manifold system (Vac-Man 96 Vacuum Manifold, Promega, Madison, WI, USA) assembled with a nylon membrane filter (0.45 µm pore size, 13 mm diameter, Whatman, Piscataway, NJ, USA) and a filter holder (Millipore, Billerica, MA, USA). The filtered cells were then washed with 2.5 mL of distilled water at room temperature. The entire process for fast filtration was completed within 1 min. The filter membrane containing the washed cells was quickly mixed with 1 mL of the pre-chilled acetonitrile/water mixture (1:1, v/v) and 100 µL of glass beads. The extraction mixture was vortexed for 3 min to disrupt the cell membrane and to extract intracellular metabolites. The extraction mixture was then centrifuged at 16,000 × g for 3 min at 4 °C, and 0.8 mL of the supernatant containing the intracellular metabolites was dried using a speed vacuum concentrator for 6 h. The samples were prepared for GC/MS analysis as mentioned in the analytical methods section.
Metabolome data analysis
The raw data obtained from the GC/MS analysis were processed using an automated mass spectral deconvolution and identification system (AMDIS) software for peak detection and deconvolution of mass spectra (Stein 1999). The processed data were uploaded to SpectConnect (http://spectconnect.mit.edu) for peak alignment and generating the data matrix with the Golm Metabolome Database (GMD) mass spectral reference library (Kopka et al. 2005; Styczynski et al. 2007). The normalized abundance values for each metabolite were obtained by dividing the peak intensities with dry cell weight. For statistical analysis, such as PCA analysis and hierarchical cluster analysis representing as a heat map, Statistica (version 7.1; StatSoft, Tulsa, OK, USA), MetaboAnalyst and MultiExperiment Viewer software were used, respectively (Howe et al. 2010),(Chong et al. 2019). GraphPad Prism 6 (GraphPad, San Diego, CA, USA) was used for plotting intensity graphs.
Analytical methods
Prior to analysis, culture samples were centrifuged, and the supernatant was passed through a 0.22-μm polyethersulfone syringe filter. Glucose, xylose, and acetate concentrations were measured using a Shimadzu high-performance liquid chromatography system (Shimadzu, Kyoto, Japan) equipped with a cation H micro-guard cartridge (Bio-Rad Laboratories Hercules, CA, USA) and Aminex HPX-87H carbohydrate analysis column (Bio-Rad Laboratories, Hercules, CA, USA) operated at 65 °C. Eluent was 5 mM H2SO4 at a constant flow rate of 0.6 mL/min. Compounds were monitored using a RID-10A refractive index detector (Shimadzu, Kyoto, Japan) and were quantified using calibration curves built using authentic standards.
The metabolome samples were derivatized by methoxyamination and trimethylsilylation as described previously (Kim et al. 2013; Yun et al. 2018). For methoxyamination, 10 µL of 40 mg/mL methoxyamine chloride in pyridine (Sigma-Aldrich, St. Louis, MO, USA) was added to the samples and incubated for 90 min at 30 °C and 200 rpm. The samples were then trimethylsilylated by adding 45 µL of N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich, St. Louis, MO, USA) for 30 min at 37 °C and 200 rpm. For GC/MS, the derivatized metabolite samples were applied to an Agilent 7890A GC/5975C MSD system (Agilent Technologies, Wilmington, DE, USA) equipped with a RTX-5Sil MS capillary column (30 m × 0.25 mm, 0.25 µm film thickness; Restek, Bellefonte, PA, USA) and an additional 10 m long integrated guard column. One microliter of the derivatized sample was injected into the GC inlet in splitless mode. The oven temperature was initially set at 150 °C for 1 min, after which the temperature was increased to 330 °C at 20 °C/min, where it was held for 5 min. The mass spectra were recorded in a scan range 85–500 m/z at an electron impact of 70 eV, and the temperatures of the ion source and transfer line were 230 °C and 280 °C, respectively.
Results
Growth profiles in different carbon source
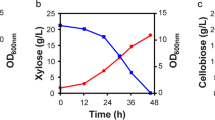
R. toruloides was grown on YP medium alone or on YP medium containing one of the following substrates: glucose (YPG), xylose (YPX), acetate (YPA), or soybean oil (YPS). YP was chosen as the base medium, because it can support robust, vegetative growth. We note that R. toruloides can grow on YP medium alone. In addition, since this medium contains sufficient nutrients, the cells do not enter the lipogenic phase where lipids accumulate within the cell. The rationale for choosing this medium was that we were focused solely on exploring the metabolic pathways for utilizing these substrates rather than on identifying the pathways involved in lipid production. As shown in Fig. 1, R. toruloides achieved highest cell densities on glucose, followed by acetate, xylose, and soybean oil. Growth on soybean oil was similar to YP medium alone. In addition, R. toruloides was able to completely utilize 20 g/L glucose, xylose, and acetate after 48 h of growth. Consistent with the growth data, glucose was utilized at a rate faster than either xylose or acetate. Unfortunately, we were unable to accurately measure the utilization of soybean oil.
Growth profiles of R. toruloides IFO0880. Growth on different carbon sources at 20 g/L in rich medium: glucose (a), xylose (b), acetate (c), soybean oil (d), and yeast peptone (e). Solid diamonds (♦) are used to denote sugar concentrations. Solid circles (●) are used to denote the optical density (OD600). For each plot (a–c), sugar concentration is plotted on the left y-axis, and OD600 values are plotted on the right y-axis
Identification of genes associated with substrate utilization
We used RNA-Seq to measure differences in gene expression during growth on the different substrates. RNA was isolated for three biological replicates during the exponential growth phase (24 h for YPG, YPX, YPA, and YP medium; and 16 h for YPS medium). A total of 229 million raw reads were obtained from fifteen samples. Approximately 88% of the reads mapped to a unique location on the R. toruloides IFO0880 genome (Supplementary Fig. S1 and Supplementary Table S1). Principal component analysis demonstrated that these growth changes led to clearly distinct expression patterns (Fig. 2). Only during growth on YPS medium did we not observe a large change from growth on YP medium alone. This may reflect that growth on both was similar (Fig. 1).
We next identified genes with altered expression during utilization of the different substrates. We compared growth on YPG with growth on YP to identify the pathways involved in uptake of glucose. We chose the same timepoint (24 h) for RNA extraction for both YPG and YP, with the intention of emphasizing the gene expression resulting from glucose uptake. For analyzing the gene expression patterns during growth on xylose, acetate, and soybean oil, growth on glucose was chosen as the control. YPG, YPX, and YPA were sampled at 24 h, which was mid-exponential and close to 50% substrate utilization. However, YPS was sampled at 16 h, at mid-exponential growth (Fig. 1d).
R. toruloides IFO0880 has 8490 predicted genes. Among these genes, 3582 were upregulated and 3627 were downregulated during growth on the four substrates as compared to YP medium alone (Supplementary Fig. S2). We also compared differential expression during growth on glucose, xylose, acetate, and soybean oil. The 1488 genes have significantly higher expression, and another 1419 genes have significantly lower expression during growth on glucose as compared to YP. The expression of 882, 1297, and 1296 genes was significantly higher during growth on xylose, acetate, and soybean oil, respectively, as compared to glucose. Likewise, the expression of 537, 1693, and 1392 genes was significantly lower during growth on xylose, acetate, and soybean oil as compared to YPG. For comparative gene expression, fold change > 2 and adjusted p value < 0.05 was considered significant. Details are provided in Supplementary Dataset S1.
Changes in intracellular metabolites during growth on different substrates
We used gas chromatography-mass spectrometry (GC–MS) to measure changes in the concentrations of 55 intracellular metabolites as described previously (Jagtap et al. 2019; Jagtap and Rao 2018a,b). Among the 55 metabolites analyzed (Supplementary Dataset S2), significant differences in the concentrations of 44 metabolites were observed during growth on glucose, xylose, acetate, or soybean oil as compared to YP (Fig. 3). Significance was evaluated using two sample t-tests, and the cutoffs for significance were relative metabolite concentration > 2 and adjusted p value < 0.05. The 16 metabolites had higher concentration and another 10 metabolites had lower concentrations during growth on glucose, as compared to YP. The concentrations of 7, 9, and 7 metabolites were higher during growth on xylose, acetate, and soybean oil, respectively, as compared to glucose. Likewise, the concentrations of 17, 11, and 16 metabolites were lower during growth on xylose, acetate, and soybean oil, respectively, as compared to glucose.
Heat map of intracellular metabolites from glycolysis (a), fatty acid biosynthesis (b), TCA cycle (c), and xylose utilization (d) of R. toruloides IFO0880 grown on glucose, xylose, acetate, soybean oil, and yeast peptone. All experiments were performed with six replicates. Color key represents the z-score for each metabolite (normalized for all growth conditions)
Growth on glucose
We first explored glucose metabolism in R. toruloides by comparing gene expression on YP medium containing glucose (YPG) versus YP medium alone. We chose the same timepoint (24 h) for analysis in order to isolate changes in gene expression resulting from glucose consumption. Overall, 1488 genes were significantly upregulated, and another 1419 genes were significantly downregulated during growth on YPG versus YP alone (fold change > 2 and p < 0.05 was considered significant).
We focused on the specific genes involved in central carbon metabolism (Figs. 4 and 5). With regard to glycolysis, namely the Embden–Meyerhof–Parnas pathway, we found that the genes involved in the lower half of the pathway were upregulated at least twofold during growth on YPG versus YP. However, the expression of hexokinase (HXK2) and phosphofructokinase (PFK) was reduced during growth on YPG. We did not observe any significant changes in the expression of the genes involved in the pentose phosphate pathway except for 6-phosphogluconolactonase (SOL3), whose expression was increased during growth on YPG. In addition, the expression of the alpha and beta subunits of pyruvate dehydrogenase (PDA1 and PDB1) was increased during growth on YPG. Invertase expression (SUC2) was significantly reduced (32-fold) during growth on YPG, which is consistent with results from S. cerevisiae (Lutfiyya and Johnston 1996; Mormeneo and Sentandreu 1982).
Transcriptional changes in pathways associated with TCA cycle (a), glycolysis (b), pentose phosphate pathway (c), xylose utilization (d), acetate utilization (e), and lipid biosynthesis (f) in R. toruloides IFO0880 grown on glucose (YPG), xylose (YPX), acetate (YPA), soybean oil (YPS), and yeast peptone (YP). RNAseq data was collected in triplicate for each condition. Color key represents the z-score for each gene (normalized for all growth conditions)
Significant changes were also observed in the expression of the genes involved in the TCA cycle. The expression of citrate synthase (CIT1), malate dehydrogenase (MDH1), and fumarate hydratase (FUM1) was increased during growth on YPG. However, the expression of succinate dehydrogenase (membrane anchor; SDH4) and α-ketoglutarate dehydrogenase (KGD1) was reduced. In addition, the expression of the cytosolic isocitrate lyase (ICL1) and malate synthase (MLS1) was reduced.
Aside from glucose metabolism, expression of the genes involved in lipid catabolism was significantly reduced. Expression of the genes involved in lipid biosynthesis was increased. These included ATP citrate lyase (ACL1), which provides cytosolic acetyl-CoA for lipid biosynthesis, and acetyl-CoA carboxylase, which generates malonyl-CoA. In addition, the expression of glutamate dehydrogenase (GDH1) was greatly increased (115-fold). However, the expression of NAD-dependent glutamate dehydrogenase GDH2 was reduced. The expression of general amino-acid GAP1 permeases (protein ID: 14,229 and 15,074) was also reduced. However, the expression of DIP5 dicarboxylic amino-acid transporters (protein ID: 8962, 9319, and 9322) and K16261 YAT amino-acid transporter (protein ID: 12,758) was increased. We also observed the increased expression of several putative sugar transporters and the decreased expression of a few other putative sugar transporters (Supplementary Table S2). Whether these transporters are specific for glucose or other sugars is not known.
With respect to intracellular metabolites (Fig. 3 and Supplementary Fig. S4), the concentrations of glucose, 3-phosphoglycerate, and pyruvate were increased during growth on YPG versus YP, which is consistent with their role in glycolysis. In addition, galactose concentrations were also increased. We also observed increased concentration of ornithine-1,5-lactam (more commonly known as 3-aminopiperidin-2-one), a delta lactam of ornithine. Consistent with the increase in expression of the genes involved in lipid synthesis, we observed increased concentrations of 2-hexadecenoic-acid, tetradecanoic acid, n-heptadecan-1-ol, and n-docosan-1-ol. The intracellular concentration of several amino acids — phenylalanine, lysine, threonine, and tyrosine — increased during growth on YPG. Interestingly, the concentration of asparagine decreased along with 2-amino-adipic acid and cystathionine, which are intermediates in lysine and cysteine biosynthesis, respectively. In addition, the concentration of tyramine, a byproduct of tyrosine, also decreased. Finally, we observed decreased concentration of the sugar alcohols: threitol, xylitol, and mannitol.
Growth on xylose
We next explored xylose metabolism in R. toruloides by comparing gene expression during growth in YP medium containing xylose (YPX) versus YP medium containing glucose (YPG). Overall, 882 genes were significantly upregulated and 537 genes were significantly downregulated during growth on YPX versus YPG. The gene expression profile on xylose was the closest to growth on glucose, in comparison to the other substrates we tested.
Consistent with a mechanism where growth on xylose induces expression of the xylose metabolic genes, expression of xylose reductase (XR) and xylitol dehydrogenase (XDH) was increased during growth on YPX versus YPG. However, expression of xylulokinase (XKS1) was identical during growth on YPX and YPG. Further exploration of the data revealed that XKS1 was not expressed during growth on either YPG or YPX. These results are consistent with recent observations (Kim et al. 2021). This would suggest an alternate mechanism for xylose utilization. One possibility is the conversion of xylulose to arabitol, catalyzed by arabitol dehydrogenase (ARD1). Indeed, significant arabitol production was previously observed during growth on xylose in rich medium (Jagtap and Rao 2018a,b). In support of this mechanism, expression of arabitol dehydrogenase (ARD1) was significantly higher (42-fold) during growth on YPX as compared to YPG. This enzyme converts xylulose to arabitol using NADH as the cofactor (Ingram and Wood 1965; Wong et al. 1993). This enzyme can also convert arabitol to ribulose using NAD as the cofactor (Wong et al. 1993). Ribulose is then phosphorylated by ribulose kinase (RK), forming ribulose-5-P. This metabolite is a substrate for the non-oxidative pentose phosphate pathway. Consistent with this mechanism, expression of the ribulose kinase gene (RK) was increased during growth on YPX as compared to YPG. Why R. toruloides employs this more circuitous route for xylose metabolism is not known.
Aside from enzymes directly involved in xylose utilization, we observed a few significant changes in the expression of genes involved in central carbon metabolism. Oxoglutarate dehydrogenase (KGD1b) was 27-fold upregulated. Pyruvate carboxylase (PYC1), succinate dehydrogenase (SDH4), malic enzyme (ME1), isocitrate lyase (ICL1, ICL2), malate synthase (MLS1), and glucose-6-phosphate dehydrogenase (ZWF1) were all upregulated at least twofold during growth on YPX as compared to YPG. Acetyl-CoA carboxylase (ACC1), ATP: citrate lyase (ACL1), and fatty-acyl-CoA synthase (FAS1 and FAS2) were downregulated 2- to threefold.
From the metabolomics analysis (Fig. 3; Supplemental Dataset 2), large increases in the concentrations of xylose (77-fold), xylitol (122-fold), and xylulose (threefold) were observed, which is consistent with the XR/XDH pathway for xylose utilization. In addition, we also observed an increase in the concentration of xylonic acid (threefold). This compound is formed from xylose by glucose oxidase (Toivari et al. 2012). However, we could not identify any gene associated with glucose oxidase activity. The concentrations of many intracellular acids decreased during growth on YPX, which is consistent with the downregulation of genes in the fatty acid biosynthesis pathway. In addition, we observed decreased concentrations of the storage carbohydrate trehalose, which is linked to glucose metabolism, as well as galactose and glycerol.
Growth on acetate
We next explored acetate metabolism in R. toruloides by comparing gene expression during growth on YP medium containing acetate (YPA) versus YPG medium. Overall, 1297 genes were significantly upregulated, and 1693 genes were significantly downregulated during growth on YPA versus YPG.
Acetate enters the central metabolism through the action of acetyl-CoA synthetase (ACS1) (Fig. 4). This enzyme converts acetate and ATP to acetyl-CoA, AMP, and pyrophosphate. Consistent with this mechanism, the expression of ACS1 was increased during growth on YPA as compared to YPG. In addition, we observed significantly higher expression of isocitrate lyase (ICL1, 154-fold) and malate synthase (MLS1, 24-fold). Both these enzymes are involved in the glyoxylate cycle and are predicted to reside in the cytosol. While acetate can cross the plasma membrane, many organisms also utilize transporters for acetate uptake (Casal et al. 2008). The expression of two predicted permeases (protein ID: 11,570 and 10,804) was increased almost 150 and 145-fold, respectively, during growth on YPA versus YPG. We also observed increased expression of acetyl-CoA hydrolase (ACH1). Increased expression of this enzyme is consistent with a detoxification mechanism.
We also observed increased expression of the genes involved in glycolysis, the pentose phosphate pathway, the TCA cycle, and the electron transport chain during growth on YPA versus YPG (Fig. 5). Fructose-1,6-biphosphatase (FBP1) and phosphoenolpyruvate carboxylase (PCK1) were both upregulated during growth on growth, suggesting increased flux through gluconeogenesis (Supplementary Fig. S3). In addition, we observed reduced expression of the genes involved in lipid production. Fatty acid biosynthesis genes including acetyl-CoA carboxylase (ACC1), and fatty-acyl-CoA synthase (FAS1 and FAS2), were downregulated. Interestingly, the expression of ATP-citrate lyase (ACL1) also decreased. Likely, reduced expression of ACL1 reflects the high concentration of cytoplasmic acetyl-CoA arising from acetate as the main carbon source. However, pathways involved in carbon storage and triacylglycerol (TAG) synthesis, including the glucan branching enzyme (GLG3), glycogen synthase (GSY1), myo-inositol-1-phosphate synthase (INO1), diacylglycerol O-acyltransferase (DGA1), acylglycerone-phosphate reductase (AYR1), glycerol 3-phosphate dehydrogenase (GPD1, GPD2, and GUT1), were upregulated. Taken together, these results indicate that the genes involved in acetate utilization, including the glyoxylate cycle, are induced by acetate. Interestingly, we also found that the genes involved in central metabolism, namely glycolysis, were also upregulated.
During growth on YPA, we did not observe many significant changes in the concentration of intracellular metabolites as compared to growth on YPG. We mostly observed increased concentrations for several amino acids: alanine, tyramine, and threonine (Fig. 3; Supplemental Dataset 2). Only the concentration of lysine decreased. In agreement with the increased expression of carbon storage pathways, we observed an increase in the concentration of glycerol, mannitol, and myo-inositol, and a decrease in the concentration of glycerol-3-phosphate. Lastly, the concentrations of intracellular long-chain fatty acids decreased, in alignment with the reduced expression of fatty acid biosynthesis.
Growth on oil
We lastly explored fatty-acid metabolism in R. toruloides by comparing gene expression during growth on YP medium containing soybean oil (YPS) versus YPG medium (Fig. 6). Overall, 1296 genes were significantly upregulated, and 1392 genes were significantly downregulated during growth on YPS versus YPG.
Transcriptional changes in pathways associated with fatty acid utilization in R. toruloides IFO0880 grown on glucose (YPG), xylose (YPX), acetate (YPA), soybean oil (YPS), and yeast peptone (YP). RNAseq data was collected in triplicate for each condition. Color key represents the z-score for each gene (normalized for all growth conditions)
Lipases catalyze the hydrolysis of fats and oils. R. toruloides has 10 predicated lipases. During growth on YPS, expression of two secreted lipases (sLIP1 and sLIP2) and one intracellular lipase (TGL1), predicted to reside in the peroxisome, increased. Interestingly, the expression of the secreted sLIP3 lipase decreased, which hydrolyses triacylglycerol into free fatty acids and glycerol (Fickers et al. 2011).
Not surprisingly, we observed increased expression of the genes involved in fatty-acid catabolism and decreased expression of the genes involved in fatty-acid synthesis, when comparing growth on YPS versus YPG (Fig. 6). Fatty-acid ligases (FAA) are involved in the activation of imported long-chain fatty acids (C12–C18) by forming the associated acyl-CoAs (Færgeman et al. 2001; Johnson et al. 1994). Expression of FAA1-5 was increased during growth on YPS as compared to YPG. FAA2 is involved in the activation of medium-chain fatty acids directed towards peroxisomal β-oxidation, whereas FAA3 is more active towards fatty acids with chain lengths longer than 18 (Færgeman et al. 2001). We also observed increased expression of the FAT1 fatty-acid transporter.
Fatty-acid catabolism is a multistep process and requires four different types of enzymes (Hiltunen et al. 2003). In the peroxisome, the first reaction is catalyzed by acyl-coenzyme A oxidase (ACOX) and acyl-CoA dehydrogenase (ACAD), which converts acyl-CoA to enoyl-CoA. R. toruloides has three ACOX’s (ACOX1-3) and six different ACAD’s (ACAD1-6) (Coradetti et al. 2018). Multiple ACOX’s isozymes in a single organism often have different specificities, including ones active against short-chain fatty acids, long-chain fatty acids, or both (Wang et al. 1999a, 1999b). Expression of ACOX1 and ACOX3 was increased during growth on YPS. Among the six ACAD’s, expression of ACAD3 and ACAD4 was significantly increased (> 15 fold). The expression of the other three ACAD’s, ACAD2, ACAD5 and ACAD6, were more moderately increased (~ ninefold). The second and third steps are catalyzed by enoyl-CoA hydratase (ECH) and 3-hydroxyacyl-CoA dehydrogenase (HADH), which converts trans-enoyl-CoA to β-keto-acyl-CoA. The expression of both ECH genes (ECH1 and ECH2) and HADH1 was increased. The expression of FOX2, the peroxisomal multifunctional enzyme, which catalyzes both steps 2 and 3 (Hiltunen et al. 1992), was also increased. In the fourth step, 3-ketoacyl-CoA thiolase (POT1) catalyzes the thiolytic cleavage of the 3-ketoacyl-CoA intermediate, yielding one acetyl-CoA and an acyl-CoA molecule shortened by two carbon atoms (Igual et al. 1991). We observed an almost tenfold increase in the expression of POT1. We also observed increased expression of the genes involved in the glyoxylate cycle: malate synthase (MLS1), isocitrate dehydrogenase (IDP1), and isocitrate lyase (ICL1).
The peroxisome membrane is impermeable to the acyl-CoA’s generated during the oxidation of long-chain fatty acids. The carnitine shuttle uses carnitine to channel shorter acyl groups from coenzyme A to form acetyl carnitine, which can be shuttled across membranes of the peroxisomes and mitochondria (Elgersma et al. 1995; Strijbis and Distel 2010; Swiegers et al. 2001). Carnitine acetyltransferase (CAT2), localized in the peroxisome, transfers an acyl group from acyl-CoA to carnitine. We observed increased expression of CAT2 during growth on YPS versus YPG. Acyl-carnitine is then transported into mitochondria by acylcarnitine translocase (CRC) (Palmieri et al. 1999). We did not observe significant upregulation of CRC. The reverse transfer of the acyl group from acyl-carnitine to coenzyme A is catalyzed by mitochondrial CAT2, whose expression was also increased. This process regenerates acyl-CoA and carnitine inside the mitochondria.
Short-chain acyl-CoA that are carried through the carnitine shuttle, or activated in the cytosol by FAA2, enter the mitochondria for further oxidation. The mitochondrial oxidation also comprises of four enzymatic steps, similar to peroxisomal β-oxidation (Elgersma et al. 1995; Strijbis and Distel 2010; Swiegers et al. 2001). R. toruloides has mammalian orthologs of mitochondrial short-chain and branched chain acyl-CoA dehydrogenases (ACADSB) and medium-chain acyl-CoA dehydrogenases (ACADM). The first reaction is catalyzed by ACADSB and ACADM, which converts long- and short-chain acyl-CoA’s to trans-enoyl-CoA. We observed a significant (50-fold) increase in the expression of ACADSB. ACADM expression was also increased, albeit more moderately (twofold). We also observed increased expression of enoyl-CoA hydratase (ECHS1) and 3-hydroxyacyl-CoA dehydrogenase (HADH1 and HADH2), which convert trans-enoyl-CoA to β-keto-acyl-CoA. Finally, the expression of acetyl-CoA acetyltransferase (ACAA1 and ACAA2), which catalyze the conversion of β-keto-acyl-CoA into one acetyl-CoA and an acyl-CoA molecule shortened by two carbon atoms, was also increased. Overall, these changes are consistent with the utilization of oil.
Finally, we observed increased expression of many genes involved in the TCA and glyoxylate cycle. Increased expression of the glycolytic enzymes, phosphofructokinase (PFK1/2), pyruvate carboxylase (PYC1), phosphoenolpyruvate carboxykinase (PCK1), and hexokinase (HXK), was also observed. However, expression of pyruvate kinase (PYK1), pyruvate dehydrogenase (PDB1) and glyceraldehyde-3-phosphate dehydrogenase (GADPH) was decreased. Since the respiratory chain and TCA cycle are metabolically connected with mitochondrial β-oxidation, these changes in expression are likely due to increased β-oxidation.
When comparing intracellular metabolites during growth on YPS versus YPG, we observed increased concentrations of mannitol and xylitol (Fig. 3; Supplemental Dataset 2). The origin for these differences is not immediately evident. Interestingly, we observed a reduction in the concentration of many fatty-acid derived molecules, which likely reflects the fact that the cells are degrading fatty acids rather than making them. In addition, both the concentrations of ornithine and ornithine-1,5-lactam decreased. Likely, the cells are using fatty acids for energy and the amino acids in YPS medium for biomass, unlike the case during growth on YPG.
Discussion
R. toruloides can grow on multiple carbon sources, including glucose, xylose, acetate, galactose, arabinose, mannose, fructose, sucrose, and oil. However, little is known about the governing metabolic pathways and their regulation. To better understand these pathways, we measured gene expression and intracellular metabolites during growth of R. toruloides IFO0880 on glucose, xylose, acetate, and oil. These substrates were chosen because they are being explored as potential substrates for conversion by R. toruloides into more valuable fuels and chemicals. All the samples were collected at exponential phase of YPG, YPX, YPA, and YPS medium in a baffled shake flask. The sampling point for the YP control was late exponential. R. toruloides can grow well on YP medium without additional carbon source. We selected the 24-h sampling time point for the YP control to ensure a sufficiently high OD and also used a similar time as the glucose, xylose, and acetate experiments. The sample point for oil was chosen earlier because it already reached an OD of 10 after 16 h of growth. Significant differences were observed in gene expression and metabolite concentrations during growth on the different substrates (Figs. 2 and 3). To better understand these differences, we mapped them onto the central carbon metabolic pathways of R. toruloides (Fig. 4). This analysis required us to identify and annotate many of the genes governing these pathways (Supplementary Dataset S1).
Most of the changes in gene expression and metabolite concentrations were consistent with the metabolism of these substrates. During growth on glucose, expression of the genes involved in lower glycolysis and the TCA cycle was increased. Expression of GDH1 was also increased by more than 100-fold. GDH1 catalyzes the reversible conversion of ammonia and α-ketoglutarate to form glutamate using NADPH as the cofactor. Likely, the large increase in GDH1 expression reflects the need of R. toruloides to scavenge nitrogen from amino acids in the growth medium. With respect to intracellular metabolites, the concentrations of glycolytic metabolites, galactose, and ornithine were higher during growth on YPG versus YP. Glucose can be converted to galactose, which is likely used to synthesize polysaccharides, glycolipids, and glycoproteins. In addition, the concentration of ornithine, which is involved in the urea cycle, increased. This likely reflects increased utilization of amino acids for production of energy during growth on glucose.
Like many yeasts, R. toruloides employs the xylose reductase (XR)/xylitol dehydrogenase (XDH) pathway for xylose utilization (Fig. 4). The first enzyme, xylose reductase, converts xylose to xylitol using NADPH as the cofactor. The second enzyme, xylitol dehydrogenase, converts xylitol to xylulose using NAD as the cofactor. In most yeasts, xylulokinase (XKS) converts xylulose to xylulose-5-phosphate, which then enters the non-oxidative branch of the pentose phosphate pathway (Jagtap and Rao 2018a). Growth on xylose principally increases the expression of genes involved in the non-canonical xylose utilization pathway in R. toruloides and has only a minor effect on other genes involved in central metabolism, though it does affect the expression of many other genes. Growth on xylose resulted in increased expression of the genes involved in xylose metabolism, with the notable exception of xylulokinase. This suggests that there may be xylose-specific transcriptional regulators. However, the identity of these regulators and associate processes are unknown.
One interesting finding concerns xylose metabolism. Previously, R. toruloides was found to produce relatively large amounts of arabitol during growth on xylose (Jagtap and Rao 2018b). This behavior is puzzling, because it does not have any obvious benefit to the cell. Our analysis suggests that this behavior is due to poor expression of xylulokinase, resulting in a bypass through arabitol dehydrogenase. Even stranger is that xylose reduces xylulokinase expression. One possibility is that this is a strain-specific mutation and unique to R. toruloides IFO0880. However, many yeasts produce arabitol during growth on xylose, suggesting that it is an adaptive strategy of unknown benefit (Pinheiro et al. 2020; Quarterman et al. 2017). Regardless, these results provide a potential strategy for improving the growth on R. toruloides on xylose, which natively is very poor, by constitutively expressing xylulokinase.
In the case of acetate, expression of the genes involved in TCA cycle, gluconeogenesis, and pentose phosphate pathway were increased. Acetate enters the central metabolism through the action of acetyl-CoA synthetase (Fig. 4). This enzyme converts acetate and ATP to acetyl-CoA, AMP, and pyrophosphate. Increased expression of two permeases (protein ID: 11,570 and 10,804) suggests that they may facilitate acetate uptake. However, more detailed studies are required before proving this claim. Collectively, these results are consistent with acetate inducing the expression of acetate utilization genes. Acetyl-CoA hydrolase converts acetyl-CoA into acetate. However, increased expression of this enzyme would result in a futile cycle that consumes ATP by hydrolyzing the acetyl-CoA formed by acetyl-CoA synthetase. Researchers have previously shown that this enzyme in many fungi is a CoA-transferase involved in the detoxification of intracellular acetate by transforming acetate and succinyl-CoA into acetyl-CoA and succinate (Buu et al. 2003; Fleck and Brock 2009).
Lastly, during growth on soybean oil, we observed increased expression of many secreted lipases and putative fatty-acid transporters. Fatty acid transporter (FAT1) has been reported to play a pivotal role in the import of long-chain fatty acids in Saccharomyces cerevisiae and Yarrowia lipolytica (Choi and Martin 1999; Watkins et al. 1998). In another study, it was reported that FAT1 is another member of the yeast long-chain acyl-CoA synthetase family (Watkins 2008; Watkins et al. 2007). In vitro analysis has shown that FAT1 has CoA ligase activity towards very long-chain fatty acids (C22-C26) (Coe et al. 1999). These results suggest that, in addition to transporter activity, FAT1 can also activate long-chain fatty acids (LCFAs) by forming the associated acyl-CoA’s.
Following the import aided by FAA’s and FAT1, fatty acids are degraded by β-oxidation. We observed increased expression of the genes involved in β-oxidation in both the mitochondria and peroxisomes. In R. toruloides, both mitochondrial and peroxisomal fatty acid β-oxidation pathways have been reported (Coradetti et al. 2018; Zhu et al. 2012). Long-chain fatty acids are shortened in the peroxisome and then transferred via the carnitine shuttle to the mitochondria, where short-chain fatty acids are oxidized to acetyl-CoA. Both peroxisomal and mitochondrial beta-oxidation are necessary for robust growth on fatty acids (Chegary et al. 2009; Swigonová et al. 2009).
In conclusion, these results further our understanding of central carbon metabolism in R. toruloides. These results and the associated data sets may help metabolic engineers further exploit the ability of R. toruloides to make diverse fuels and chemicals. In addition, they may also aid in the development of genome-scale models (Castañeda et al. 2018; Dinh et al. 2019; Kim et al. 2021; Tiukova et al. 2019). Future work is focused on understanding the mechanisms governing lipid production during nutrient starvation and identifying the regulatory mechanisms governing metabolism.
Data availability
RNA sequencing reads are uploaded to a NCBI bioproject (Accession number: PRJNA660884). Additional datasets are uploaded at the research group github page: https://github.com/raogroupuiuc/rt88_growth. Other datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom
Blighe K LA (2021) PCAtools: everything principal components analysis. R package version 240
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Buu LM, Chen YC, Lee FJ (2003) Functional characterization and localization of acetyl-CoA hydrolase, Ach1p, in Saccharomyces cerevisiae. J Biol Chem 278(19):17203–17209. https://doi.org/10.1074/jbc.M213268200
Casal M, Paiva S, Queiros O, Soares-Silva I (2008) Transport of carboxylic acids in yeasts. FEMS Microbiol Rev 32(6):974–994. https://doi.org/10.1111/j.1574-6976.2008.00128.x
Castañeda MT, Nuñez S, Garelli F, Voget C, De Battista H (2018) Comprehensive analysis of a metabolic model for lipid production in Rhodosporidium toruloides. J Biotechnol 280:11–18. https://doi.org/10.1016/j.jbiotec.2018.05.010
Chegary M, Ht B, Ruiter JPN, Wijburg FA, Stoll MSK, Minkler PE, van Weeghel M, Schulz H, Hoppel CL, Wanders RJA, Houten SM (2009) Mitochondrial long chain fatty acid β-oxidation in man and mouse. Biochim Biophys Acta 1791(8):806–815. https://doi.org/10.1016/j.bbalip.2009.05.006
Choi J-Y, Martin CE (1999) The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J Biol Chem 274(8):4671–4683. https://doi.org/10.1074/jbc.274.8.4671
Chong J, Wishart DS, Xia J (2019) Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinformatics 68(1):e86. https://doi.org/10.1002/cpbi.86
Coe NR, Smith AJ, Frohnert BI, Watkins PA, Bernlohr DA (1999) The fatty acid transport protein (FATP1) is a very long chain acyl-CoA synthetase. J Biol Chem 274(51):36300–36304. https://doi.org/10.1074/jbc.274.51.36300
Coradetti ST, Pinel D, Geiselman GM, Ito M, Mondo SJ, Reilly MC, Cheng Y-F, Bauer S, Grigoriev IV, Gladden JM, Simmons BA, Brem RB, Arkin AP, Skerker JM (2018) Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. eLife 7:e32110. https://doi.org/10.7554/eLife.32110
Cui JD, Qiu JQ, Fan XW, Jia SR, Tan ZL (2014) Biotechnological production and applications of microbial phenylalanine ammonia lyase: a recent review. Crit Rev Biotechnol 34(3):258–268
Dinh HV, Suthers PF, Chan SHJ, Shen Y, Xiao T, Deewan A, Jagtap SS, Zhao H, Rao CV, Rabinowitz JD, Maranas CD (2019) A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data. Metab Eng Commun 9:e00101. https://doi.org/10.1016/j.mec.2019.e00101
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2012) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. https://doi.org/10.1093/bioinformatics/bts635
Elgersma Y, van Roermund CW, Wanders RJ, Tabak HF (1995) Peroxisomal and mitochondrial carnitine acetyltransferases of Saccharomyces cerevisiae are encoded by a single gene. EMBO J 14(14):3472–3479
Færgeman NJ, Black PN, Zhao XD, Knudsen J, DiRusso CC (2001) The acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J Biol Chem 276(40):37051–37059. https://doi.org/10.1074/jbc.M100884200
Fei Q, O’Brien M, Nelson R, Chen X, Lowell A, Dowe N (2016) Enhanced lipid production by Rhodosporidium toruloides using different fed-batch feeding strategies with lignocellulosic hydrolysate as the sole carbon source. Biotechnol Biofuel 9(1):130. https://doi.org/10.1186/s13068-016-0542-x
Fernández-Lafuente R, Rodriguez V, Guisán JM (1998) The coimmobilization of D-amino acid oxidase and catalase enables the quantitative transformation of D-amino acids (D-phenylalanine) into α-keto acids (phenylpyruvic acid). Enzyme Microb Technol 23(1):28–33. https://doi.org/10.1016/S0141-0229(98)00028-3
Fickers P, Marty A, Nicaud JM (2011) The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol Adv 29(6):632–644. https://doi.org/10.1016/j.biotechadv.2011.04.005
Fillet S, Gibert J, Suárez B, Lara A, Ronchel C, Adrio JL (2015) Fatty alcohols production by oleaginous yeast. J Ind Microbiol Biotechnol 42(11):1463–1472. https://doi.org/10.1007/s10295-015-1674-x
Fillet S, Ronchel C, Callejo C, Fajardo M-J, Moralejo H, Adrio JL (2017) Engineering Rhodosporidium toruloides for the production of very long-chain monounsaturated fatty acid-rich oils. Appl Microbiol Biotechnol 101(19):7271–7280. https://doi.org/10.1007/s00253-017-8461-8
Fleck CB, Brock M (2009) Re-characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA-transferases are involved in acetic acid detoxification. Fungal Genet Biol 46(6–7):473–485. https://doi.org/10.1016/j.fgb.2009.03.004
Geiselman GM, Zhuang X, Kirby J, Tran-Gyamfi MB, Prahl J-P, Sundstrom ER, Gao Y, Munoz Munoz N, Nicora CD, Clay DM, Papa G, Burnum-Johnson KE, Magnuson JK, Tanjore D, Skerker JM, Gladden JM (2020) Production of ent-kaurene from lignocellulosic hydrolysate in Rhodosporidium toruloides. Microb Cell Fact 19(1):24. https://doi.org/10.1186/s12934-020-1293-8
Gilbert HJ, Clarke IN, Gibson RK, Stephenson JR, Tully M (1985) Molecular cloning of the phenylalanine ammonia lyase gene from Rhodosporidium toruloides in Escherichia coli K-12. J Bacteriol 161(1):314–320
Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42(Database issue):D699-704. https://doi.org/10.1093/nar/gkt1183
Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A (2003) The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 27(1):35–64. https://doi.org/10.1016/S0168-6445(03)00017-2
Hiltunen JK, Wenzel B, Beyer A, Erdmann R, Fossa A, Kunau WH (1992) Peroxisomal multifunctional beta-oxidation protein of Saccharomyces cerevisiae. Molecular analysis of the fox2 gene and gene product. J Biol Chem 267(10):6646–53
Hoskins J, Jack G, Peiris RD, Starr DT, Wade H, Wright E, Stern J (1980) Enzymatic control of phenylalanine intake in phenylketonuria. Lancet 315(8165):392–394
Howe E, Holton K, Nair S, Schlauch D, Sinha R, Quackenbush J (2010) MeV: MultiExperiment Viewer. In: Ochs MF, Casagrande JT, Davuluri RV (eds) Biomedical informatics for cancer research. Springer, US, pp 267–277
Hu C, Zhao X, Zhao J, Wu S, Zhao ZK (2009) Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour Technol 100(20):4843–4847. https://doi.org/10.1016/j.biortech.2009.04.041
Huang X-F, Liu J-N, Lu L-J, Peng K-M, Yang G-X, Liu J (2016) Culture strategies for lipid production using acetic acid as sole carbon source by Rhodosporidium toruloides. Bioresour Technol 206:141–149. https://doi.org/10.1016/j.biortech.2016.01.073
Igual JC, Matallana E, Gonzalez-Bosch C, Franco L, Perez-Ortin JE (1991) A new glucose-repressible gene identified from the analysis of chromatin structure in deletion mutants of yeast SUC2 locus. Yeast 7(4):379–389. https://doi.org/10.1002/yea.320070408
Ingram JM, Wood WA (1965) Enzymatic basis for D-arbitol production by Zygosaccharomyces rouxii. J Bacteriol 89(5):1186–1194. https://doi.org/10.1128/jb.89.5.1186-1194.1965
Jagtap SS, Bedekar AA, Liu J-J, Jin Y-S, Rao CV (2019) Production of galactitol from galactose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Biotechnol Biofuel 12(1):250. https://doi.org/10.1186/s13068-019-1586-5
Jagtap SS, Rao CV (2018a) Microbial conversion of xylose into useful bioproducts. Appl Microbiol Biotechnol 102(21):9015–9036. https://doi.org/10.1007/s00253-018-9294-9
Jagtap SS, Rao CV (2018b) Production of D-arabitol from D-xylose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Appl Microbiol Biotechnol 102(1):143–151. https://doi.org/10.1007/s00253-017-8581-1
Johnson DR, Knoll LJ, Levin DE, Gordon JI (1994) Saccharomyces cerevisiae contains four fatty acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J Cell Biol 127(3):751–762. https://doi.org/10.1083/jcb.127.3.751
Kim J, Coradetti ST, Kim Y-M, Gao Y, Yaegashi J, Zucker JD, Munoz N, Zink EM, Burnum-Johnson KE, Baker SE, Simmons BA, Skerker JM, Gladden JM, Magnuson JK (2021) Multi-omics driven metabolic network reconstruction and analysis of lignocellulosic carbon utilization in Rhodosporidium toruloides. Front Bioeng Biotechnol 8(1484) https://doi.org/10.3389/fbioe.2020.612832
Kim S, Lee DY, Wohlgemuth G, Park HS, Fiehn O, Kim KH (2013) Evaluation and optimization of metabolome sample preparation methods for Saccharomyces cerevisiae. Anal Chem 85(4):2169–2176. https://doi.org/10.1021/ac302881e
Kitahara Y, Yin T, Zhao X, Wachi M, Du W, Liu D (2014) Isolation of oleaginous yeast (Rhodosporidium toruloides) mutants tolerant of sugarcane bagasse hydrolysate. Biosci Biotechnol Biochem 78(2):336–342. https://doi.org/10.1080/09168451.2014.882746
Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, Willmitzer L, Fernie AR, Steinhauser D (2005) GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21(8):1635–1638. https://doi.org/10.1093/bioinformatics/bti236
Lee JJL, Chen L, Cao B, Chen WN (2016) Engineering Rhodosporidium toruloides with a membrane transporter facilitates production and separation of carotenoids and lipids in a bi-phasic culture. Appl Microbiol Biotechnol 100(2):869–877. https://doi.org/10.1007/s00253-015-7102-3
Lee JJL, Chen L, Shi J, Trzcinski A, Chen W-N (2014) Metabolomic profiling of Rhodosporidium toruloides grown on glycerol for carotenoid production during different growth phases. J Agric Food Chem 62(41):10203–10209. https://doi.org/10.1021/jf502987q
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930. https://doi.org/10.1093/bioinformatics/btt656
Liu D, Geiselman GM, Coradetti S, Cheng Y-F, Kirby J, Prahl J-P, Jacobson O, Sundstrom ER, Tanjore D, Skerker JM, Gladden J (2020) Exploiting nonionic surfactants to enhance fatty alcohol production in Rhodosporidium toruloides. Biotechnol Bioeng 117(5):1418–1425. https://doi.org/10.1002/bit.27285
Lutfiyya LL, Johnston M (1996) Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol 16(9):4790–4797. https://doi.org/10.1128/mcb.16.9.4790
Mormeneo S, Sentandreu R (1982) Regulation of invertase synthesis by glucose in Saccharomyces cerevisiae. J Bacteriol 152(1):14–18
Nora LC, Wehrs M, Kim J, Cheng J-F, Tarver A, Simmons BA, Magnuson J, Harmon-Smith M, Silva-Rocha R, Gladden JM, Mukhopadhyay A, Skerker JM, Kirby J (2019) A toolset of constitutive promoters for metabolic engineering of Rhodosporidium toruloides. Microb Cell Fact 18(1):117. https://doi.org/10.1186/s12934-019-1167-0
Palmieri L, Lasorsa FM, Iacobazzi V, Runswick MJ, Palmieri F, Walker JE (1999) Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett 462(3):472–476. https://doi.org/10.1016/s0014-5793(99)01555-0
Pilone MS, Pollegioni L (2002) D-Amino acid oxidase as an industrial biocatalyst. Biocatal Biotransfor 20(3):145–159
Pinheiro MJ, Bonturi N, Belouah I, Miranda EA, Lahtvee P-J (2020) Xylose metabolism and the effect of oxidative stress on lipid and carotenoid production in Rhodotorula toruloides: insights for future biorefinery. Frontiers in Bioengineering and Biotechnology 8(1008) https://doi.org/10.3389/fbioe.2020.01008
Quarterman J, Slininger PJ, Kurtzman CP, Thompson SR, Dien BS (2017) A survey of yeast from the Yarrowia clade for lipid production in dilute acid pretreated lignocellulosic biomass hydrolysate. Appl Microbiol Biotechnol 101(8):3319–3334. https://doi.org/10.1007/s00253-016-8062-y
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (oxford, England) 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Singh G, Jawed A, Paul D, Bandyopadhyay KK, Kumari A, Haque S (2016) Concomitant production of lipids and carotenoids in Rhodosporidium toruloides under osmotic stress using response surface methodology. Front Microbiol 7(1686) https://doi.org/10.3389/fmicb.2016.01686
Stein SE (1999) An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J Am Soc Mass Spectrom 10(8):770–781. https://doi.org/10.1016/s1044-0305(99)00047-1
Strijbis K, Distel B (2010) Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot Cell 9(12):1809–1815. https://doi.org/10.1128/ec.00172-10
Styczynski MP, Moxley JF, Tong LV, Walther JL, Jensen KL, Stephanopoulos GN (2007) Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal Chem 79(3):966–973. https://doi.org/10.1021/ac0614846
Su S, Law CW, Ah-Cann C, Asselin-Labat ML, Blewitt ME, Ritchie ME (2017) Glimma: interactive graphics for gene expression analysis. Bioinformatics 33(13):2050–2052. https://doi.org/10.1093/bioinformatics/btx094
Swiegers JH, Dippenaar N, Pretorius IS, Bauer FF (2001) Carnitine-dependent metabolic activities in Saccharomyces cerevisiae: three carnitine acetyltransferases are essential in a carnitine-dependent strain. Yeast 18(7):585–595. https://doi.org/10.1002/yea.712
Swigonová Z, Mohsen AW, Vockley J (2009) Acyl-CoA dehydrogenases: dynamic history of protein family evolution. J Mol Evol 69(2):176–193. https://doi.org/10.1007/s00239-009-9263-0
Tiukova IA, Prigent S, Nielsen J, Sandgren M, Kerkhoven EJ (2019) Genome-scale model of Rhodotorula toruloides metabolism. Biotechnol Bioeng 116(12):3396–3408. https://doi.org/10.1002/bit.27162
Toivari MH, Nygård Y, Penttilä M, Ruohonen L, Wiebe MG (2012) Microbial D-xylonate production. Appl Microbiol Biotechnol 96(1):1–8. https://doi.org/10.1007/s00253-012-4288-5
Wang H, Le Dall MT, Wache Y, Laroche C, Belin JM, Nicaud JM (1999a) Cloning, sequencing, and characterization of five genes coding for acyl-CoA oxidase isozymes in the yeast Yarrowia lipolytica. Cell Biochem Biophys 31(2):165–174. https://doi.org/10.1007/BF02738170
Wang HJ, Le Dall MT, Wach Y, Laroche C, Belin JM, Gaillardin C, Nicaud JM (1999b) Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol 181(17):5140–5148. https://doi.org/10.1128/JB.181.17.5140-5148.1999
Wang Y, Zhang S, Zhu Z, Shen H, Lin X, Jin X, Jiao X, Zhao ZK (2018) Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol Biofuels 11:148. https://doi.org/10.1186/s13068-018-1134-8
Warnes GR (2011) Gplots: various R programming tools for plotting data.
Watkins PA (2008) Very-long-chain acyl-CoA synthetases. J Biol Chem 283(4):1773–1777. https://doi.org/10.1074/jbc.R700037200
Watkins PA, Lu J-F, Steinberg SJ, Gould SJ, Smith KD, Braiterman LT (1998) Disruption of the Saccharomyces cerevisiae FAT1 gene decreases very long-chain fatty acyl-CoA synthetase activity and elevates intracellular very long-chain fatty acid concentrations. J Biol Chem 273(29):18210–18219. https://doi.org/10.1074/jbc.273.29.18210
Watkins PA, Maiguel D, Jia Z, Pevsner J (2007) Evidence for 26 distinct acyl-coenzyme A synthetase genes in the human genome. J Lipid Res 48(12):2736–2750. https://doi.org/10.1194/jlr.M700378-JLR200
Wehrs M, Gladden JM, Liu Y, Platz L, Prahl J-P, Moon J, Papa G, Sundstrom E, Geiselman GM, Tanjore D, Keasling JD, Pray TR, Simmons BA, Mukhopadhyay A (2019) Sustainable bioproduction of the blue pigment indigoidine: expanding the range of heterologous products in R. toruloides to include non-ribosomal peptides. Green Chem 21(12):3394–3406. https://doi.org/10.1039/C9GC00920E
Wen Z, Zhang S, Odoh CK, Jin M, Zhao ZK (2020) Rhodosporidium toruloides — a potential red yeast chassis for lipids and beyond. FEMS Yeast Res 20(5) https://doi.org/10.1093/femsyr/foaa038
Wiebe MG, Koivuranta K, Penttilä M, Ruohonen L (2012) Lipid production in batch and fed-batch cultures of Rhodosporidium toruloides from 5 and 6 carbon carbohydrates. BMC Biotechnol 12(1):26. https://doi.org/10.1186/1472-6750-12-26
Wong B, Murray JS, Castellanos M, Croen KD (1993) D-arabitol metabolism in Candida albicans: studies of the biosynthetic pathway and the gene that encodes NAD-dependent D-arabitol dehydrogenase. J Bacteriol 175(19):6314–6320. https://doi.org/10.1128/jb.175.19.6314-6320.1993
Wu S, Zhao X, Shen H, Wang Q, Zhao ZK (2011) Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour Technol 102(2):1803–1807. https://doi.org/10.1016/j.biortech.2010.09.033
Yaegashi J, Kirby J, Ito M, Sun J, Dutta T, Mirsiaghi M, Sundstrom ER, Rodriguez A, Baidoo E, Tanjore D, Pray T, Sale K, Singh S, Keasling JD, Simmons BA, Singer SW, Magnuson JK, Arkin AP, Skerker JM, Gladden JM (2017) Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels 10(1):241. https://doi.org/10.1186/s13068-017-0927-5
Yun EJ, Oh EJ, Liu J-J, Yu S, Kim DH, Kwak S, Kim KH, Jin Y-S (2018) Promiscuous activities of heterologous enzymes lead to unintended metabolic rerouting in Saccharomyces cerevisiae engineered to assimilate various sugars from renewable biomass. Biotechnol Biofuels 11(1):140. https://doi.org/10.1186/s13068-018-1135-7
Zhang S, Ito M, Skerker JM, Arkin AP, Rao CV (2016a) Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl Microbiol Biotechnol 100(21):9393–9405. https://doi.org/10.1007/s00253-016-7815-y
Zhang S, Jagtap SS, Deewan A, Rao CV (2019) pH selectively regulates citric acid and lipid production in Yarrowia lipolytica W29 during nitrogen-limited growth on glucose. J Biotechnol 290:10–15. https://doi.org/10.1016/j.jbiotec.2018.10.012
Zhang S, Skerker JM, Rutter CD, Maurer MJ, Arkin AP, Rao CV (2016b) Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol Bioeng 113(5):1056–1066. https://doi.org/10.1002/bit.25864
Zhu Z, Ding Y, Gong Z, Yang L, Zhang S, Zhang C, Lin X, Shen H, Zou H, Xie Z, Yang F, Zhao X, Liu P, Zhao ZK (2015) Dynamics of the lipid droplet proteome of the oleaginous yeast Rhodosporidium toruloides. Eukaryot Cell 14(3):252–264. https://doi.org/10.1128/EC.00141-14
Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, Zhou YJ, Jin G, Ye M, Zou H, Zhao ZK (2012) A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun 3:1112. https://doi.org/10.1038/ncomms2112
Acknowledgements
We thank Hoang Dinh for helpful discussions. We thank Dr. Christopher J. Fields and Dr. Jenny Drnevich, affiliated with High-performance Computing in Biology (HPCBio) at University of Illinois at Urbana-Champaign, for their help with the initial RNAseq data analysis. This material is based upon the work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Numbers DE-SC0018420 and DE-SC0018260.
Author information
Authors and Affiliations
Contributions
SJ, YJ, and CR conceived the project. SJ, AD, JL, EY, and HW designed and performed experiments. AD, SJ, JL, EY, and CR analyzed the data. SJ, AD, and CR wrote the manuscript and designed the figures. All authors discussed the results and contributed to the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors have given their consent for the publication of this work.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sujit Sadashiv Jagtap and Anshu Deewan are Equal Contribution
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jagtap, S.S., Deewan, A., Liu, JJ. et al. Integrating transcriptomic and metabolomic analysis of the oleaginous yeast Rhodosporidium toruloides IFO0880 during growth under different carbon sources. Appl Microbiol Biotechnol 105, 7411–7425 (2021). https://doi.org/10.1007/s00253-021-11549-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11549-8