Abstract
Gut microbiota have a significant impact on host physiology and health, and host genetics and diet are considered as two important factors, but it is difficult to discriminate the influence of each single factor (host or diet) on gut microbiota under natural conditions. Moreover, current studies of avian microbiota mainly focus on domestic or captive birds, and it is still uncertain how host and diet take part in changing avian gut microbiota composition, diversity, and function in the wild. Here, high-throughput sequencing of 16S rRNA was used to identify the gut microbiota communities for sympatric wintering Great Bustards and Common Cranes at different diets. The results showed that 8.87% operational taxonomic units (OTUs) were shared among all sampling birds; in contrast, 39.43% of Kyoto Encyclopedia of Genes and Genomes (KEGG) functional pathways were common among all individuals, indicating the existence of gut microbiota conservatism both in microbiota structure and function. Microbiota abundance and diversity differed between Great Bustards and Common Cranes in a specific wintering site, and microbiota variation was detected for the same host species under two different sites, suggesting that the change of gut microbiota was induced by both host and diet. Furthermore, we found that changes of both microbial communities and functional pathways were larger between hosts than those between diets, which revealed that host might be the dominant factor determining microbiota characteristics and function, while diet further drove the divergence of gut microbiota. Gut microbiota functions appeared to be more conserved than bacterial community structure, indicating that different bacteria may function in a similar way, while microbiota OTU diversity might not be necessarily associated with functional diversity. With diet shifting, gut microbiota changed both in terms of microbial communities and functional pathways for the sympatric birds, which implies that avian habitats and their physiological microbiota would be influenced by different farmland management regimes.
Key points
• Gut microbiota can be shaped by both diets and hosts in sympatric species.
• Host was the dominant factor shaping the gut microbiota communities and functional pathways.
• Gut microbiota were conservative both in structure and in function, but more conservative in function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gut microbiota have a significant impact on host physiology and health among a variety of animals. Microbes can intervene in digestion, metabolism, pathogen protection, immunity, and even organ development (Al-Asmakh et al. 2014; AlHilli and Bae-Jump 2020; Xue et al. 2020). For birds, the composition and characteristics of gut microbiota are different from other animals due to their unique and complex diets, physiological traits, and migration strategies (Song et al. 2020). The study of microbial diversity and function contributes to understanding of avian physiology and adaptive capacity to variable environments (McWhorter et al. 2009), and there is a growing shift towards understanding microbiome variation in wild animals.
Gut microbial structure and function are determined by variable factors. Human activities, sex, age, habitat environment, diet, host, season, and even migration can impact bacterial composition and function (Bermingham et al. 2018; Chang et al. 2016; Hu et al. 2018; Luo et al. 2017; Wu et al. 2018). In addition, researchers have found that land use change has host-specific effects on gut microbial communities among six avian species due to the interaction between diet changes and hosts (San Juan et al. 2020). In some studies, host genetics is the most key factor influencing host bacterial composition, while geographic distribution has little impact (Hird et al. 2015), and gut microbiota has more strong response on host species than habitat type (San Juan et al. 2020).
Foraging habitats and niches are similar or dissimilar among sympatric animals, which exist in the same region. Although sympatric species consume common diet resources, their gut microbiota are quite different both in communities and diversity (Perofsky et al. 2019; Xiang et al. 2019). The divergence of bacterial communities in sympatric overwintering birds caused by diets and hosts was found in Grus monacha and Anser anser domesticus (Fu et al. 2020b). By investigating the gut microbiota of sympatric animals, it will help to control interfering factors (e.g., host, diet, season, habitat) and facilitate differentiating impacts for a given factor.
Eastern population of Great Bustard (Otis tarda dybowskii) is a typical steppe bird, as well as a long-distance migration bird, flying 2000 kilometers from Mongolia to China for overwintering (Kessler et al. 2013). The population size of eastern Great Bustard is approximately 1600–2200 (Liu et al. 2017) and is still struggling to survive with a decreasing trend (Alonso and Palacin 2010; Collar et al. 2017). Almost all eastern Great Bustards winter in northern and central China and spend 4 months of their history life in scattered wintering habitats (Kessler et al. 2013; Mi et al. 2014). A percentage of 40.55% eastern Great Bustards feed and roost in the farmland (Li et al. 2021), meaning they largely dependent on farmland, and a series of potential risks influence their wintering security, including crop planting uncertainty, pesticide residues, poor concealment of overwintering grounds, reduced food diversity, and even nutritional stress, as well as power line collisions and irregular human interference near overwintering grounds (Jiang 2004; Liu et al. 2018; Tian and Zhang 2006).
Besides those threatening factors, eastern Great Bustards have to compete with sympatric birds for space and food, and the obvious competitor is Common Cranes (Grus grus) during the wintering season, where it can be often seen that Great Bustards and Common Cranes feed together in the same farmland (Fig. 1). Common Cranes are widely distributed, with a population size of 21,632 to 22,401 in China, and conduct an enormous distance migration in eastern China (Kanai et al. 2002; Youhui and Hong 2003). Although Common Cranes are wetland birds, they also have to rely on farmlands for food because of severe wetland loss and degradation (Li et al. 2020). Both Great Bustards and Common Cranes rely heavily on farmlands to overwinter by consuming the agricultural food and roosting in the farmland habitats (Liu et al. 2018; Mi et al. 2016; Yu et al. 2008; Zhan et al. 2007). Migrating birds are able to adapt to the local environment by regulating their gut microbiota in response to the diet variations following their arrival to the wintering grounds (Davidson et al. 2020a; Wang et al. 2019; Wu et al. 2018); however, how the sympatric overwintering birds respond in gut microbiota is well worth exploring.
In this study, we aimed to analyze the composition and diversity of gut microbial communities of sympatric overwintering Great Bustards and Common Cranes, and further tested the impacts of diet and host on the gut microbiota of both farmland-dependent birds. Finally, the metabolism and function profiles of the gut microbiome were investigated for each species at different diets, and the relationship between microbiota structure changes and function alterations has been examined.
Materials and methods
Study area and sample collection
Fecal samples of Great Bustards and Common Cranes were gathered from wintering sites in the Jinzhou, Liaoning province, and Xinxiang, Henan province, in December 2017 (Fig. 1). In the Jinzhou wintering area, peanut and rice farmlands provide the main food for Great Bustards and Common Cranes, while in the Xinxiang wintering area, wheat seedlings and corn are the main food. The food composition and structure were kept stable across patches of farm fields in each specific sampling region. According to the wintering survey of bird population size, there were 52 and 285 Great Bustards in Jinzhou and Xinxiang, respectively, and 51 and 2000 Common Cranes in Jinzhou and Xinxiang, respectively. The fresh fecal samples were collected after the birds flew away, and only fecal samples with a minimum distance interval of 5 m were collected in order to avoid recollecting fecal samples from the same individual. We only focused on the same bird flock and collected the fecal samples from individuals living in the same flock. In addition, we only collected the aboveground part of feces in order to avoid collecting the soil and shedding leaves on the ground, which might be potential environmental sources of bacteria. For individuals, we classified the sample type by host and diet, for example, Great Bustard with wheat diet (GB_W), Great Bustard with peanut diet (GB_P), Common Crane with wheat diet (CC_W), and Common Crane with peanut diet (CC_P). Notably, agricultural grains in harvested farmlands are the main and stable food for them to overwinter, though they also eat other wild grasses (Liu et al. 2018). Forty-eight fecal samples were collected in total, and each group had twelve samples. All samples were immediately frozen and stored at −20°C in the wild and later stored at −80°C in the laboratory.
DNA isolation, amplification, and sequencing
Fecal DNA isolation was carried out using the QIAamp DNA Stool Mini Kit (Valencia, Germany). Two blank DNA extractions were included to detect cross-contamination during the DNA isolation process. The highly variable V3-V4 region of bacterial 16S rRNA was chosen and amplified using primers 515F (5’- GTGCCAGCMGCCGCGGTAA -3’) and 806R (5’- GGACTACHVGGGTWTCTAAT -3’) (Caporaso et al. 2012). The polymerase chain reaction (PCR) reactions were performed in a total of 50 μl volume, using 0.5 μl AmpliTaq Gold DNA Polymerase (Waltham, MA, USA) and 3 μl of DNA template. The PCR cycling conditions were set as follows: firstly, an initial denaturing at 95°C for 5 min; then followed by 30 cycles of 95°C for 35s, 56°C for 30s, and 72°C for 35s; and finally finished with an extension of 72°C for 10 min. After quantifying the PCR products using NanoDrop 2000 (Waltham, MA, USA), PCR products were pooled in equal-molar concentrations, and purified PCR products were sequenced using the Illumina MiSeq sequencing platform (San Diego, CA, USA) at Shanghai Sangon Biotech Co. Ltd (Shanghai, China).
Statistical analysis
Reads quality checking, filtering, and demultiplexing were done using the standard operating procedure with QIIME version 2 (Bolyen et al. 2019). UCHIME2 was used to remove amplicon sequencing chimeras (Edgar et al. 2011). The clean sequences were clustered into operational taxonomic units (OTUs) with a sequence similarity threshold of 97% and assigned to taxa using the SILVA v132 bacterial taxonomy database (Quast et al. 2013). By checking the taxonomy assignment results, OTUs identified as chloroplasts, mitochondria, or archaea were removed from the dataset.
In order to examine the sequencing effectiveness, rarefaction curves were made with observed unique amplicon sequencing variants and Shannon index for each sample in R package BiodiversityR (Kindt and Kindt 2019). Three indicators of microbiome diversity and richness, Shannon index, inverse Simpson index, and Chao1 index (Chao 1984; Magurran 1988; Simpson 1949), were calculated, and the difference was tested between hosts for a given diet using phyloseq (McMurdie and Holmes 2013) and ggplot2 (Ginestet 2011). The standard deviation of each indicator was shown. For indicators that did not follow the normal distribution after a Kolmogorov-Smirnov test (p < 0.05), the non-parametric Kruskal-Wallis test was conducted in the R package ggpubr (Kassambara 2020). Based on relative abundance, the Mann-Whitney test was applied to test the abundance difference of core phyla between groups in SPSS version 21 (Cor 2012).
Hierarchical clustering heatmap was built with the Ward clustering algorithm based on Bray-Curtis distance for the top 100 genera. To measure inter-sample diversity, beta diversity was analyzed between samples based on the Bray-Curtis distance using Microbiome (Ernst et al. 2020) and phyloseq (McMurdie and Holmes 2013). Statistical significance of host and diet differences was assessed through analysis of similarities (ANOSIM) with R package vegan (Oksanen et al. 2013). In order to visualize the gut microbiota profile between hosts and between diets, principal coordinates analysis (PCoA) was performed using Microbiome Analyst (Chong et al. 2020; Dhariwal et al. 2017).
The core microbiota shared by most samples were identified using Microbiome (Ernst et al. 2020). The core microbiota was dominant OTUs at over 1% relative abundance level and shared among at least 25% of the samples. Venn diagrams were plotted using VennDiagram to visualize the amount of OTUs shared by groups and associated with host and diet (Shade and Handelsman 2012).
Microbiota taxa significantly associated with host and diet was also tested using Linear discriminant analysis Effect Size (LEfSe) (Segata et al. 2011) within Microbiome Analyst (Chong et al. 2020; Dhariwal et al. 2017), which applies both the factorial Kruskal-Wallis test and linear discriminant analysis (LDA) to estimate effect size (Thorsen et al. 2016). Alpha value for the test among classes was set as 0.05, and threshold on the logarithmic LDA score for discriminative features was set as 2.0, which can be interpreted as the degree of consistent difference in relative abundance between features in the two classes of analyzed microbial communities (Segata et al. 2011).
The molecular functions of each OTU were predicted by identifying gene copy numbers using PICRUSt (Langille et al. 2013). Metabolism and functions of the avian gut microbiome were annotated based on the KEGG database, which was used to inform the biological system and comprehend the microbiome functions (Kanehisa et al. 2014). Molecular functions were predicted and summarized into KEGG pathway functions and 41 level II pathway categories. Based on KEGG pathway functions, the alpha diversity and its significance (Mann-Whitney test) between groups were calculated by using phyloseq (McMurdie and Holmes 2013) and ggplot2 (Ginestet 2011). To observe the difference between groups, hierarchical clustering heatmap was made with the Ward clustering algorithm for level II pathway categories and statistical significance was also assessed through ANOSIM analysis by using R package vegan (Oksanen et al. 2013).
Results
Gut bacterial communities and relative abundance
The total abundance was 1,913,151 across all 48 samples occupying all OTUs, with 39,857 reads per sample on average. A total of 7063 OTUs were identified and assigned, belonging to 25 phyla, 49 classes, 74 orders, 175 families, and 462 genera. Great Bustards had 22 phyla, 40 classes, 66 orders, 143 families, and 329 genera, and Common Cranes contained 22 phyla, 47 classes, 71 orders, 168 families, and 372 genera. The rarefaction curves uncovered that the sequencing depth of each sample reached saturation, meaning the microbiome communities were well represented (Supplemental Fig. S1).
Venn diagram showed that 27.60% OTUs were shared by CC_P and CC_W, followed by 23.86% OTUs between GB_P and CC_P, 23.37% OTUs in GB_W and CC_W, and 21.57% OTUs in GB_P and GB_W (Fig. 2a). Based on the flower diagram, the percentages of unique OTUs belonging to GB_P, GB_W, CC_P, and CC_W were 11.27% (796), 7.28% (514), 21.21% (1498), and 16.52% (1167), respectively, with an average percent of 14.07% (Fig. 2b). All groups shared 626 OTUs (8.87%), including the genera Akkermansia, Lactobacillus, Planococcus, Pseudomonas, Rhizobium, Alistipes, Subdoligranulum, Barnesiella, Gemmiger, and Coprococcus. Based on the threshold of prevalence (25%) and detection (0.1%), the 61 most frequent core OTUs were identified and shared among four groups, including the five phyla Firmicutes (49.15% OTUs), Proteobacteria (20.34% OTUs), Verrucomicrobia (10.17% OTUs), Bacteroidetes (6.78% OTUs), and Actinobacteria (5.08% OTUs).
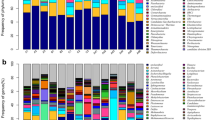
a The Venn diagram of the OTUs in all samples among four groups. b The unique OTUs belonging to each group. c The boxplot of phylum composition based on relative abundance among four groups. CC_P, Common Crane with peanut diet; CC_W, Common Crane with wheat diet; GB_P, Great Bustard with peanut diet; GB_W, Great Bustard with wheat diet
At the phylum level, 20, 19, 20, and 20 phyla were identified in GB_P, GB_W, CC_P, CC_W, respectively. Firmicutes, Proteobacteria, Verrucomicrobia, and Actinobacteria were the dominant phyla in all groups, occupying 84.74–97.37% of the overall bacterial community (Fig. 2c). Furthermore, the three enriched phyla were Firmicutes, Proteobacteria, and Verrucomicrobia in Great Bustards, while in Common Cranes, they were Firmicutes, Proteobacteria, and Actinobacteria. No significant difference was found in Firmicutes to Bacteroidetes ratio between diets for any species; however, Great Bustard had a significantly lower Firmicutes to Bacteroidetes ratio (7.60) than Common Cranes (82.11) (N = 48, Z = −5.938, p = 0.000) (Table 1). The relative abundance of Verrucomicrobia differed both between hosts and between diets, and it was higher in Great Bustard (26.97%) than that in Common Crane (2.00%) (N = 48, Z = −5.918, p=0.000), and higher in peanut (21.11%) than that in wheat (7.86%) (N = 48, Z = −2.103, p=0.035).
At the genus level, 462 genera were found across 48 samples, and genus composition was dissimilar among four groups. The abundant genus Akkermansia was significantly decreased in GB_W (13.82%) compared to that in GB_P (39.65%) (N = 24, Z = −3.406, p=0.001). In contrast, the dominant genus Lactobacillus was not significantly higher in CC_W (40.69%) than that in CC_P (19.54%) (N = 24, Z = −1.386, p = 0.166). GB_P and GB_W had 304 and 242 genera, only 24 (7.89%) and 28 (11.57%) genera with a relative abundance larger than 0.005; however, CC_P and CC_W harbored 315 and 287 genera, with 18 (5.7%) and 15 (5.2%) genera having a relative abundance larger than 0.005. The top 50 genera with the highest abundance uncovered a large various microbial composition in each group, ranging from 75.81 to 85.74%, and common genera shared by all groups were Lactobacillus, Akkermansia, and Catellicoccus.
Effects of host and diet on microbiota diversity
Alpha diversity revealed that the microbiota diversity varied between different diets even for the sympatric species (Fig. 3a, Table 2). In Great Bustards, higher inverse Simpson was yielded in wheat diet (30.78 ± 11.86) compared to that in peanut diet (9.18 ± 7.18) (p = 0.0001), and Shannon index showed the consistency (wheat: 4.26 ± 0.30, peanut: 3.15 ± 0.81, p = 0.0001), but no significant difference was observed for Chao1 index. However, it was almost the opposite in Common Cranes, with wheat diet being significantly lower than peanut diet in inverse Simpson (CC_W: 3.24 ± 1.48, CC_P, 6.72 ± 3.41, p = 0.0001), Shannon (CC_W: 1.98 ± 0.38, CC_P, 3.04 ± 0.40, p = 0.0001) and Chao1 index (CC_W: 794.55 ± 227.08, CC_P, 1324.11 ± 142.08, p = 0.0001). For a given wheat diet, Great Bustards had higher Shannon (4.26 ± 0.30) than Common Cranes (1.98 ± 0.38) (p = 0.0001), and inverse Simpson index showed a similar trend (GB_W: 30.78 ± 11.86, CC_W, 3.24 ± 1.48, p = 0.0001), but no difference was observed for Chao1 index. On the other hand, no difference was found in the inverse Simpson index between Great Bustards and Common Cranes for peanut diet, but the Chao1 index was significantly lower in GB_P (848.97 ± 174.85) compared to that in CC_P (1324.11 ± 142.08) (p = 0.0001).
a The boxplot of three alpha niches about four groups based on OTUs. b The boxplot of alpha diversity based on KEGG pathway functions. The asterisk (*) on the top means the significance between groups (****p = 0.0001, ***p = 0.001, **p = 0.01, *p = 0.05, NS. indicates no significance, Mann-Whitney test). CC_P, Common Crane with peanut diet; CC_W, Common Crane with wheat diet; GB_P, Great Bustard with peanut diet; GB_W, Great Bustard with wheat diet
The ANOSIM analysis showed significant clustering of samples according to both host and diet, with a relatively higher difference between hosts for any diet than between diets for any host (Table 3). In agreement, PCoA based on Bray-Curtis distance revealed that different bacterial composition was characterized by host and diet (Fig. 4). PCoA1 and PCoA2 explained 29.8% and 13.8% of the total variation. Furthermore, the heatmap (based on Bray-Curtis distance) within the top 100 genera supported the gut microbial community was separated between host groups, and then divided by different diets (Supplemental Fig. S2).
Specific gut bacterial taxa related to host and diet were identified based on LEfSe analysis. Great Bustards were significantly associated with 4 phylum-level taxa (Verrucomicrobia, Firmicutes, Bacteroidetes, and Synergistetes) and 8 class-level taxa (Clostridia, Verrucomicrobiae, Bacteroidia, Betaproteobacteria, Synergistia, Negativicutes, Erysipelotrichia, and Deltaproteobacteria), while 2 phyla (Proteobacteria and Actinobacteria) and 5 classes (Bacilli, Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, and Epsilonproteobacteria) were characterized significantly associated with Common Cranes. Bacteria in 3 phyla (Verrucomicrobia, Actinobacteria, and Synergistetes) and 7 classes (Bacilli, Verrucomicrobiae, Alphaproteobacteria, Actinobacteria, Betaproteobacteria, Synergistia, and Deltaproteobacteria) were significantly related to peanut diet, while wheat diet was significantly linked to 3 phyla (Proteobacteria, Firmicutes, and Bacteroidetes) and 6 classes (Clostridia, Gammaproteobacteria, Bacteroidia, Epsilonproteobacteria, Negativicutes, and Erysipelotrichia) (Supplemental Fig. S3).
Effects of host and diet on gut microbiome functions
A total of 6909 KEGG pathway functions were identified based on the KEGG database, ranging from 6262 in CC_P to 5197 in GB_W. Of these KEGG pathways, 2469 (39.43%) were shared in all groups. At level II KEGG functional category, the dominant functional categories identified included membrane transport (13.61%), carbohydrate metabolism (10.49%), and amino acid metabolism (9.86%).
Significantly different abundances of KEGG functional pathways were observed in gut metagenomes of Great Bustards and Common Cranes (p = 0.001). For the same species, diet has a significant effect on the abundance of KEGG functional pathways (p = 0.029) (Table 4). By comparing the functional diversity between hosts or between diets, neither inverse Simpson nor Shannon index showed a significant difference between diets, revealing no functional diversity change due to diet shift (Fig. 3b). In contrast, a significant difference in functional diversity was found between hosts in terms of the same diet (Fig. 3b). The heatmap of the 41 level II KEGG pathway categories showed that the metabolic pathways were different among groups (Fig. 5). In particular, compared with the GB_P group, the GB_W group was more significantly enriched in the digestive system, and the CC_P was significantly abundant in cardiovascular diseases, cell communication, cellular processes, and signaling, digestive system, sensory system, and xenobiotics biodegradation and metabolism (Fig. 5; Supplemental Table S1). Such functional pathway separation by host and diet was also supported by ANOSIM analysis, revealing that 80.49% level II pathway categories were significantly different between Great Bustards and Common Cranes for peanut diet, and 92.68% for wheat diet, and only 63.41% level II pathway categories were significantly different between peanut and wheat diets (Supplemental Table S1). Specifically, CC_P and CC_W differed significantly in amino acid metabolism (p = 0.008), and energy metabolism (p = 0.002), but not between GB_P and GB_W (Supplemental Table S1). In contrast, GB_P was significantly different from GB_W with regard to the signaling molecules and interaction (p = 0.002) and lipid metabolism (p = 0.001). The results indicated that specific KEGG metabolic pathways were related to both host and diet (Supplemental Table S1).
Discussion
Exploring the microbiota difference of sympatric host species would help discriminate the effects of environmental factors, such as diet or geographical location. In previous studies, some have focused on a single species in multiple habitats (Davidson et al. 2020a, b; Liu et al. 2020) or different species in varied environments (Risely et al. 2018). However, the similarity of foraging preferences could result in the microbiota similarity for species inhabiting in different environments, which means the environmental effects might be masked (Fu et al. 2020a; Mikaelyan et al. 2015). In this study, the microbiota profiles and functional pathways were explored in sympatric wintering Great Bustards and Common Cranes for the first time.
The results revealed that 8.87% OTUs were shared by sympatric wintering Great Bustards and Common Cranes (Fig. 2a, b), and this percentage was almost consistent with other microbiome studies in the avian species (Grond et al. 2017; Hird et al. 2015; Liu et al. 2020), supporting the existence of microbiota conservatism. At the phylum level, Firmicutes, Proteobacteria, Verrucomicrobia, and Actinobacteria were the dominant phyla among all four groups, accounting for 84.74–97.37% of the overall bacterial community (Fig. 2c), which was in accordance with studies on other birds, including turkeys (Wilkinson et al. 2017), great tits (Davidson et al. 2020b), neotropical birds (Hird et al. 2015), and 74 bird species from Equatorial Guinea (Capunitan et al. 2020). The microbiota conservatism among birds was also supported in Grus monacha and Anser anser domesticus (Fu et al. 2020b), Anser cygnoides (Wu et al. 2018), Dumetella carolinensis (Lewis et al. 2016), and some neotropical birds (Hird et al. 2015). This might be a long-term evolutionary consequence of microbiota and host species, because these microbiota communities play an important role in digestion, energy intake, and other vital functions (Flint et al. 2008; Fujio-Vejar et al. 2017; He et al. 2019; Speirs et al. 2019).
Microbiota diversity was different within sympatric species between diets, as well as being different between hosts, suggesting that both host and diet took part in shaping microbiota diversity. The ANOSIM analysis indicated both host and diet significantly affected microbiota relative abundance, and the statistic R was higher for host compared to that for diet (Table 3), suggesting host is the dominant factor influencing microbiota communities, and diet promoted a further divergence based on the diet niche difference for the sympatric species. This finding was further supported by PCoA and heatmap of the gut microbial community of the top 100 genera (Fig. 4 and Supplemental Fig. S2). However, it did not show the same pattern in OTU diversity and richness. Compared to the significance between other groups (p = 0.0001), lower significance of inverse Simpson (p > 0.05) and Shannon index (p = 0.01) was found between hosts with peanut diet. In addition, the trends of the two indexes (inverse Simpson and Shannon index) between hosts are completely opposite (Great Bustard: wheat diet was significantly higher than peanut diet; Common Cranes: peanut diet was significantly higher than wheat diet, p = 0.0001). Chao1 index changes showed no consistency between hosts and between diets (only those of Common Cranes and peanut diet were significant, p = 0.0001) (Fig. 3a). These results revealed that changes in intestinal microbiota diversity and richness were diverse among hosts, implying that peanut and wheat diets have different effects on the intestinal microbiota of different hosts, and it may be caused by the various adaptations of different hosts to different diets.
Firmicutes to Bacteroidetes ratio was significantly lower for Great Bustards (7.60) than that for Common Cranes (82.11), suggesting a dissimilar adaption to the environment for Great Bustards and Common Cranes. Firmicutes to Bacteroidetes ratio contributed to absorb calories efficiently and defend intestinal pathogens (He et al. 2019), and the increased ratio was associated with gut dysbiosis (De Angelis et al. 2015). The species-specific adaptive strategy to the environment was also supported by the KEGG functional pathways, with Great Bustards having higher abundance of environmental adaptation pathway than Common Cranes (p < 0.01), but this situation was not found between diets (Supplemental Table S1) (Fig. 5). Moreover, the Verrucomicrobia phylum was enriched in Great Bustards (Supplemental Fig. S3a) and higher abundance of the Akkermansia genus (belonging to Verrucomicrobia) was observed in Great Bustards (N = 48, Z = −5.918, p = 0.000), which indicated Great Bustards had healthier guts than Common Cranes (Fujio-Vejar et al. 2017). The results suggested that Common Cranes might develop different defense abilities from Great Bustards, and diet cannot change host pathogen resistance. Additionally, Great Bustards had healthier bacterium and stronger environmental adaptation than Common Cranes. One reason for this may be the fact that Great Bustards had overwintered in farmland for centuries because of the destruction of grassland, while Common Cranes had not been overwintering in farmland for long (Palacín et al. 2012; Yan 1982).
The KEGG pathway categories heatmap, not strictly divided by hosts, was different from that based on the top 100 genera (Fig. 5, Supplemental Fig. S2, and Supplemental Fig. S4), indicating that functional communities were more stable than microbiota abundance and diversity. Such a disparity may have resulted from the fact that different OTUs perform similar or identical functions. In addition, functional differences were also related to diet. Peanuts have higher lipid content than wheat, while wheat is richer in carbohydrate than peanuts (Zhao 2020; Zhou et al. 2020). Functional pathway abundance, such as amino acid metabolism, metabolism of other amino acids, energy metabolism, and lipid metabolism, was significantly higher for peanut diet than that for wheat diet in Common Cranes; in contrast, it was significantly lower in glycan biosynthesis and metabolism. The ANOSIM analysis based on level II pathway categories suggested 53.66% level II pathway categories were significantly different between diets, but 75.61% between hosts (Supplemental Table S1), to some extent suggesting host might pose a stronger influence on bacterial functions than diet. However, it cannot be ignored that some pathways were not significant in specific diet or host (e.g., amino acid metabolism, infectious diseases, lipid metabolism), which suggested specific diet and host may have week impacts on specific functions, indicating researchers may need to carefully balance the importance and effect of diet and host on specific physiological function both in the wild and in the lab. For migratory birds, life history characteristics at different stages may pose selection pressures on the gut microbiota (Turjeman et al. 2020), and the intrinsic and extrinsic factors impacting gut microbiota need to be investigated in the future.
In conclusion, the effects of host and diet on gut microbiota abundance, diversity, and functional pathways were investigated in sympatric wintering Great Bustards and Common Cranes. The effect of host has been discriminated to some extent from diet, suggesting host is the dominant factor determining the microbiota communities, and diet is another driver to further cause gut microbiota difference. The microbiota function appeared to be more conserved than OTU community compositions, and different bacteria may achieve the same function in a similar way, while microbiota OTU diversity might not be necessarily associated with functional diversity. The effects of land use–type change on wildlife are still difficult to be measured because diet did not show a strong influence on gut microbiome function, though bacterial community alterations were significantly different. The sympatric overwintering birds, Great Bustards and Common Cranes, have similar migratory routes and feeding habits; however, the response of gut microbiome to diet change was different, and it can be inferred that species-specific defense ability to intestinal pathogens may be developed.
Data availability
The original sequencing data of gut microbiota can be found in NCBI, under accession number PRJNA698750.
Code availability
Not applicable.
References
Al-Asmakh M, Stukenborg J-B, Reda A, Anuar F, Strand M-L, Hedin L, Pettersson S, Söder O (2014) The gut microbiota and developmental programming of the testis in mice. PLoS One 9(8):e103809. https://doi.org/10.1371/journal.pone.0103809
AlHilli MM, Bae-Jump V (2020) Diet and gut microbiome interactions in gynecologic cancer. Gynecol Oncol 159(2):299–308. https://doi.org/10.1016/j.ygyno.2020.08.027
Alonso JC, Palacin C (2010) The world status and population trends of the Great Bustard. Chinese Birds 01(2):141–147. https://doi.org/10.5122/cbirds.2010.0007
Bermingham EN, Young W, Butowski CF, Moon CD, Maclean PH, Rosendale D, Cave NJ, Thomas DG (2018) The fecal microbiota in the domestic cat (Felis catus) is influenced by interactions between age and diet; a five year longitudinal study. Front Microbiol 9(1231). https://doi.org/10.3389/fmicb.2018.01231
Bolyen E, Rideout JR, Dillon MR, Bokulich N, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo JR, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang LJ, Kaehler BD, Bin Kang K, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, vander Hooft JJJ, Vargas F, Vazquez Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan YH, Wang MX, Warren J, Weber KC, CHD W, Willis AD, Xu ZZ, Zaneveld JR, Zhang YL, Zhu QY, Knight R, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8):852–857. https://doi.org/10.1038/s41587-019-0209-9
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6(8):1621–1624. https://doi.org/10.1038/ismej.2012.8
Capunitan DC, Johnson O, Terrill RS, Hird SM (2020) Evolutionary signal in the gut microbiomes of 74 bird species from Equatorial Guinea. Mol Ecol 29(4):829–847
Chang CW, Huang BH, Lin SM, Huang CL, Liao PC (2016) Changes of diet and dominant intestinal microbes in farmland frogs. BMC Microbiol 16(1):33. https://doi.org/10.1186/s12866-016-0660-4
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 29:265–270. https://doi.org/10.1080/02664760220136221
Chong J, Liu P, Zhou G, Xia J (2020) Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 15(3):799–821. https://doi.org/10.1038/s41596-019-0264-1
Collar NJ, Baral HS, Batbayar N, Bhardwaj GS, Brahma N, Burnside RJ, Choudhury AU, Combreau O, Dolman PM, Donald PF, Dutta S, Gadhavi D, Gore K, Goroshko OA, Hong C, Jathar GA, Jha RRS, Jhala YV, Koshkin MA, Lahkar BP, Liu G, Mahood SP, Morales MB, Narwade SS, Natsagdorj T, Nefedov AA, Silva JP, Thakuri JJ, Wang M, Zhang Y, Kessler AE (2017) Averting the extinction of bustards in Asia. Forktail (33):1–26
Cor I (2012) Released. IBM SPSS Statistics for Windows, version 21.0. In.: Armonk, NY: IBM Corp.
Davidson G, Wiley N, Cooke AC, Johnson CN, Fouhy F, Reichert MS, Hera I, Crane JM, Kulahci IG, Ross RP (2020a) Diet-induced changes to host gut microbiota are linked to foraging innovation in a wild bird. bioRxiv:827741. https://doi.org/10.1101/827741
Davidson GL, Wiley N, Cooke AC, Johnson CN, Fouhy F, Reichert MS, Hera I, Crane JMS, Kulahci IG, Ross RP, Stanton C, Quinn JL (2020b) Diet induces parallel changes to the gut microbiota and problem solving performance in a wild bird. Sci Rep 10(1):13. https://doi.org/10.1038/s41598-020-77256-y
De Angelis M, Francavilla R, Piccolo M, De Giacomo A, Gobbetti M (2015) Autism spectrum disorders and intestinal microbiota. Gut Microbes 6(3):207–213. https://doi.org/10.1080/19490976.2015.1035855
Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia JG (2017) MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45(W1):W180–WW88. https://doi.org/10.1093/nar/gkx295
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Ernst FGM, Shetty SA, Huang R, Braccia DJ, Bravo HC, Lahti LM (2020) MicrobiomeExperiment and the emerging R ecosystem for microbiome research. F1000Research 9:1459. https://doi.org/10.7490/f1000research.1118445.1
Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA (2008) Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6(2):121–131. https://doi.org/10.1038/nrmicro1817
Fu H, Zhang L, Fan C, Liu C, Li W, Cheng Q, Zhao X, Jia S, Zhang Y (2020a) Environment and host species identity shape gut microbiota diversity in sympatric herbivorous mammals. Microb Biotechnol. https://doi.org/10.1111/1751-7915.13687
Fu R, Xiang X, Dong Y, Cheng L, Zhou L (2020b) Comparing the intestinal bacterial communies of sympatric wintering Hooded Crane (Grus monacha) and Domestic Goose (Anser anser domesticus). Avian Res 11(1). https://doi.org/10.1186/s40657-020-00195-9
Fujio-Vejar S, Vasquez Y, Morales P, Magne F, Vera-Wolf P, Ugalde JA, Navarrete P, Gotteland M (2017) The gut microbiota of healthy Chilean subjects reveals a high abundance of the phylum Verrucomicrobia. Front Microbiol 8(1221). https://doi.org/10.3389/fmicb.2017.01221
Ginestet C (2011) ggplot2: elegant graphics for data analysis. Journal of the Royal Statistical Society: Series A (Statistics in Society) 174(1):245–246. https://doi.org/10.1111/j.1467-985X.2010.00676_9.x
Grond K, Lanctot RB, Jumpponen A, Sandercock BK (2017) Recruitment and establishment of the gut microbiome in arctic shorebirds. FEMS Microbiol Ecol 93(12):9. https://doi.org/10.1093/femsec/fix142
He S, Zhang Z, Sun H, Zhu Y, Cao X, Ye Y, Wang J, Cao Y (2019) Potential effects of rapeseed peptide Maillard reaction products on aging-related disorder attenuation and gut microbiota modulation in d-galactose induced aging mice. Food Funct 10(7):4291–4303. https://doi.org/10.1039/c9fo00791a
Hird SM, Sánchez C, Carstens BC, Brumfield RT (2015) Comparative gut microbiota of 59 neotropical bird species. Front Microbiol 6:1403. https://doi.org/10.3389/fmicb.2015.01403
Hu X, Liu G, Li Y, Wei Y, Lin S, Liu S, Zheng Y, Hu D (2018) High-throughput analysis reveals seasonal variation of the gut microbiota composition within forest musk deer (Moschus berezovskii). Front Microbiol 9(1674). https://doi.org/10.3389/fmicb.2018.01674
Jiang JS (2004) The status of resource and conservation of Great Bustard in China. Dissertation. Northeast Forestry University, Harbin, China.
Kanai Y, Ueta M, Germogenov N, Nagendran M, Mita N, Higuchi H (2002) Migration routes and important resting areas of Siberian cranes (Grus leucogeranus) between northeastern Siberia and China as revealed by satellite tracking. Biol Conserv 106(3):339–346. https://doi.org/10.1016/s0006-3207(01)00259-2
Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42(D1):D199–D205. https://doi.org/10.1093/nar/gkt1076
Kassambara A (2020) Package ‘ggpubr’ version 0.4.0.
Kessler A, Batbayar N, Natsagdorj T, Batsuur D, Smith A (2013) Satellite telemetry reveals long-distance migration in the Asian great bustard Otis tarda dybowskii. J Avian Biol 44(4):311–320. https://doi.org/10.1111/j.1600-048X.2013.00072.x
Kindt R, Kindt MR (2019) Package ‘BiodiversityR’. Package Community Ecol Suitability Analysis 2:11–12
Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821. https://doi.org/10.1038/nbt.2676
Lewis WB, Moore FR, Wang S (2016) Characterization of the gut microbiota of migratory passerines during stopover along the northern coast of the Gulf of Mexico. J Avian Biol 47(5):659–668. https://doi.org/10.1111/jav.00954
Li L, Hu R, Huang J, Bürgi M, Zhu Z, Zhong J, Lü Z (2020) A farmland biodiversity strategy is needed for China. Nature Ecology Evolution 4(6):772–774. https://doi.org/10.1038/s41559-020-1161-2
Li C, Zhou JY, Gong MH, Tian XH, Meng DR, Liu CG, Dang DP, Liu G (2021) Current distribution of Asian Great Bustard (Otis tarda dybowskii) in China. Chinese Journal of Ecology, 40(6):1793-1801. doi:10.13292/j.1000-4890.202106.009
Liu G, Shafer AB, Hu X, Li L, Ning Y, Gong M, Cui L, Li H, Hu D, Qi L (2018) Meta-barcoding insights into the spatial and temporal dietary patterns of the threatened Asian Great Bustard (Otis tarda dybowskii) with potential implications for diverging migratory strategies. Ecology Evolution 8(3):1736–1745. https://doi.org/10.1002/ece3.3791
Liu G, Meng DR, Gong MH, Li HX, Wen WY, Wang YH, Zhou JY (2020) Effects of sex and diet on gut microbiota of farmland-dependent wintering birds. Front Microbiol 11:11. https://doi.org/10.3389/fmicb.2020.587873
Luo Y, Zhang L, Li H, Smidt H, Wright ADG, Zhang K, Ding X, Zeng Q, Bai S, Wang J, Li J, Zheng P, Tian G, Cai J, Chen D (2017) Different types of dietary fibers trigger specific alterations in composition and predicted functions of colonic bacterial communities in BALB/c Mice. Front Microbiol 8(966). https://doi.org/10.3389/fmicb.2017.00966
Magurran AE. (1988) Ecological diversity and its measurement. Princeton university press.
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217. https://doi.org/10.1371/journal.pone.0061217
McWhorter TJ, Caviedes-Vidal E, Karasov WH (2009) The integration of digestion and osmoregulation in the avian gut. Biol Rev 84(4):533–565. https://doi.org/10.1111/j.1469-185X.2009.00086.x
Mi CR, Huettmann F, Guo YM (2014) Obtaining the best possible predictions of habitat selection for wintering Great Bustards in Cangzhou, Hebei Province with rapid machine learning analysis. Chin Sci Bull 59(32):4323–4331. https://doi.org/10.1007/s11434-014-0445-9
Mi C, Falk H, Guo Y (2016) Climate envelope predictions indicate an enlarged suitable wintering distribution for Great Bustards (Otis tarda dybowskii) in China for the 21st century. PeerJ 4:e1630. https://doi.org/10.7717/peerj.1630
Mikaelyan A, Dietrich C, Köhler T, Poulsen M, Sillam-Dussès D, Brune A (2015) Diet is the primary determinant of bacterial community structure in the guts of higher termites. Mol Ecol 24(20):5284–5295. https://doi.org/10.1111/mec.13376
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Package ‘vegan’. Community Ecology Package Version 2(9):1–295
Perofsky AC, Lewis RJ, Meyers LA (2019) Terrestriality and bacterial transfer: a comparative study of gut microbiomes in sympatric Malagasy mammals. ISME J 13(1):50–63. https://doi.org/10.1038/s41396-018-0251-5
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(D1):D590–DD96. https://doi.org/10.1093/nar/gks1219
Risely A, Waite DW, Ujvari B, Hoye BJ, Klaassen M (2018) Active migration is associated with specific and consistent changes to gut microbiota in Calidris shorebirds. J Anim Ecol 87(2):428–437. https://doi.org/10.1111/1365-2656.12784
San Juan PA, Hendershot JN, Daily GC, Fukami T (2020) Land-use change has host-specific influences on avian gut microbiomes. The ISME Journal 14(1):318–321. https://doi.org/10.1038/s41396-019-0535-4
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12(6):R60. https://doi.org/10.1186/gb-2011-12-6-r60
Shade A, Handelsman J (2012) Beyond the Venn diagram: the hunt for a core microbiome. Environ Microbiol 14(1):4–12. https://doi.org/10.1111/j.1462-2920.2011.02585.x
Simpson EH (1949) Measurement of diversity. Nature 163(4148):688–688. https://doi.org/10.1038/163688a0
Song SJ, Sanders JG, Delsuc F, Metcalf J, Amato K, Taylor MW, Mazel F, Lutz HL, Winker K, Graves GR, Humphrey G, Gilbert JA, Hackett SJ, White KP, Skeen HR, Kurtis SM, Withrow J, Braile T, Miller M, McCracken KG, Maley JM, Ezenwa VO, Williams A, Blanton JM, McKenzie VJ, Knight R (2020) Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. Mbio 11(1):14. https://doi.org/10.1128/mBio.02901-19
Speirs LBM, Rice DTF, Petrovski S, Seviour RJ (2019) The phylogeny, biodiversity, and ecology of the Chloroflexi in activated sludge. Front Microbiol 10:28. https://doi.org/10.3389/fmicb.2019.02015
Thorsen J, Brejnrod A, Mortensen M, Rasmussen MA, Stokholm J, Al-Soud WA, Sørensen S, Bisgaard H, Waage J (2016) Large-scale benchmarking reveals false discoveries and count transformation sensitivity in 16S rRNA gene amplicon data analysis methods used in microbiome studies. Microbiome 4(1):62. https://doi.org/10.1186/s40168-016-0208-8
Tian XH, Zhang BL (2006) Progress in Researches of Great Bustard (Otis tarda) in China and its Conservation Strategy. Chinese Wildlife (03):32–37
Turjeman S, Corl A, Wolfenden A, Tsalyuk M, Lublin A, Choi O, Kamath PL, Getz WM, Bowie RCK, Nathan R (2020) Migration, pathogens and the avian microbiome: a comparative study in sympatric migrants and residents. Mol Ecol 29(23):4706–4720. https://doi.org/10.1111/mec.15660
Wang W, Zheng S, Li L, Yang Y, Liu Y, Wang A, Sharshov K, Li Y (2019) Comparative metagenomics of the gut microbiota in wild greylag geese (Anser anser) and ruddy shelducks (Tadorna ferruginea). MicrobiologyOpen 8(5):e00725. https://doi.org/10.1002/mbo3.725
Wilkinson TJ, Cowan AA, Vallin HE, Onime LA, Oyama LB, Cameron SJ, Gonot C, Moorby JM, Waddams K, Theobald VJ, Leemans D, Bowra S, Nixey C, Huws SA (2017) Characterization of the microbiome along the gastrointestinal tract of growing turkeys. Front Microbiol 8:1–11. https://doi.org/10.3389/fmicb.2017.01089
Wu Y, Yang Y, Cao L, Yin H, Xu M, Wang Z, Liu Y, Wang X, Deng Y (2018) Habitat environments impacted the gut microbiome of long-distance migratory swan geese but central species conserved. Sci Rep 8(1):13314. https://doi.org/10.1038/s41598-018-31731-9
Xiang XJ, Zhang F, Fu R, Yan SF, Zhou LZ (2019) Significant differences in bacterial and potentially pathogenic communities between sympatric hooded crane and greater white-fronted goose. Front Microbiol:10. https://doi.org/10.3389/fmicb.2019.00163
Xue JJ, Ajuwon KM, Fang RJ (2020) Mechanistic insight into the gut microbiome and its interaction with host immunity and inflammation. Animal Nutrition 6(4):421–428. https://doi.org/10.1016/j.aninu.2020.05.007
Youhui W, Hong W (2003) Distribution and number of common crane in China. Sichuan Journal of Zoology 22(1):35–38
Yu GH, Zou CI, Sun XW, Yang BB, Zhang XJ (2008) Wintering population of Otis tarda near Dagang area and the ecological observation. Jilin Forestry Sci Technol 4
Zhan YJ, Chen W, Hu D, Wu XS, Zhang JG (2007) Food selection of wintering common crane (Grus grus) in the Wetland of Beijing. Wetland Science 5(1):45–50
Zhao ZH (2020) High oleic acid peanut oil supplementation attenuates metabolic syndrome and its mechanisms. Dissertation. Chinese Academy of Agricultural Sciences.
Zhou WX, Li YH, Chen Y, Liu FM, Zhao L, Hu ZY, Wang K (2020) Comparsion of nutrition and processing characteristics of whole wheat flours with different particle sizes. Modern Food Sci Technol 36(9):172–180. https://doi.org/10.13982/j.mfst.1673-9078.2020.9.0151
Acknowledgements
We are particularly grateful to Lijuan Cui, Xiaoming Kang, and Chunyi Li for their help during the study. Great thanks to Mr. Lian Yu, Mr. Keming Song, and Mr. Yinguo Xiao for helping with sample collection in the field.
Funding
This study was funded with Fundamental Research Fund of Chinese Academy of Forestry (Grant No. CAFYBB2018QB010), and the Fundamental Research Funds for the Central Non-profit Research Institution of CAF (Grant No. CAFYBB2018GB001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
CL analyzed the data, prepared figures and tables, authored or reviewed drafts of the paper, and approved the final draft. YL, MHG, CMZ, and CLZ collected samples and provided basic data about the Great Bustard wintering sites and analyzed the data. HXL, WYW, and YHW authored or reviewed drafts of the paper. GL conceived and designed the experiments, analyzed the data, prepared figures and tables, authored or reviewed drafts of the paper, and approved the final draft.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 901 kb)
Rights and permissions
About this article
Cite this article
Li, C., Liu, Y., Gong, M. et al. Diet-induced microbiome shifts of sympatric overwintering birds. Appl Microbiol Biotechnol 105, 5993–6005 (2021). https://doi.org/10.1007/s00253-021-11448-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11448-y