Abstract
The acyl-CoA dehydrogenases (ACADs) are enzymes that catalyze the α,β-dehydrogenation of acyl-CoA esters in fatty acid and amino acid catabolism. Eleven ACADs are now recognized in the sequenced human genome, and several homologs have been reported from bacteria, fungi, plants, and nematodes. We performed a systematic comparative genomic study, integrating homology searches with methods of phylogenetic reconstruction, to investigate the evolutionary history of this family. Sequence analyses indicate origin of the family in the common ancestor of Archaea, Bacteria, and Eukaryota, illustrating its essential role in the metabolism of early life. At least three ACADs were already present at that time: ancestral glutaryl-CoA dehydrogenase (GCD), isovaleryl-CoA dehydrogenase (IVD), and ACAD10/11. Two gene duplications were unique to the eukaryotic domain: one resulted in the VLCAD and ACAD9 paralogs and another in the ACAD10 and ACAD11 paralogs. The overall patchy distribution of specific ACADs across the tree of life is the result of dynamic evolution that includes numerous rounds of gene duplication and secondary losses, interdomain lateral gene transfer events, alteration of cellular localization, and evolution of novel proteins by domain acquisition. Our finding that eukaryotic ACAD species are more closely related to bacterial ACADs is consistent with endosymbiotic origin of ACADs in eukaryotes and further supported by the localization of all nine previously studied ACADs in mitochondria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The acyl-CoA dehydrogenases are mitochondrial flavoenzymes involved in fatty acid and amino acid catabolism. The degradation of fatty acids, also known as β-oxidation, is a spiral reaction of four enzymatic steps that takes place in mitochondria and peroxisomes. The first and rate-limiting step is the desaturation of the acyl-CoA esters that is catalyzed by enzymes from two protein families, the acyl-CoA dehydrogenases (ACADs) and the acyl-CoA oxidases (ACOXs). In mitochondria, ACADs transfer electrons from their corresponding CoA ester substrates to the electron transferring protein (ETF), which in turn transfers the electrons to ETF dehydrogenase. The latter is coupled with the electron transport chain via Coenzyme Q to CoQH2-cytochrome c oxidoreductase (Complex III) for the production of ATP. In peroxisomes, the ACOXs transfer electrons directly to molecular oxygen producing H2O2. All of these enzymes utilize flavin adenine dinucleotide (FAD) as a cofactor and are structurally similar. While the importance of the mitochondrial β-oxidation in plants is controversial, the existence of both β-oxidation systems in animals is well recognized. Animals seem to have different purposes for the two β-oxidation pathways. While the primary function of mitochondrial β-oxidation is energy production, the peroxisomal β-oxidation serves primarily in the chain shortening of a wide variety of lipophilic compounds for a variety of metabolic processes (Kunau et al. 1995). Although ACADs have been identified in bacteria (Klein 1973) and the mitochondria of animals (Hall 1978) and fungi (Kionka and Kunau 1985; Valenciano et al. 1996), the distribution of these enzymes across the tree of life is unknown.
The human genome contains 11 members of the ACAD family with a range of substrate specificity and tissue expression profiles. Five of them, SCAD (short-chain acyl-CoA dehydrogenase), MCAD (medium-chain acyl-CoA dehydrogenase), LCAD (long-chain acyl-CoA dehydrogenase), VLCAD (very long-chain acyl-CoA dehydrogenase), and ACAD9 are involved in β-oxidation of fatty acids, catalyzing the α,β-dehydrogenation of substrate with the production of a trans-enoyl-CoA. Four other members are involved in catabolism of amino acids: IVD (isovaleryl-CoA dehydrogenase) for leucine, SBCAD (short/branched-chain acyl-CoA dehydrogenase, also known as 2-methyl branched-chain acyl-CoA dehydrogenase) for isoleucine, IBD (isobutyryl-CoA dehydrogenase) for valine, and GCD (glutaryl-CoA dehydrogenase) for lysine and tryptophan. Genome searches and annotations have led to identification of two additional putative ACADs, the ACAD10 (Ye et al. 2004) and ACAD11 (Strausberg et al. 2002) but their function remains unknown. All nine characterized ACADs are encoded in the nucleus, synthesized in the cytosol as precursors that are 2–4 kDa larger than their mature form, imported into mitochondria, and processed into mature forms by a proteolytic mechanism (Ikeda et al. 1987).
The crystal structures of pig MCAD (PDB IDs: 3MDD, 3MDE; Kim et al. 1993), human MCAD (1EGD, 1UDY, and 2A1T; Lee et al. 1996; Toogood et al. 2004), human IVD (1IVH; Tiffany et al. 1997), Megasphaera elsdenii SCAD (1BUC; Djordjevic et al. 1995), rat SCAD (1JQI; Battaile et al. 2002), Thermus thermophilus ACAD (1UKW; Hamada et al. 2004), human IBD (1RXO; Battaile et al. 2004), human GCD (1SIQ and 1SIR; Fu et al. 2004), and VLCAD (2UXW; unpublished) have been determined. Their three-dimensional structures have highly similar tertiary fold consisting of three major domains: an NH2-terminal domain of α-helices, a central domain of β-sheets, and a COOH-terminus domain of α-helices. The major differences between the different ACADs are in the structure of their binding cavity determining their substrate specificity. All of the characterized ACADs except the VLCAD and ACAD9 are tetrameric molecules of about 43 kDa exhibiting tetrahedral symmetry that can be viewed as a dimer of dimers. VLCAD and ACAD9 are homodimers with a subunit mass of 73 kDa. A molecule of FAD is noncovalently bound to each monomer with the flavin moiety positioned at the active site between the middle β-sheet and the carboxyl-terminal α-helix domain of one subunit, with the adenine moiety of the FAD extending into a pocket between the carboxyl-terminal α-helices domains of the neighboring subunits. Highly conserved residues constituting the adenine binding pocket are important confirmatory determinants for identification of putative ACADs.
The availability of whole genome sequences provides an unprecedented opportunity to study evolution of protein families, to investigate the correlation between protein structure and function, and to provide insight into the coevolution of enzymes and the enzymatic reactions they catalyze. Numerous studies have focused on structural and functional characteristics of several ACADs and others on mutational variability and their clinical consequences; however, a comprehensive examination of the taxonomic distribution of these enzymes and their evolutionary relationships has not been undertaken previously. The aim of this study was to expand our knowledge of the distribution of the ACADs across the tree of life, to investigate the molecular mechanisms that have influenced the evolution of this enzymatic family, and to examine the functional conservancy of ACAD homologs. In our comparative approach, we integrated homology searches of whole genomes and methods of phylogenetic reconstruction. While, in most of the cases, our results support the putative functional identity of published protein sequences, we also detected sequences that seem to deviate from the functional identity of its closely related homologs, indicating lineage-specific functional diversification of ACAD homologs with acquisition of novel function.
Materials and Methods
Outgroup Choice
We used two outgroups in our preliminary analyses to accurately assess the homology of identified ACAD sequences. One outgroup was composed of sequences of ACOX homologs. The second outgroup contained sequences of aidB genes encoding proteins involved in adaptive response to DNA damage by preventing N-methyl-N′-nitro-N-nitrosoguanidine-mediated mutagenesis (Landini et al. 1994). The ACADs, ACOXs, and aidB proteins exhibit a significant sequence and structural similarity, indicating their descent from a common ancestor (Krasko et al. 1998; Nandy et al. 1996). Several additional criteria considering residues important for acyl-CoA and FAD binding were used in homology assessment (below). These preliminary analyses recovered highly supported monophyletic clades corresponding to the aidB, ACOX, and ACAD superfamilies (bootstrap values 98–100, not shown). To allow an increase in taxon sampling of the ingroup and to minimize the computational time, we limited the number of outgroup sequences in the data by using a reduced set of ACOX sequences in our further analyses. To optimize the speed of analytical runs, regions composed of more than 25% of missing data were generally excluded from the alignment, except several specific cases when the presence/absence of a homolog in a genome was questioned.
Database Search
BLASTP searches (Altschul et al. 1997) were performed to screen the nonredundant protein databases of the National Center for Biotechnology Information (NCBI), the UniProt/SwissProt protein database, the Joint Genome Institute (http://www.jgi.doe.gov/), and the TIGR Microbial Database for homologs of all 11 human ACADs. In addition, we searched for homologs of ACOXs and aidB genes. Using results of phylogenetic analyses, we performed additional homology searches in which we used identified ACAD homologs of bacterial and archaeal taxa as query sequences to increase the taxon sampling (Supplementary Table 2). Sequences that exhibited more that 45% identity with identified ACADs (from phylogenetic analyses) are color coded in Supplementary Tables 2 and 3.
Sequence Alignment
Sequences were aligned using ClustalW (Thompson et al. 1994) as implemented in MacVector 7.2.2 package (Accelrys Inc.) and further manually edited with reference to crystal structure of rat peroxisomal acyl-CoA oxidase 2 (1IS2), human IVD (1IVH_A), and pig MCAD (3MDEA). See supplementary material for the sequence alignments. Partial sequences of ACAD homologs were included only to confirm the presence/absence status of a gene in an organism. Spliced isoforms were excluded since they are not informative at the level of family phylogeny.
Homology Criterion
All sequences considered as ACAD homologs based on sequence identity were required to meet additional criteria to be included in the phylogenetic analyses. These criteria considered conserved residues important for the acyl-CoA and FAD binding. The glutamate at position 375 (IVD numbering) is the catalytic residue in all described ACADs except IVD and LCAD where it is located at position 254 (IVD numbering). Other conserved residues considered included S142, R255, R280, F283, G351, and G352 (numbering as in the mature form of IVD), all of which are invariant in all described ACADs. R255 is important for isovaleryl-CoA binding in IVD, the F238 is involved in isovaleryl-CoA/FAD binding at the second ACAD subunit, and R280 and G351 are important for FAD binding at the second subunit. Sequences that violated several of these conserved residues were excluded from the analyses (see text).
Phylogenetic Analysis
Sequences with >90% amino acid sequence identities were excluded in the combined analysis (analysis including all ACAD species) to reduce computational time. At the initial step of each phylogenetic analysis, sequences that failed the χ 2 homogeneity test for deviation of amino acid frequencies as implemented in TREE-PUZZLE version 5.2 (Schmidt et al. 2002) were excluded from further analyses since the current methods of phylogenetic reconstructions cannot deal with a strong compositional bias in the data (Foster and Hickey 1999). Phylogenetic analyses included maximum parsimony (MP), maximum likelihood, and Bayesian likelihood analyses. All MP were performed using version 4.0b10 of PAUP* (Swofford 2000). The most parsimonious trees were estimated using heuristic searches with 100 or 1,000 random stepwise addition replicates with tree bisection-reconnection branch-swapping algorithm and ACCTRAN options selected. Branch support in the resulting trees was examined by nonparametric bootstrapping (Felsenstein 1985) with 10 or 100 pseudo-replicates with 10 random-addition heuristic searches in each pseudo-replicate.
Maximum likelihood of all ACAD sequences (combined analysis) was performed using PHYML (Guindon and Gascuel 2003) with Whelan and Goldman (WAG) substitution model (Whelan and Goldman 2001), shape parameter of gamma distribution estimated from the data, using NJ as the initial tree, and bootstrap analysis with 100 replicates. Bayesian likelihood analyses were performed using MrBayes 3.1.2 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). Prior to the analyses, we examined the contribution of each of the amino acid models to the estimated phylogeny by adjusting the prior settings to the mixed amino acid model, allowing model jumping between the different amino acid models (Ronquist and Huelsenbeck 2003). By examining the table of posterior probabilities of each model, we found that the WAG substitution model (Whelan and Goldman 2001) contributed the most and was used in all analyses. To account for site rate variation, we used TREE-PUZZLE to estimate the shape parameter of gamma distribution (α) and the proportion of invariable sites (P inv). Since the estimated proportion of invariable sites was insignificant, all analyses were performed with all sites potential to vary. Therefore each final Bayesian analysis was performed with a WAG substitution model and a mixed eight-category discrete-gamma model of among-site rate variation (WAG + Γ). In all Bayesian analyses, the starting tree was generated randomly in each run with all starting trees equally probable. For each analysis, two Monte Carlo Chains with 1,000,000 cycles were run. A tree was saved into a file each 1,000 generations. The plot for summary parameters was used to allocate the appropriate burn-in. The resulting trees from the two chains were pooled together and a majority-rule consensus tree was used to generate approximations of the posterior probability of each clade on the tree topology. The consensus tree was visualized with Treeview (Page 1996), and the phylogram was exported as a picture file for consecutive modifications to the presented form using Adobe Ilustrator CS. Pairwise sequence divergence was calculated as uncorrected pairwise distances using matrices of sequence alignments using PAUP*, excluding sites with large length heterogeneity.
Results
ACAD Family
Preliminary analysis of homologs of human ACADs from completely sequenced genomes of 3 archaeal, 3 bacterial, and 11 eukaryotic taxa (Supplementary Table 1), revealed that the origin of most of the members of the ACAD family could be traced to the root of the tree of life. Most strongly supported clades contained bacterial and eukaryotic homologs, while archaeal sequences were generally found to arise from the basal polytomy. Among the ACAD species, two pairs of ACADs (ACAD10 and ACAD11; VLCAD and ACAD9) shared more recent common ancestors. To further investigate whether bacterial ACADs are indeed closer to eukaryotic ACADs, we extended the dataset for homologs of additional archaeal and bacterial species. Since inclusion of these sequences enormously increased the dataset, eukaryotic sequences were restricted to taxa with preference for basal lineages (Supplementary Table 1). Maximum likelihood of the extended dataset recovered results consistent with our preliminary analysis. Sequences recovered in highly supported clades corresponded to particular species of ACADs with the exception of the more recent duplication events yielding the ACAD10/11 and VLCAD/ACAD9 paralogs (Fig. 1). Three groups of archaeal sequences were recovered in highly supported clades, specifically within the GCD and IVD clades and basally to ACAD10/11 clade. Other archaeal sequences were recovered in several clades; however, their placement within the phylogeny was not significantly supported and varied among different analyses.
Maximum likelihood phylogeny of acyl-CoA dehydrogenase protein family. Maximum likelihood phylogeny was estimated from a dataset of 353 amino acid positions from 205 sequences using PHYML (Guindon and Gascuel 2003). Tree was rooted (R) with ACOX and aidB homologs (not shown). Numbers at internodes refer to maximum likelihood bootstrap values. Only bootstrap values of >50% are shown. Gray bar shows the basal tree topology that differed in various analyses and exhibited low bootstrap support (<50%). Thick branches represent well-supported clades corresponding to human ACAD species (marked by circles) that were consistently recovered in all analyses performed. Arrows point to human ACAD homologs. Sequence accession numbers are in parentheses
To examine each of the ACAD clades in more detail, we have searched for homologs from additional organisms (e-values similar to human ACAD species and to the homologs defining each of the clades in Fig. 1) and performed clade-specific phylogenetic analyses. From all the high-scoring sequences only those were selected that increased the taxon sampling (mostly at the order level) of the diversity of archaeal, bacterial, and eukaryotic domains and would be informative in presented trees (therefore many homologs, particularly from the crown of the eukaryotic tree, were omitted). Detected homologs from additional taxa that were not included in the phylogenetic analyses were noted. Complete list of taxa considered in the study is presented in Supplementary Tables 2 and 3.
Glutaryl-CoA Dehydrogenase
Glutaryl-CoA Dehydrogenase (GCD) is a unique member of the ACAD family because of its dual enzymatic function. In addition to the α,β-dehydrogenation common to all ACADs, it also catalyzes a decarboxylation step, resulting in the overall oxidative decarboxylation of glutaryl-CoA to crotonyl-CoA and CO2 (Goodman et al. 1995; Lenich and Goodman 1986). Homologs to human GCD were found in all three domains of life (Fig. 2a). Archaeal GCDs share on average 51 ± 4% residues with bacterial sequences (excluding or including clade B3) and 50 ± 2% residues with eukaryotic GCD sequences (excluding clade P). The bacterial sequences nested within eukaryotic lineages (B3) share on average 64 ± 5% residues with eukaryotic sequences (E) but only 49 ± 6% residues with other bacterial sequences (B1 and B2). Fungi GCDs share on average 59 ± 3% and 60 ± 3% of residues with Proteobacteria (B3) and Eukaryota (E), respectively. Topological placement of the B3 clade within the eukaryotes indicates that an LGT event occurred from eukaryotes to Proteobacteria, resulting in “eukaryotic-like GCD” in this particular group of bacteria. This LGT most likely occurred after divergence of fungi and before the radiation of higher eukaryotes.
Bayesian phylogeny of glutaryl-CoA dehydrogenase (a) and ACAD10/11 (b) protein subfamilies. Unrooted phylograms were estimated from 377 and 402 amino acids, respectively. Numbers at internodes show posterior probabilities for Bayesian consensus tree. Bars (right to a) refer to clades or groups: A = archaeal taxa, B1–B3 = bacterial taxa, P and E = eukaryotic taxa
Interestingly, the GCD homologs of the P group (Fig. 2a) were consistently recovered as part of the GCD clade in all combined analyses. They share on average only 35 ± 2% residues with other GCD sequences. The presence of long internodes is indicative of acceleration in substitution rate and possible data saturation (Felsenstein 1985), although the χ 2 homogeneity test for deviation in amino acid frequencies was not significant for these sequences. Analysis performed with exclusion of the entire P clade resulted in identical topology to that presented in Fig. 2a. The active sites of Tetrahymena thermophila and the plant homologs are not consistent with GCD function since residues equivalent to those that are key to the decarboxylation function including R94, E87, and S95 are different in these sequences. In addition, the last 3′ triplet of amino acids of the plant sequences represent putative peroxisomal targeting signal (ARL/SRL) indicating its localization to peroxisome. Indeed, the homolog of Arabidopsis thaliana (at3g51840) was previously reported to function as short-chain acyl-CoA oxidases and localize to peroxisomes (Hayashi et al. 1999).
It is also interesting to note that the social amoeba Dictyostelium discoideum contains two GCD homologs (accession numbers XP_643912 and XP_639044). The former, which has an active site structure consistent with an acyl-CoA oxidase (exhibited base composition bias), was recovered in preliminary analyses basally to the plant sequences (not shown), while the other, XP_639044 with an active site that matches GCD, was consistently nested within the E clade (“animal-like GCD”, Fig. 2a). Since the basal eukaryotic protozoan pathogen Trypanosoma cruzi, as well as other eukaryotic genomes including plants and fungi, contain only one GCD homolog, the topological placement of the two GCD homologs of D. discoideum suggests a gene gain event predating the diversification of Mycetozoa and green plants. Nesting of the plant GCD-like oxidase sequences within the GCD bacterial sequences suggests the possibility that the plant GCD-like oxidase gain event could have been a lateral gene transfer (LGT) from a bacterial donor (gene xenologous to the “animal-like” GCD). Furthermore, while both xenologs are preserved in the genome of D. discoideum, the prokaryotic xenolog must have been fixed in the plant lineage while the animal-like GCD was lost. In contrast, the ancestral “animal-like” GCD was retained in the Metazoan lineage and the “plant-like” protozoan xenolog was lost. Testing this hypothesis further will require availability of additional completely sequenced genomes.
ACAD10 and ACAD11
The combined analysis (Fig. 1) indicated that ACAD10 and ACAD11 share more recent common ancestor with each other than with any other ACADs. Unlike other ACADs, the transcript map and exon structure of human ACAD10 and ACAD11 shows presence of multiple domains. Both transcripts contain additional sequences at the 5′-end that exhibit similarity to an aminoglycoside phosphotransferase domain (APH; E-value of 5 × 10−35). The human ACAD10 contains a second additional domain at its most 5′-terminus that shows similarity to a haloacid dehalogenase-like hydrolase domain (HAD; E-value of 2 × 10−14).
Homology searches were performed using full-length transcripts as well as each domain separately as a query. Genome mapping of fungi sequences revealed HAD, APH, and ACAD homologs in unlinked loci, in most cases on different chromosomes (not shown). Furthermore, genomes of bacteria, fungi, and the diatom Thalassiosira pseudonana have only one ACAD10/11 homolog, consisting only of an ACAD domain. A single homolog was also identified in plants; however, these consisted of both APH and ACAD domains. Homologs to both human ACAD10 and ACAD11 were found in genomes of other eukaryotic organisms with the most basal taxon of the cnidarian Nematostella vectensis (Fig. 2b). Contrary to other animals, the genomes of the nematodes Caenorhabditis elegans and C. brigssae contain only a single homolog encoding all three domains. Arthropods completely lack an ACAD10/11 homolog. The archaeal sequence of Natromonas pharaonis, recovered at the base of the ACAD10/11 clade in the combined phylogeny (Fig. 1), differs at residues that are conserved across all eukaryotic and bacterial ACAD10/11 homologs and was excluded from further analysis. Only sequences corresponding to ACAD domain were used in phylogenetic analyses (Fig. 2b) because spatially disassociated domains may evolve under different selective constraints. Bacterial sequences share on average 56 ± 5% residues with eukaryotic homologs, whereas the two eukaryotic clades corresponding to ACAD10 and ACAD11 homologs share 54 ± 3% residues. The single homologs of fungi and the marine diatom Thalassiosira pseudonana share on average 52 ± 3% and 48 ± 3% residues, respectively, with both eukaryotic ACAD10 and ACAD11. The single homolog of Tetrahymena thermophila exhibits 55(±2)% and 57(±2)% similarity to eukaryotic ACAD10 and ACAD11, respectively.
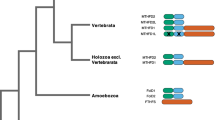
Based on our phylogenetic analysis, we have reconstructed the chronology of the formation of the multidomain structure of ACAD10 and ACAD11 (Fig. 3). A single ancestral gene consisted only of an ACAD domain as exemplified in bacterial species. Early in the eukaryotic lineage the ancestral ACAD10/11 acquired an APH domain, most likely before the divergence of plants. The lack of an APH domain in fungi and diatom Thalassiosira pseudonana homologs together with their phylogenetic nesting within bacterial clade (Fig. 2b) indicate that the presence of ACAD10/11 in these genomes is the result of an LGT from a bacterial donor (most likely proteobacterial) that resulted in orthologous replacement of the ancestral homolog. ACAD10 likely resulted from a duplication event of an already dual-domain ACAD10/11 before the divergence of Cnidaria that was coupled with acquisition of an additional HAD domain. Lineage-specific deletions occurred later in Nematoda (deletion of ACAD11) and before radiation of Arthropoda (deletion of both ACAD10 and ACAD11).
Inferred order of events in evolution of ACAD10/11 subfamily. Bars represent protein domains: long bar = ACAD, medium bar = APH, and short bar = HAD domains. The protein subfamily originated from an ancestral single-domain ACAD10/11, as exemplified in bacterial genomes. In eukaryotes, this single-domain ACAD10/11 acquired a new domain, the APH, and became a two-domain protein as exhibited in plant genomes. Later, most likely before diversification of major nonplant eukaryotic lineages, a duplication event resulted in two paralogs, the ACAD10 and ACAD11. ACAD10 acquired a second domain, the HAD. The single-domain homolog in fungi is most likely the result of an LGT from a bacterial donor that resulted in orthologous replacement of the ancestral ACAD10/11 homolog
Sequence analysis of the terminal amino acid triplet of ACAD10 and 11 homologs across 26 metazoan sequences, including 3 plant, 6 fungi, 1 diatom, 1 ciliate, 2 cnidarian, 1 nematode, and 12 vertebrate sequences, revealed a conserved peroxisomal targeting signal (Fig. 4), suggesting that this novel protein may be targeted to peroxisomes rather than to mitochondria. Recent studies of the mouse proteome are in agreement with this finding (Kikuchi et al. 2004).
Sequence logo of the last triplet of C-terminal amino acids of eukaryotic ACAD10 and ACAD11 homologs. The data used to generate this terminal motive contained 26 metazoan sequences: 3 plant, 6 fungi, 1 diatom, 1 ciliate, 2 cnidarian, 1 nematode, and 12 vertebrate sequences. The abundance of a residue is directly related to its size. The logo was generated by the weblogo server at http://weblogo.berkeley.edu/ using default settings (Crooks et al. 2004)
IVD
Homologs of human IVD were found widely present in eukaryotes, but bacterial homologs were found only in Proteobacteria (Fig. 5a). Sequence from the archaean species Thermoplasma acidophilum and Picrophilus toridus were recovered in combined analyses within the IVD clade; however, both sequences exhibited deviation from homogeneity of amino acid frequencies in the dataset consisting of only IVD homologs and were excluded from clade-specific analysis. The IVD homologs of Trypanosoma cruzi, Thalassiosira pseudonana, Oceanicola batsensis, and Syntrophobacter fumaroxidans were recovered in a monophyletic clade exhibiting long internodes despite homogeneous amino acid composition. Analysis with exclusion of these taxa resulted in identical topology. Pairwise amino acids comparison showed that Archaea share about 43 ± 2% and 40 ± 2% of residues with Proteobacteria and Eukaryota, respectively. Excluding the four taxa exhibiting long internodes, proteobacterial and eukaryotic IVDs share on average 64 ± 3% of residues.
Bayesian phylogeny of isovaleryl (a), very long-chain and ACAD9 (b), long-chain (c), and isobutyryl-CoA (d) dehydrogenase subfamilies. Unrooted phylograms were estimated from 384, 544, 375, and 373 amino acids, respectively. Numbers at internodes show posterior probabilities for Bayesian consensus tree. Small triangle in b refers to gene duplication event
VLCAD and ACAD9
VLCAD and ACAD9 exhibit overlapping specificity with long-chain unsaturated substrates from C14 to C20, with optima for palmitoyl-CoA (C16:0-CoA) and stearoyl-CoA (C18:0-CoA) (Ensenauer et al. 2005; Souri et al. 1998b; Zhang et al. 2002). While both VLCAD and ACAD9 are ubiquitously expressed in human tissues with high levels in liver, heart, and skeletal muscle, ACAD9 has been recently reported to also be significantly expressed in human brain (Ensenauer et al. 2005), including embryonic and fetal brain (Oey et al. 2006). This finding suggests a specific role for ACAD9 in developing brain not paralleled by VLCAD. Although homologs of VLCAD/ACAD9 could be identified from bacterial domain, genomes of the examined plants, fungi, and basal eukaryotes including Trypanosoma cruzi, Leishmania major, D. discoideum, Thalassiosira pseudonana, and Tetrahymena thermophila do not contain a VLCAD or ACAD9 homolog. This implies that the origin of VLCAD/ACAD9 in the eukaryotic domain is the result of an LGT introduction from a bacterial donor to the ancestor of Animalia. The genomes of N. vectensis, C. elegans, Tribolium castaneum, Drosophila melanogaster, and Anopheles gambiae contain only a single homolog that was recovered within the VLCAD clade with consecutive bootstrap support of 100 (Fig. 5b), suggesting that a duplication event yielding VLCAD and ACAD9 paralogs predated the divergence of Cnidaria. ACAD9 subsequently was lost in several lineages, specifically Cnidaria, Pseudocoelomata—Nematoda and Coelomata—Protostomia—Arthropoda. Pairwise sequence comparison revealed that bacterial sequences share more than 41 ± 3% residues. Bacterial homologs share on average 38 ± 3% residues with eukaryotic homologs. Eukaryotic sequences encompassed by the VLCAD and ACAD9 clades share on average 49 ± 4% residues.
LCAD
Although LCAD overlaps with MCAD and VLCAD in the ability to utilize saturated substrates of C8–C16 carbon chain, it has been recognized to play a key role in degradation of unsaturated fatty acids, specifically those with double bonds at positions 4,5 and 5,6, and branched-chain substrates (Lea et al. 2000). However, the physiological role of LCAD in humans still remains obscure. Maximum likelihood analysis of combined data recovered human LCAD as a sister taxon to a bacterial sequence with bootstrap support of 100 (Fig. 1); however, basal placement of C. elegans and N. vectensis sequences exhibited small bootstrap values. Maximum parsimony of combined data, however, revealed a clade consisting of human and bacterial LCAD together with sequences of C. elegans and N. vectensis with a bootstrap of 90 (not shown). Analysis of data with increased taxon sampling showed topology of two clades (Fig. 5c). One consisted of sequences from cnidarian, nematodal, and fungal genomes (“LCAD-like”), while the other clade contained sequences from genomes belonging to Deuterostomia with bacterial sequences at the base (“LCAD”). Pairwise sequence comparison showed that bacterial and eukaryotic LCADs share 53 ± 3% residues while both share only 30 ± 5% residues with eukaryotic “LCAD-like” sequences. The recovered topology indicates that the “LCAD-like” and LCAD genes are not true orthologs since the LCAD gene in Deuterostomia is most likely the result of an LGT from a proteobacterial donor. From the functional point of view, our analyses suggest that the “LCAD-like” gene may represent a novel ACAD that is not paralleled in the human and higher eukaryotic genomes.
IBD
IBD in human and rodents is specific to valine catabolism. Bacterial IBDs form a monophyletic clade (Fig. 5d) and on average they share 56 ± 4% residues with eukaryotic IBDs. Within the eukaryotes, the ciliate Tetrahymena thermophila exhibited base composition heterogeneity in the dataset composed of IBD homologs and was excluded from clade-specific phylogenetic estimations. Patchy distribution of IBD homologs within the eukaryotic domain is significant and is also noticeable in the bacterial domain (Supplementary Table 2). The absence of an IBD homolog in euglenozoa, viridiplantae, fungi, arthropoda, and echinodermata seem to be result of parallel losses since eukaryotic sequences form clearly a monophyletic clade (Fig. 5d) and exhibit significant sequence identity (72 ± 11%). Alternative hypothesis of independent introduction of IBD to different eukaryotic lineages would demand a similar source to every independent introduction and is not very probable.
SBCAD, SCAD, and MCAD
Human SBCAD is specific to isoleucine catabolism. SCAD with substrate specificity for butyryl-CoA (C4:0) represents the final stage of carbon chain shortening in fatty acid oxidation. MCAD acts on straight medium-chain acyl-CoA substrates with carbon backbone length of C4–C16 (Finocchiaro et al. 1987). All eukaryotic SBCAD, SCAD, and MCAD homologs were recovered in strongly supported monophyletic clades (bootstrap of 100, posterior probability of 100), clearly indicating their descent from single ancestral eukaryotic genes (Fig. 1). These three eukaryotic ACAD species, however, arise from the basal polytomy from which numerous bacterial sequences arise as well. This unresolved topology prevented identification of closely related bacterial homologs of the three ACADs. To examine this further, we screened additional bacterial and eukaryotic taxa for homologs of these genes and then performed combined analyses. Since this analysis yielded similar topology, we performed separate analyses of each of the three ACAD species in which we used all the bacterial sequences together with additional bacterial ACADs as outgroups. The resulting phylogenies differ in the topology of the bacterial sequences with respect to eukaryotic clades, indicating a weak signal for candidates of bacterial homologs (Fig. 6). Future taxon sampling of additional genomes may improve the basal resolution and possibly uncover additional events of LGT as currently exemplified by the SCAD homolog of Aedes aegypti (Fig. 6b).
Bayesian phylogeny of short/branched-chain (a), short-chain (b), and medium-chain (c) acyl-CoA dehydrogenase subfamilies. Unrooted phylograms were estimated from datasets of 377, 379, and 376 amino acids, respectively. Numbers at internodes show posterior probabilities for Bayesian consensus tree. Small circles refer to gene duplication events
Novel ACADs with Unknown Function
Sequence comparison has revealed a few intriguing sequences that require further examination. One is an ACAD-like sequence, YP_386244 (Fig. 1), present in Geobacter metallireducens. The hypothetical catalytic residue of this putative protein is a threonine, T363, rather than a glutamate. Another is a short, perhaps incomplete sequence of Cryptococcus neoformans, XP_567619 (Fig. 6c), where a crucial residue that comes within contact distance of the reactive atoms in the active site is a cysteine rather than a tyrosine (the equivalent position in MCAD is Y373 in the mature protein). A thiol group at such a position is expected to be reactive and likely to perform some novel function. In addition, W166 (mature MCAD numbering), which is a crucial residue for flavin binding and enzyme reaction mechanism, is a glycine in this same hypothetical protein. A glycine at this position is also found in a hypothetical protein with homology to MCAD from C. elegans, NP_495066 (Fig. 6a). A signature feature for accommodating branching at the substrate acyl C-3 position, a tyrosine to glycine substitution found only in IVD proteins, is found in three hypothetical proteins with homology to SCAD (Fig. 6b) in Acidobacteria bacterium (ABF41370), Bacillus cereus (NP_978856), and Geobacillus kaustophilus (YP_147450). With the exception of the active site catalytic residue position and the residue equivalent to human IVD Y371, which is distant in the active site, other residues are the same as the ones in IVD, leading us to speculate that these hypothetical proteins would utilize isovaleryl-CoA as well as other substrates.
Discussion
Our sequence analyses demonstrate that the origin of the ACAD family can be traced to the common ancestor of Archaea, Bacteria, and Eukaryota, suggesting its essential role in metabolism of early life. The emergence of the family was likely associated with gene duplication of the prehistoric ancestor gene even before the emergence of major life domains. The most ancient ACADs are ancestral GCD, ACAD10/11, and IVD. Additionally, domain and lineage-specific duplications and losses and LGT events then occurred after the divergence of Archaea, Bacteria, and Eukaryota resulting in an overall patchy distribution of human ACAD homologs across the tree of life. Additional mechanisms shaping the evolution of the ACAD family include alteration of cellular localization and evolution of novel proteins by domain acquisition.
Lineage-Specific Duplications and Losses of ACADs
Enormous variability in the number of ACADs is evident in Archaea and Bacteria. In the archaeal domain there are genomes with as low as 4 ACADs (such as Pyrobaculum torridus) and as many as 14 ACADs (Archaeoglobus fulgidus). This range may not be fully representative due to limited sources of fully sequenced genomes. The bacterial domain exhibits even larger variability in ACADs (Fig. 1; Supplementary Table 2), suggesting that the ancestor of bacteria most likely contained many more species of ACADs and that lineage-specific retention of these ancestral ACADs as different bacteria moved to various environments contributed to the high degree of patchiness detected throughout the bacterial domain. An extreme example is provided by the genome of the pathogenic bacterium Clostridium tetani (Firmicutes) that possesses six ACAD homologs five of which are recent paralogs, indicating a loss of most other ACAD species.
Except for the few instances discussed below, all eukaryotic sequences were recovered as monophyletic clades corresponding to specific ACAD species with bacterial homologs as their closest relatives. This indicates that at least seven ACAD members were present in the ancestor of all eukaryotes. The most profound reduction in the number of ACADs occurred in the Viridiplantae that retain only the three most ancient ACADs: the GCD, IVD, and ACAD10/11 homologs. Other eukaryotic lineages also seem to have experienced secondary losses of different ACAD species. It seems that diatoms have lost IBD and MCAD, fungi have lost IBD and SCAD, ciliates have lost SCAD, MCAD, and SBCAD, and arthropods have lost ACAD10/11 and IBD. This excludes VLCAD/ACAD9 and LCAD from consideration since they resulted from a more recent LGT event from the bacterial to eukaryotic domain.
Lineage-specific duplications and retention of functional paralogs are usually associated with functional diversification. As an example, the genomes of the nematodes C. elegans and C. briggsae seem to have lost SCAD but contain multiple copies of MCAD and SBCAD that are apparently functional (Fig. 6). On the other hand, complete losses of ACADs may be attributable to the life cycle of some organisms. The parasitic eukaryote Trypanosoma cruzi exemplifies the process of pseudogene formation that may ultimately lead to gene loss. The increased rate of substitution detected in several Trypanosoma cruzi ACADs, loss of nucleotide conservation at highly conserved sites, and significant departure from homogeneity of amino acid composition illustrate the genomic consequence of environmental and physiological constrains on parasitic species.
Lateral Gene Transfer Within ACAD Family
Although LGT is undoubtedly an essential part of bacterial evolution and certainly effected evolution of ACADs in this domain, we did not pursue to investigate the level of its contribution to ACAD evolution in bacteria. In this study, we have concentrated on the eukaryotic taxa. One example of an LGT comes from the GCD subfamily, where an LGT event resulted in eukaryotic-like GCD in Proteobacteria (Fig. 2a). Another example is provided by the LCAD subfamily, where the “LCAD-like” homologs of lower eukaryotes, including fungi, cnidaria, and nematodes are not true orthologs to the “LCAD” homologs of higher animals since the “LCAD” homolog is the result of an LGT from a Proteobacterial donor to ancestor of Deuterostomata (Fig. 5c). An important implication of this finding is that extensive phylogenetic analysis should precede any considerations to use animal models for human studies. This specific example demonstrates that C. elegans would be a poor choice as a model organism to study biology of LCAD and its deficiency.
Evolution by Domain Acquisition
The first example comes from the VLCAD/ACAD9 subfamily. Unlike other ACADs, human VLCAD and ACAD9 contain an extension of the C-terminus suggested to be involved in intramitochondrial membrane binding as well as acting as the equivalent of the second dimer in the tetrameric ACADs (Andresen et al. 1996; Souri et al. 1998a). The presence of this extension in bacterial and other eukaryotic VLCAD/ACAD9 homologs indicate its acquisition early in the evolution of this subfamily. Homology searches of the most conserved region of the C-terminal extension (Fig. 7) against the NCBI Conserved Domain database recovered similarity to an ACAD domain (e-value of 0.001), suggesting its possible origin from an ACAD paralog with subsequent sequence divergence and chimerization with the ancestral VLCAD/ACAD9. The lack of this extension in the Brucella suis homologs (NP699419 and NP70024, not shown) and in Danio rerio (AAH95385) must, therefore, be a secondary loss. Additionally, the active site structure of these last three sequences does not conform to a VLCAD or ACAD9 function.
Conserved region of the C-terminal extension of VLCAD and ACAD9 exhibiting similarity to ACAD domain (e = 0.001). Alignment of 9 bacterial and 23 eukaryotic VLCAD and ACAD9 homologs. Conserved residues are shaded. Arrows show residues that are invariant or highly conserved across the 32 sequences compared. While Q522 is involved in binding the adenine moiety of the FAD, R544 forms a salt bridge with the invariant E354, apparently supporting the tertiary structure integrity. The K513, W566, and E569 seem to stabilize the tertiary structure as well. In the tetrameric ACADs, interactions across the dimer–dimer interface help stabilize the protein quaternary structure
Second example of chimeric proteins are ACAD10 and ACAD11. The existence of multidomain transcripts has been experimentally documented for human ACAD10 and ACAD11, as well as for the C. elegans and A. thaliana homologs. Our analyses suggest that two consecutive chimerization events occurred in the evolution of the ACAD10/11 subfamily (Fig. 3). The first preceded the divergence of plants and ciliates, when an ancestral homolog containing only the ACAD domain acquired an APH domain, as seen in Ostreococcus lucimarinus, A. thaliana, Oryza sativa, and Tetrahymena thermophila (Fig. 2). The second chimerization occurred before the divergence of the major animal lineages when the already two-domain ACAD10/11 homolog duplicated and one of the resulting paralogs (ACAD10) acquired the HAD domain, as exemplified by ACAD10 and ACAD11 homologs from cnidarian N. vectensis, echinodermate Strongylocentrotus purpuratus, and vertebrate taxa. Moreover, the presence of a low sequence conservation region between the APH and ACAD domains that does not overlap with either APH or ACAD sequences of bacteria and fungi, suggests its intronic origin and further implies that ACAD10 and ACAD11 are transcription-induced chimeras rather than the results of trans-splicing.
Mitochondrial β-Oxidation in Plants
In contrast to a well-characterized peroxisomal β-oxidation pathway, the existence of mitochondrial β-oxidation in higher plants has been a subject of controversy. Experimental studies using respiratory-chain inhibitors to inhibit mitochondrial β-oxidation (Dieuaide et al. 1993; Gerhardt et al. 1995) made a strong argument for the existence of plant ACADs. Purified mitochondria from carbohydrate-starved maize root tips and sunflower embryos exhibited ACAD activity (Bode et al. 1999); however, several of the putative plant ACADs based on sequence similarity with mammalian ACADs (Bode et al. 1999) were identified later as peroxisomal acyl-CoA oxidases (De Bellis et al. 2000; Hayashi et al. 1999). The report of a single plant ACAD, putative IVD, identified from mitochondria of A. thaliana (Daschner et al. 1999) was later followed by identification of an IVD from Solanum tuberosum (Faivre-Nitschke et al. 2001). Measurement of [1-14C] palmitate oxidation in cotyledons, plumules, and radicles of Pisum sativum clearly demonstrated not only the presence of a mitochondrial β-oxidation system in higher plants but also that its activity paralleled high biosynthetic activity during root and leaf development (Masterson and Wood 2001). The most current view of β-oxidation in plants, advanced by analysis of completely sequenced genome of A. thaliana, states that straight-chain fatty acid catabolism is carried via β-oxidation in peroxisomes, whereas mitochondria play an important role in catabolism of branched-chain amino acids (Graham and Eastmond 2002). Our findings clearly identify three ACADs in plants, the GCD and IVD, and a two-domain ACAD10/11 homolog but their roles in mitochondrial β-oxidation remains to be elucidated. An additional circumstantial evidence in support of the presence of ACADs in plants is the presence of ETF, which is the ACAD electron transfer partner that shuttles electrons to the electron transport chain.
Functional Differentiation of Lineage-Specific Paralogs
In general, retention of gene duplicates anticipates functional differentiation among the paralogs. Two functionally diverged IVD paralogs that share 84% residues were previously found in S. tuberosum (Faivre-Nitschke et al. 2001). While one of the two IVD paralogs was found to function as an IVD, the other paralog had maximum activity with 2-methylbutyryl-CoA as substrate, corresponding to human SBCAD (Goetzman et al. 2005). Since plants do not contain SBCAD homologs, this is an example of functional substitution for a lost enzyme.
Detailed analysis of MCAD homologs revealed the presence of three copies of the MCAD gene in the genomes of the nematodes, C. elegans and C. briggsae (Fig. 6c). Although two of the C. elegans genes are paralogs, exhibiting 60% sequence identity to the third C. elegans gene, they still significantly differ in their amino acid sequences (77% identical). Previously, two genes encoding MCAD homologs were reported from a close relative, a parasitic nematode Ascaris suum (Komuniecki et al. 1985) whose genome has not yet been fully sequenced. These were recovered in the MCAD phylogeny as a close relative to the third paralog of C. elegans and C. briggsae. The ascarid enzymes have been shown to function in the reverse direction in vivo as NADH-dependent enoyl-CoA reductases (Komuniecki et al. 1985). Expression of the ascarid enzyme is tissue specific and developmentally up regulated with the switch to anaerobic oxidation at the third larval instar (Duran et al. 1998). The functional modification of the MCAD homologs in Ascaris apparently resulted from ecological adaptation to a parasitic life style. C. elegans, however, is a free-living aerobic nematode that still retains multiple apparently functional copies of MCAD gene. Our analysis revealed that a duplication event producing the two ascarid genes occurred on the lineage leading to the genus Ascaris and therefore these should be considered paralogs; both are homologous to the third MCAD paralog of C. elegans (Fig. 6c). Functional studies of these enzymes will be necessary to gain further insight into the evolution of enzymatic properties of these ACADs. One of the paralogs may functionally substitute for the missing SCAD in these species or they may all have developed alternative functions.
Endosymbiotic Origin of the ACAD Family in Eukaryotes
Our comparative and phylogenetic analyses demonstrate that the origin of the ACAD family can be traced to the origin of the three domains of life: Archaea, Bacteria, and Eukaryota. The bacterial and eukaryotic ACADs are phylogenetically more closely related to each other than to any archaeal ACADs. Bacterial and eukaryotic taxa share seven ACADs (GCD, ACAD10/11, IVD, IBD, SBCAD, SCAD, and MCAD) that are consistent with endosymbiotic acquisition of ACAD genes in eukaryotes because for each of them the bacterial and eukaryotic clades are joined in the trees at the basal level. The endosymbiotic origin is also supported by cellular localization of known human ACADs in mitochondria. Although a single bacterial donor is not obvious due to a patchy distribution of ACADs across the bacterial tree, it is likely that such a donor was a proteobacterium, since those are the only bacteria that contain homologs of all ACADs. In addition, the difference in fatty acid composition in archaeal and bacterial/eukaryotic membranes should be noted. Unlike Eukaryota and Bacteria that have predominantly ester-linked fatty acids in lipids, Archaea have ether-linked lipids with C20–C40 isoprenoid (multiply-branched) chains that likely require ACADs with different substrate specificities. Nevertheless, the long branched isoprenoid hydrocarbon fatty acids are not exclusive to Archaea but are also found in Bacteria and are prominent in basal Metazoa, specifically in the Porifera. This leads us to speculate that the substrates for the unknown ancient members of the ACADs (ACAD10 and ACAD11) are also long branched-chain substrates.
Conclusion
We show that the evolutionary history of ACAD family is very dynamic. The objectives initiating our investigations were to examine taxonomic distribution of human ACAD homologs, since studies using model organisms are sought for functional investigations of undescribed members. Results of our analyses, however, open up additional intriguing questions that require further investigations. Is the patchy distribution of ACAD species in the bacterial domain in particular the result of gene duplications and secondary losses or rather result of intensive LGT? Unlike to eukaryotic taxa that contain in general only sequences homologous to human ACAD species (nine subfamilies), most of examined bacterial taxa contain additional ACAD-like sequences (Supplementary Table 3). These findings suggest that bacteria in general contain much more ACAD species than do eukaryotes. Are these homologs results of more recent duplications and thus are specific to crown bacterial taxa? Some of them undoubtedly are, one example is Clostridium tetani with six ACADs, five of which are recent paralogs. Further investigations would be required to find how many of these ACAD-like sequences are functional and how many of them are pseudogenes. Detection of pseudogene formation would support argument for gene loss while detection of functional paralogs would support gene duplications, several examples are apparent already from our analyses. In conclusion, from currently limited data, we witness that evolution of ACAD family is remarkably dynamic including LGT, gene duplications, and losses. How much each of these molecular mechanisms has contributed to the ACAD protein evolution, especially among bacteria, can be addressed only after more genomes are completely sequenced.
Abbreviations
- ACADs:
-
Acyl-CoA dehydrogenases
- ACOX:
-
Acyl-CoA oxidase
- GCD:
-
Glutaryl-CoA dehydrogenase
- IBD:
-
Isobutyryl-CoA dehydrogenase
- IVD:
-
Isovaleryl-CoA dehydrogenase
- VLCAD:
-
Very long-chain acyl-CoA dehydrogenase
- LCAD:
-
Long-chain acyl-CoA dehydrogenase
- SCAD:
-
Short-chain acyl-CoA dehydrogenase
- SBCAD:
-
Short/branched-chain acyl-CoA dehydrogenase, also known as 2-methyl branched chain acyl-CoA dehydrogenase
- MCAD:
-
Medium-chain acyl-CoA dehydrogenase
- LGT:
-
Lateral gene transfer
References
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andresen BS, Bross P, Vianey-Saban C, Divry P, Zabot MT, Roe CR, Nada MA, Byskov A, Kruse TA, Neve S, Kristiansen K, Knudsen I, Corydon MJ, Gregersen N (1996) Cloning and characterization of human very-long-chain acyl-CoA dehydrogenase cDNA, chromosomal assignment of the gene and identification in four patients of nine different mutations within the VLCAD gene. Hum Mol Genet 5:461–472
Battaile K, Molin-Case J, Paschke R, Wang M, Bennett D, Vockley J, Kim J-JP (2002) Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA; comparison with other acyl-CoA dehydrogenases. J Biol Chem 277:12200–12207
Battaile KP, Nguyen TV, Vockley J, Kim JJ (2004) Structures of isobutyryl-CoA dehydrogenase and enzyme-product complex: comparison with isovaleryl- and short-chain acyl-CoA dehydrogenases. J Biol Chem 279:16526–16534
Bode K, Hooks MA, Couee II (1999) Identification, separation, and characterization of acyl-coenzyme A dehydrogenases involved in mitochondrial beta-oxidation in higher plants. Plant Physiol 119:1305–1314
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Daschner K, Thalheim C, Guha C, Brennicke A, Binder S (1999) In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol Biol 39:1275–1282
De Bellis L, Gonzali S, Alpi A, Hayashi H, Hayashi M, Nishimura M (2000) Purification and characterization of a novel pumpkin short-chain acyl-coenzyme A oxidase with structural similarity to acyl-coenzyme A dehydrogenases. Plant Physiol 123:327–334
Dieuaide M, Couee I, Pradet A, Raymond P (1993) Effects of glucose starvation on the oxidation of fatty acids by maize root tip mitochondria and peroxisomes: evidence for mitochondrial fatty acid beta-oxidation and acyl-CoA dehydrogenase activity in a higher plant. Biochem J 296(Pt 1):199–207
Djordjevic S, Pace CP, Stankovich MT, Kim JJP (1995) Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera esdenii. Biochemistry 34:2163–2171
Duran E, Walker DJ, Johnson KR, Komuniecki PR, Komuniecki RW (1998) Developmental and tissue-specific expression of 2-methyl branched-chain enoyl CoA reductase isoforms in the parasitic nematode, Ascaris suum. Mol Biochem Parasitol 91:307–318
Ensenauer R, He M, Willard JM, Goetzman ES, Corydon TJ, Vandahl BB, Mohsen AW, Isaya G, Vockley J (2005) Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty acids. J Biol Chem 280:32309–32316
Faivre-Nitschke SE, Couee I, Vermel M, Grienenberger JM, Gualberto JM (2001) Purification, characterization and cloning of isovaleryl-CoA dehydrogenase from higher plant mitochondria. Eur J Biochem 268:1332–1339
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Finocchiaro G, Ito M, Tanaka K (1987) Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases from human liver. J Biol Chem 262:7982–7989
Foster PG, Hickey DA (1999) Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol 48:284–290
Fu Z, Wang M, Paschke R, Rao KS, Frerman FE, Kim JJ (2004) Crystal structures of human glutaryl-CoA dehydrogenase with and without an alternate substrate: structural bases of dehydrogenation and decarboxylation reactions. Biochemistry 43:9674–9684
Gerhardt B, Fischer K, Maier U (1995) Effect of palmitoylcarnitine on mitochondrial activities. Planta 196:720–726
Goetzman ES, Mohsen AW, Prasad K, Vockley J (2005) Convergent evolution of a 2-methylbutyryl-CoA dehydrogenase from isovaleryl-CoA dehydrogenase in Solanum tuberosum. J Biol Chem 280:4873–4879
Goodman SI, Kratz LE, DiGiulio KA, Biery BJ, Goodman KE, Isaya G, Frerman FE (1995) Cloning of glutaryl-CoA dehydrogenase cDNA, and expression of wild type and mutant enzymes in Escherichia coli. Hum Mol Genet 4:1493–1498
Graham IA, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41:156–181
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall CL (1978) Acyl-CoA dehydrogenases and electron-transferring flavoprotein. Methods Enzymol 53:502–518
Hamada K, Ago H, Kuramitsu S, Miyano M (2004) Crystal structure of Thermus thermophilus medium-chain acyl-CoA dehydrogenase. PDB
Hayashi H, De Bellis L, Ciurli A, Kondo M, Hayashi M, Nishimura M (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J Biol Chem 274:12715–12721
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Ikeda Y, Keese S, Fenton WA, Tanaka K (1987) Biosynthesis of four rat liver mitochondrial acyl-CoA dehydrogenases. Import into mitochondria and processing of their precursors in a cell-free system and in cultured cells. Arch Biochem Biophys 252:662–674
Kikuchi M, Hatano N, Yokota S, Shimozawa N, Imanaka T, Taniguchi H (2004) Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J Biol Chem 279:421–428
Kim JJP, Wang M, Paschke R (1993) Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with and without substrate. Proc Natl Acad Sci USA 90:7523–7527
Kionka C, Kunau WH (1985) Inducible beta-oxidation pathway in Neurospora crassa. J Bacteriol 161:153–157
Klein K (1973) Dehydrogenasen und ETF in Escherichia coli. Studien zum Fettsäureabbau. Doctoral dissertation, University of Cologne, Cologne, Germany
Komuniecki R, Fekete S, Thissen-Parra J (1985) Purification and characterization of the 2-methyl branched-chain Acyl-CoA dehydrogenase, an enzyme involved in NADH-dependent enoyl-CoA reduction in anaerobic mitochondria of the nematode, Ascaris suum. J Biol Chem 260:4770–4777
Krasko A, Schroder HC, Hassanein HM, Batel R, Muller IM, Muller WE (1998) Identification and expression of the SOS response, aidB-like, gene in the marine sponge Geodia cydonium: implication for the phylogenetic relationships of metazoan acyl-CoA dehydrogenases and acyl-CoA oxidases. J Mol Evol 47:343–352
Kunau WH, Dommes V, Schulz H (1995) Beta-oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog Lipid Res 34:267–342
Landini P, Hajec LI, Volkert MR (1994) Structure and transcriptional regulation of the Escherichia coli adaptive response gene aidB. J Bacteriol 176:6583–6589
Lea W, Abbas AS, Sprecher H, Vockley J, Schulz H (2000) Long-chain acyl-CoA dehydrogenase is a key enzyme in the mitochondrial beta-oxidation of unsaturated fatty acids. Biochim Biophys Acta 1485:121–128
Lee HJ, Wang M, Paschke R, Nandy A, Ghisla S, Kim JJ (1996) Crystal structures of the wild type and the Glu376Gly/Thr255Glu mutant of human medium-chain acyl-CoA dehydrogenase: influence of the location of the catalytic base on substrate specificity. Biochemistry 35:12412–12420
Lenich AC, Goodman SI (1986) The purification and characterization of glutaryl-coenzyme A dehydrogenase from porcine and human liver. J Biol Chem 261:4090–4096
Masterson C, Wood C (2001) Mitochondrial and peroxisomal beta-oxidation capacities of organs from a non-oilseed plant. Proc Biol Sci 268:1949–1953
Nandy A, Kuchler B, Ghisla S (1996) Molecular evolution and substrate specificity of acyl-CoA dehydrogenases: chimaeric “medium/long’ chain-specific enzyme from medium-chain acyl-CoA dehydrogenase. Biochem Soc Trans 24:105–110
Oey NA, Ruiter JP, Ijlst L, Attie-Bitach T, Vekemans M, Wanders RJ, Wijburg FA (2006) Acyl-CoA dehydrogenase 9 (ACAD 9) is the long-chain acyl-CoA dehydrogenase in human embryonic and fetal brain. Biochem Biophys Res Commun 346:33–37
Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Schmidt HA, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504
Souri M, Aoyama T, Hoganson G, Hashimoto T (1998a) Very-long-chain acyl-CoA dehydrogenase subunit assembles to the dimer form on mitochondrial inner membrane. FEBS Lett 426:187–190
Souri M, Aoyama T, Yamaguchi S, Hashimoto T (1998b) Relationship between structure and substrate-chain-length specificity of mitochondrial very-long-chain acyl-coenzyme A dehydrogenase. Eur J Biochem 257:592–598
Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA, Mammalian Gene Collection Program T (2002) Generation and initial analysis of more than 15, 000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99:16899–16903
Swofford DL (2000) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0 Beta. Sinauer, Sunderland
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen AWA, Vockley J, Kim JJP (1997) Structure of human isovaleryl-coA dehydrogenase at 2.6 angstrom resolution—basis for substrate specificity. Biochemistry 36:8455–8464
Toogood HS, van Thiel A, Basran J, Sutcliffe MJ, Scrutton NS, Leys D (2004) Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex. J Biol Chem 279:32904–32912
Valenciano S, Lucas JR, Pedregosa A, Monistrol IF, Laborda F (1996) Induction of beta-oxidation enzymes and microbody proliferation in Aspergillus nidulans. Arch Microbiol 166:336–341
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699
Ye X, Ji C, Zhou C, Zeng L, Gu S, Ying K, Xie Y, Mao Y (2004) Cloning and characterization of a human cDNA ACAD10 mapped to chromosome 12q24.1. Mol Biol Rep 31:191–195
Zhang J, Zhang W, Zou D, Chen G, Wan T, Zhang M, Cao X (2002) Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family. Biochem Biophys Res Commun 297:1033–1042
Acknowledgments
This work was supported by PHS NIH Grant R01-DK54936 and the Pennsylvania Department of Health, Tobacco Formula Funding. We thank Eric Goetzman for discussion on physiological effects of ACAD deficiencies.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below are the links to the electronic supplementary materials.
Rights and permissions
About this article
Cite this article
Swigoňová, Z., Mohsen, AW. & Vockley, J. Acyl-CoA Dehydrogenases: Dynamic History of Protein Family Evolution. J Mol Evol 69, 176–193 (2009). https://doi.org/10.1007/s00239-009-9263-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9263-0