Abstract
The herbicide butachlor has been used in huge quantities worldwide, affecting various environmental systems. Butachlor residues have been detected in soil, water, and organisms, and have been shown to be toxic to these non-target organisms. This paper briefly summarizes the toxic effects of butachlor on aquatic and terrestrial animals, including humans, and proposes the necessity of its removal from the environment. Due to long-term exposure, some animals, plants, and microorganisms have developed resistance toward butachlor, indicating that the toxicity of this herbicide can be reduced. Furthermore, we can consider removing butachlor residues from the environment by using such butachlor-resistant organisms. In particular, microbial degradation methods have attracted much attention, with about 30 kinds of butachlor-degrading microorganisms have been found, such as Fusarium solani, Novosphingobium chloroacetimidivorans, Chaetomium globosum, Pseudomonas putida, Sphingomonas chloroacetimidivorans, and Rhodococcus sp. The metabolites and degradation pathways of butachlor have been investigated. In addition, enzymes associated with butachlor degradation have been identified, including CndC1 (ferredoxin), Red1 (reductase), FdX1 (ferredoxin), FdX2 (ferredoxin), Dbo (debutoxylase), and catechol 1,2 dioxygenase. However, few reviews have focused on the microbial degradation and molecular mechanisms of butachlor. This review explores the biochemical pathways and molecular mechanisms of butachlor biodegradation in depth in order to provide new ideas for repairing butachlor-contaminated environments.
Key points
• Biodegradation is a powerful tool for the removal of butachlor.
• Dechlorination plays a key role in the degradation of butachlor.
• Possible biochemical pathways of butachlor in the environment are described.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Butachlor (N-(butoxymethyl)-2-chloro-N-2′,6′-dimethyl acetanilide) is a kind of herbicide which is mainly used to kill grass weeds and many broad-leaved weeds. It is a chloroacetamide herbicide that inhibits early plant development by inhibiting the biosynthesis of very-long-chain fatty acids (VLCFAs) in microsomes (Matthes and Böger 2002; Trenkamp et al. 2004). As a selective and systematic herbicide, it can inhibit the enzyme and inhibit the extension of the VLCFAs (Abigail et al. 2015). It has been widely used in Asia, South America, and Africa to inhibit the synthesis of protein lipids and lignin in weeds, as well as to control some broad-leaved weeds and a range of annual grasses. In agricultural production, it is commonly used as an early herbicide before or after the emergence of wheat, vegetables, peanut, barley, beet, cotton, rapeseed, and other crops (Dwivedi et al. 2010). The half-life of butachlor in non-sterile soil is 2.67–29.79 days, differing in different soil environments (Guo et al. 2010; Kaur et al. 2017; Mohanty and Jena 2019; Kaur and Goyal 2020).
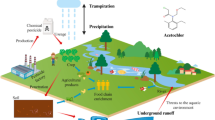
Butachlor is highly effective and low toxic, so it is widely used all over the world. However, the residual butachlor may negatively harm non-target organisms in the environment due to its extensive use (Jolodara et al. 2021; Toan et al. 2013). After extensive scientific tests, the presence of butachlor has been detected in river silt, plants, and animals all over the world (Chau et al. 2015; Kaur et al. 2018; Deknock et al. 2019; Gautam et al. 2020). It is suspected to be a carcinogen, neurotoxin, genetic toxin, which can persist in environments, thus having toxic effects on living systems (Anbumani and Mohankumar 2014; Shuman et al. 2017; Griffith et al. 2018). Butachlor not only affects lipid biosynthesis, but also has adverse effects on other metabolic processes and redox homeostasis (Agrawal et al. 2014). The bioavailability of insecticides is determined by their properties, the properties of the soil, and the absorption routes of earthworms. The dissipation of butachlor in the natural environment can lead to both abiotic and biological effects (Fig. 1) (Abigail et al. 2015; Mukherjee et al. 2018; Torabi et al. 2020). In order to deal with the risk of butachlor from the outside environment, non-target organisms may possess butachlor biotransformation mechanisms, as shown in Fig. 2 (Ademola et al. 1993; Kitamoto et al. 2011; Ou and Lin 1992). When catalyzed by cytochrome P450 and arylamidase from human skin or rat liver microsomes, butachlor was converted into 4-amino 3,5-diethylphenol (Coleman et al. 2001). Another explanation for the detoxication of butachlor is that it is finally converted into mercapturate in the liver under the catalysis of coenzyme A (coA) and other transferases (Ademola et al. 1993; Ou and Lin 1992).
To reduce butachlor pollution in the environment, previous studies have confirmed the existing physical and chemical methods that can effectively degrade butachlor, such as nanophotocatalysis, anodic Fenton treatment, and filtration and batch adsorption (Friedman et al. 2006; Hosseini and Toosi 2019; Mahmoodi et al. 2007). However, biodegradation and in situ bioremediation have attracted the attention of researchers and are more environmentally friendly methods (Bhatt et al. 2021; Birolli et al. 2018; Cycoń et al. 2017; Pang et al. 2020). Microorganisms can perform soil bioremediation more quickly, effectively, and safely, minimizing the adverse health and environmental effects of butachlor pesticides (Kaur and Goyal 2020; Singh and Kadapakkam 2018; Zhang et al. 2011). At present, there are many bacteria known to degrade butachlor, but only three strains of fungi (Chakraborty and Bhattacharyya 1991; Li et al. 2019; Liu et al. 2012; Raut and Kulshrestha 2008; Singh and Kadapakkam 2018). Some studies have explored the microbial degradation pathways of butachlor, but the degradation mechanisms of butachlor still need further systematic study. Microorganisms and their enzymes have been proven to be effective for the biodegradation of butachlor. However, few reviews have focused on the microbial degradation and molecular mechanisms of butachlor. In this review, we provide insights into the microbial degradation of butachlor, considering microbial strains, metabolic pathways, and molecular mechanisms.
Toxicity of butachlor
Due to its long and widespread use, butachlor is always detected in soil, plants, and surface water. Its residues continually damage subsequent crops, especially crops in sandy soils with low organic matter. Butachlor not only affects lipid biosynthesis, but also has adverse effects on other metabolic processes and redox homeostasis (Agrawal et al. 2014). Butachlor has been reported to be toxic to algae in aquatic systems and to have an adverse effect on nitrogen fixation (Singh and Singh 2013; Chen et al. 2020). In fact, low doses of butachlor may disrupt the growth of cyanobacteria in paddy fields and be beneficial for paddy fields with neutral to alkaline soil pH (Kumari et al. 2012). However, the excessive use of butachlor in agriculture has led to non-target hazards and environmental pollution.
Butachlor residues are commonly detected in water, so aquatic toxicity is usually the first concern and research. Butachlor is a persistent pollutant in water, having negative effects on different aquatic species, such as catfish and goldfish (Farombi et al. 2008; Karami et al. 2016). Butachlor significantly reduced the sperm quality of male rainbow trout (Oncorhynchus mykiss) and resulted in endocrine disruption (Ahmadivand et al. 2015; Ahmadivand et al. 2016). Water is the source of life, and the pollutants in water will be ingested by aquatic organisms and continuously enriched in the food chain.
Butachlor is not only harmful to aquatic life, but also highly toxic to land animals. Cane toads exposed to butachlor show a stunted growth and thyroid dysfunctions, suggesting that butachlor is harmful to amphibians (Shuman et al. 2020). Watery droppings, dullness, looseness of feathers, depression, and decreased chirping frequency were observed in male Japanese quails fed with butachlor. Significantly lower values of red blood cells, cell-specific volume percentage, and hemoglobin were recorded in male Japanese quails. These results suggest that butachlor has adverse effects on the blood and testicles of birds (Hussain et al. 2014). The toxicity of butachlor to animals makes us realize its harmfulness to human beings and reminds us to be more careful with pesticide residues.
In fact, butachlor can cause harm to our bodies, either directly or indirectly. The herbicide butachlor has been classified as having a strong neurotoxicity and genotoxicity, which poses a potential health threat to ecosystems and humans (Kim et al. 2011; Dwivedi et al. 2012; Seok et al. 2012; Takahashi et al. 2020). When exposed to butachlor, reactive oxygen species (ROS) are produced in cells, leading to mitochondrial dysfunction, oxidative DNA damage, chromosome breakage, and so on, which eventually lead to massive necrosis of peripheral blood mononuclear (PBMN) cells (Dwivedi et al. 2012). Furthermore, pollutants are bad for aquatic organisms and human health throughout the food chain (Gao et al. 2015). Therefore, the potential damage caused by residual butachlor cannot be ignored from the perspective of animals, plants, and human beings, which indicates the importance of removing it.
Degradation of butachlor
Pesticide degradation means that the structure of a pesticide becomes simpler in the environment, thus reducing or eliminating its toxic effects. The half-life of butchalor is about 640 days in sterile soil or 11.4 days in non-sterile soil, indicating that degradation is the main route of its dissipation (Zheng et al. 2012). Butachlor residues in soil or wastewater are mainly degraded by microorganisms, where the degradation rate depends on the type of pesticide and the moisture content, redox state, and microbial species of the soil. Inoculation with microbial target herbicides is an effective way to achieve the rapid degradation of herbicides in soil. Bacteria, fungi, algae, actinomycetes, and yeasts have been reported to be effective for the biodegradation of organic pollutants (Bhatti et al. 2017; Kaur and Goyal 2020; Kitamoto et al. 2011; Raut and Kulshrestha 2008). However, only bacteria and fungi have been reported as being capable of degrading butachlor and there must still be other kinds of butachlor-degrading microorganisms waiting to be discovered, as indicated in Table 1.
Butachlor degradation potential of microorganisms
The degradation of acetanilide herbicides in soil is mainly mediated by microorganisms. In order to efficiently screen for microorganisms useful for butachlor degradation, scientists often take samples in targeted contaminant enrichment sites, such as sewage plants, pharmaceutical plants, and fields (Chu et al. 2016; Lin et al. 2020).
Fusarium solani and Chaetomium globosum have been isolated from soil by an enrichment technique, which metabolized butachlor into 6 and 4 metabolites, respectively, in inorganic salt solution (Raut and Kulshrestha 2008). As the first biodegradable chloroacetamide herbicide reported at the enzyme level, Rhodococcus sp. strain B1 is able to degrade 100 mg L−1 butachlor in 5 days (Liu et al. 2012). The alkyl side chains of acetochlor and butachlor can be completely degraded by Rhodococcus sp. T3-1 at 100 mg L−1 within 6 days (Hou et al. 2014). Pseudomonas is ubiquitous in nature and plays an important role in metabolic activities, such as the degradation of exogenous pollutants, element cycling, and biogenesis. Pseudomonas putida strain ER1 is able to grow in MSM with butachlor acting as the only carbon and nitrogen source. At pH 7.0, the initial concentration of 50 mg L−1 butachlor was degraded by more than 80% within 3 days (Wang et al. 2007). In addition, many bacteria showed degradation activity to butachlor, such as Novosphingobium chloroacetimidivorans sp. nov. BUT-14T, Sphingomonas chloroacetimidivorans sp. nov., and Paracoccus sp. FLY-8 (Chen et al. 2014; Chen et al. 2015; Zhang et al. 2011). Interestingly, cyanobacterium Nostoc muscorum was found to have the potential to degrade 5 mg L−1 of butachlor for the first time (Anees et al. 2014). More highly efficient biodegradable bacteria may yet be found in the environment.
In general, mixed degradation colonies can improve the degradation efficiency of butachlor, due to the combination of their unique degradation potentials (Bhatt et al. 2021). Mycobacterium sp. J7A and Sphingobium sp. J7B worked together to degrade butachlor completely. Mycobacterium sp. J7A can take advantage of the alkyl side chain of butachlor as its carbon source and energy source to accumulate 2-chloro-N-(2,6-diethylphenyl) acetamide (CDEPA) and grow slightly on it. Then, Sphingobium sp. J7B can utilize the CDEPA to completely degrade butachlor (Kim et al. 2013). Natural butachlor catabolism bacterial Enterobacter cloacae strains FP1, FP2, and FP4 were isolated from soil contaminated by pesticide preparation unit wastewater in India, where the FP2 strain showed the highest degradation efficiency. This microbial strain can use butachlor as its only carbon and energy source in mineral salts medium (MSM), showing a biodegradation efficiency of 1.67 mg·(L h)−1 of butachlor (Mohanty and Jena 2018b). The combination of different microorganisms in the community may be a favorable opportunity to completely degrade butachlor into benign end-products. Therefore, it is necessary to examine the internal connections of the microorganism and test its joint degradation ability.
Environmental factors have a great influence on the growth of microorganisms. Therefore, various process parameters are optimized to maximize the utilization of microorganisms and thus improve the biodegradation efficiency (Bhatt et al. 2020). At optimum conditions of 30 °C and pH 7.0, Paracoccus sp. Y3B-1 degraded 65.5% of butachlor in 3 days (Ni et al. 2011). At the lowest cell density, with environment of 37 °C and pH 7, the Bacillus altitudinis strain A16 isolated from coal-tar contaminated soil degraded 90% of 50 mg L−1 butachlor, showing excellent butachlor removal ability. Through batch culture and soil inoculation, Bacillus altitudinis can be used for the bioremediation of butachlor in soil with low bioavailability (Kaur and Goyal 2020). At an initial pH of 6–9 and temperature 15–35 °C, the new strain Catellibacterium caeni sp. nov DCA-1T degraded 81.2% of 50 mg L−1 butachlor in 84 h (Zheng et al. 2012). The maximum degradation of butachlor by the Serratia Ureilytica strain AS1 was 2.08 mg·(L h)−1 under the optimal conditions of 32.5 °C culture temperature, pH 7.5, and 10% (V/V) inoculation (Mohanty and Jena 2018a). In general, the response surface methodology (RSM) is used to determine the optimal culture conditions for pollutant biodegradation.
Butachlor biodegradation and its resource utilization
The vegetation type also has a great influence on the biodegradation of herbicides in soil. Compared to soil without rhizosphere, the biodegradation rate of butachlor was significantly higher in the rhizosphere soil of Phragmites australis and Acorus calamus (Yang et al. 2013). In wheat rhizomes, especially those inoculated with a bacterial community capable of degrading butachlor, the degradation of butachlor was greatly enhanced (Yu et al. 2003). Interestingly, Stenotrophomonas acidaminiphila JS-1 not only can grow effectively on mineral salt medium with a concentration range of 0.32–9.6 mmol L−1 of butachlor, but also has the unique auxiliary properties of plant growth stimulation (Dwivedi et al. 2010). Combined plant–microbe remediation is a very worthwhile line to study, providing benefits to both plant growth and pesticide degradation.
Interestingly, Si has been reported to reduce the toxicity of butachlor to rice seedlings. Butachlor has toxic effects by inducing and increasing the accumulation of free proline, while Si reverses this process and plays a detoxification role (Tripthi et al. 2020). Therefore, the application of Si fertilizer in a paddy field can reduce the harmful impact of butachlor herbicide pollution, which is an important direction in the removal of butachlor poison.
At present, microbial electrochemical technologies, such as microbial fuel cells (MFCs), provide an inexhaustible supply of solid anodes as electron receptors, which are able to efficiently and rapidly degrade environmental pollutants in anoxic environments while generating electricity from electrically active microorganisms (Bhatt et al. 2019). In addition, MFCs with butachlor as the only accompanying carbon source have power generation capacity. In MFCs, Paracoccus, Geobacter, and Thauera butanivorans can proliferate with butachlor and sodium acetate as accompanying carbon sources. These three species can oxidize different substituents of butachlor, thus having important prospects in the bioremediation of organic compounds (Li et al. 2019). In constructed single-chamber MFCs, with Paracoccus and Geobacter microbes using butachlor as the sole carbon source and Thauera butanivorans using butachlor as the accompanying carbon source, the butachlor was efficiently degraded by 90% within 1 day (Li et al. 2019). If harmful substances can be converted into useful substances, this will be an effective way to remove pollution from the environment.
Pseudomonas sp. But2 can form a good biofilm and use butachlor as the only source of carbon, nitrogen, and energy and can completely degrade 50 mg L−1 of butachlor within 30 h (Duc et al. 2020). The Acinetobacter baumannii strain, DT, can efficiently utilize butachlor and propanil as the only carbon and nitrogen sources in a biofilm-batch reactor (Duc et al. 2020; Oanh et al. 2020). Research has shown that the “Gene Drives” method can persistently promote the degradation of pollutants by local microorganisms with minimal disruption to the ecosystem, which is a long-term and effective genetic technology (French et al. 2020). Therefore, it can be inferred that the biofilm reactor may also be an interesting direction for butachlor bioremediation.
Molecular mechanisms of butachlor biodegradation
Bacterial degradation mechanisms
The process of bacterial degradation of butachlor has been observed to occur by hydrolysis, including two pathways: deacylation and dealkylation (Fig. 3). In the first pathway, butachlor was first induced by deacylation and loss of acetyl group to form N-(butoxymethyl)-N-(2-chloroethyl)-2,6-diethylaniline. Further, the continuous catalytic formation of N-(butoxymethyl)-2,6-diethyl-N-propylaniline and N-(butoxymethyl)-2-ethylaniline occurs, due to the hydrolysis of chlorine atoms and dealkylation of nitrogenous alkyl chains (Kaur and Goyal 2020; Singh and Kadapakkam 2018). In addition, it has been found that dechlorination may form (2,6-diethylphenyl) (ethoxymethyl) carbamic acid before deacylation (Zheng et al. 2012).
Dealkylation plays an important role in the process of bacterial degradation of butachlor. In the second pathway, different sequences of dealkylation at different locations lead to different degradation products and degradation directions. In this pathway, the most common products are 2-chloro-N-(2,6-diethylphenyl) acetamide and butoxymethanol, as the C–N bond of the branch chain of butachlor is unstable and most easily hydrolyzed (Kaur and Goyal 2020; Kim et al. 2013; Liu et al. 2012). Importantly, several functional enzymes have been isolated and shown to act in the N-dealkylation of butachlor; namely, ChlH, CndA, CndB1, and CndC1 (Chen et al. 2014; Liu et al. 2012). In contrast, the enzyme Dbo was isolated and demonstrated to be capable of degrading the C–O bond of butachlor (Gao et al. 2015). Therefore, butachlor can be continuously produced to 2-chloro-N-(2,6-diethylphenyl)-N-methylacetamide, 2-chloro-N-(2,6-diethylphenyl)acetamide, and 2,6-diethlaniline by Dbo (Gao et al. 2015). In addition, when dealkylation occurs in the benzene ring of butachlor, it can generate N-(butoxymethyl)-2-chloro-N-(2-ethylphenyl)acetamide, followed by N-(butoxymethyl)-2-ethylaniline (Kaur and Goyal 2020). In the degradation pathway of Catellibacterium caeni sp. nov DCA-1T, the novel product N-(2,6-diethylphenyl)-N-(hydroxymethyl) formamide was detected, which could be converted to 2,6-dilaniline in a short time (Zheng et al. 2012). In the end, all of the above intermediates can be degraded into aniline, catechol, and muconic acid respectively, which will be converted into water and carbon dioxide eventually. Among the above steps, two oxygenases extracted from Paracoccus sp. fly-8 played important catalytic roles, aniline dioxygenase and catechol 1,2-dioxygenase (Zhang et al. 2011). Many butachlor degradation bacteria and their degradation products have been studied in great depth, but many more degrading enzymes and genes deserve to be isolated and identified.
Fungal degradation mechanisms
At present, only 3 strains of fungi are known to degrade butachlor; however, the degradation products of fungi have been studied in detail in the literature. In this paper, the previous knowledge is summarized into the following degradation pathways, as shown in Fig. 4 (Chakraborty and Bhattacharyya 1991; Raut and Kulshrestha 2008).
The processes of fungal degradation of butachlor mainly involve de-chlorination, hydroxylation, dehydrogenation, debutoxymethylation, dealkylation, and cyclization. The products of de-chlorination and dealkylation are basically the same as the bacterial degradation pathway 2, all of which include the products, 2-chloro-N-(2,6-diethylphenyl) acetamide and 2,6-diethylaniline. However, 2-chloro-N-(2,6-diethylphenyl) acetamide is broken down by bacteria to 2,6-diethlaniline, but a dual ring compound is produced in fungi. Another difference was that the compound 2,6-diethylaniline was further degraded by oxidative decomposition in bacteria, while 1-ethyl-3-methylbenzene was formed in fungus, which was not further degraded. Compared with the bacterial degradation process, the products of butachlor degradation by fungi are characterized by cyclization. Multiple rings are produced in various products in fungi, which does not seem to make the products more easily degradable. Many related products have been found in the fungal degradation processes of butachlor, but there is no mineralization process (Chakraborty and Bhattacharyya 1991; Raut and Kulshrestha 2008). In addition, more butachlor degrading-related fungal enzymes and genes remain to be discovered.
Enzymes and encoding genes related to butachlor biodegradation
The reason why microorganisms can be widely used in pesticide degradation treatment is that strong catalytic biochemical enzymes are produced by microorganisms (Mishra et al. 2020). When pesticides are mixed with microorganisms, the microorganisms can rapidly increase the number of enzymes they contain or form enzymes with degradation effects due to rapid genetic adaptations (Satish et al. 2017). Under the action of enzymes, pesticides can be degraded into simple inorganic compounds. In addition, the risk of environmental threat can be reduced if microbial enzymes are used to treat pesticide residues, rather than directly using microbial strains.
A powerful tool for studying the reactions of cells to toxins is proteomic analysis, which has led to the discovery of new domestication mechanisms. As a key separation technique in proteome analysis, two-dimensional gel electrophoresis has the advantages of simultaneously separating and revealing thousands of proteins at a time. Therefore, two-dimensional gel electrophoresis has become a common determination method (Wang et al. 2007). The enzymes of butachlor degrading bacteria and the resistant proteins of animals are summarized in Table 2, and their evolutionary relationship is shown in Fig. 5.
A phylogenetic tree of the key functional enzymes involved in the biodegradation of butachlor. The code after the name of enzymes is NCBI accession number. CndC1 was isolated from Sphingomonads DC-6 (Chen et al. 2014). Red1 was isolated from Sphingomonads DC-2 (Chen et al. 2014). FdX1 was isolated from Sphingomonads DC-2 (Chen et al. 2014). FdX2 was isolated from Sphingomonads DC-2 (Chen et al. 2014). CndB1 was isolated from Sphingomonads DC-6 (Chen et al. 2014). CndB2 was isolated from Sphingomonads DC-6 (Chen et al. 2014). Dbo was isolated from Bacillus sp. strain hys-1 (Gao et al. 2015)
In three species of Anabaena sp. PCC 7120, Anabaena Doliolum, and Anabaena L31.75, a cluster of high-abundance proteins was found to be related to high resistance to butachlor; these are atpA, groEL, OR, AGTase, Alr0803, Alr0806, Alr3090, Alr3199, All4050, and All405 (Agrawal et al. 2014). Senior researchers have identified the 744 bp debutoxylase, named Dbo, which shows the highest activity to degrade butachlor. Dbo had the highest activity of butachlor degradation at a pH of 6.5 and temperature of 30 °C, where metal ions play an important role in Dbo activity (Gao et al. 2015). The hydrolase ChlH can hydrolyze the dealkylation of the side chain of butachlor. The best working conditions of ChlH are pH 7.0–7.5 and 30 °C, which has been shown to catalyze the dealkylation of other chloroacetamide herbicides (Liu et al. 2012). AKR17A1 not only can metabolize butachlor, but can also withstand various stresses and use NADPH as a cofactor of enzyme activity (Agrawal et al. 2015). Aniline dioxygenase and catechol 1,2-dioxygenase play important roles in the degradation of butachlor, being catalytic enzymes of aniline (Zhang et al. 2011).
The Sphingomonads DC-6 and DC-2 have been developed in depth. Reductase and Ferredoxin components separated from Sphingomonads DC-6 and DC-2 played an important role in the degradation of butachlor. For example, CndB1 and CndC1 showed high N-dealkylase activities in this process (Chen et al. 2014). Otherwise, genes encoding degrading enzymes have been identified in Sphingomonads DC-6 and DC-2, which are cndC1, red1, fdx1, and fdx2 (Chen et al. 2014). Most of the butachlor-degrading enzymes and genes remain to be isolated and identified, and the enzymes that have been discovered need to be used more efficiently.
Conclusions and future prospects
Butachlor plays an extremely important role in agricultural production, and its wide application inevitably causes environmental impacts. Recently, the impact of butachlor residues on non-target organisms in the environment has aroused concern about environmental and human safety. Compared with the more expensive physical and chemical methods, such as advanced oxidation, scientists have been actively trying to use resistant organisms to remove or convert butachlor. Animals, plants, and microorganisms that are resistant to butachlor have been identified. However, at present, the most well-understood method is microbial degradation, while combined plant–microbial repair methods are well worth further exploration. In addition, the microorganisms used for butachlor research need to be further developed and modified, through the use of technologies including genetic engineering, metagenomics, proteomics, and immobilization techniques. The development and utilization of degrading enzymes is also expected to be an important factor in promoting the biodegradation of butachlor. The recent development of high-throughput next-generation sequencing technologies could be helpful to explore the routes of butachlor degradation in the future.
References
Abigail MEA, Samuel SM, Ramalingam C (2015) Addressing the environmental impacts of butachlor and the available remediation strategies: a systematic review. Int J Environ Sci Technol 12:4025–4036
Ademola JI, Wester RC, Maibach HI (1993) Absorption and metabolism of 2-chloro-2,6-diethyl-N-(butoxymethyl)acetanilide (Butachlor) inhuman skin in vitro. Toxicol Appl Pharmacol 121:78–86
Agrawal C, Sen S, Yadav S, Rai S, Rai LC (2015) A novel aldo-keto reductase (AKR17A1) of Anabaena sp. PCC 7120 degrades the rice field herbicide butachlor and confers tolerance to abiotic stresses in E. coli. PLoS One 10:e0137744
Agrawal C, Sen S, Singh S, Rai S, Singh PK, Singh VK, Rai LC (2014) Comparative proteomics reveals association of early accumulated proteins in conferring butachlor tolerance in three N2-fixing Anabaena spp. J Proteome 96:271–290
Ahmadivand S, Farahmand H, Mirvaghefi A, Eagderi S, Zargar A (2015) Effects of (Anti) androgenic endocrine disruptors (DEHP and Butachlor) on immunoglobulin M (IgM) and leukocytes counts of male rainbow trout (Oncorhynchus mykiss). Bull Environ Contam Toxicol 94:695–700
Ahmadivand S, Farahmand H, Teimoori-Toolabi L, Mirvaghefi A, Eagderi S, Geerinckx T, Shokrpoor S, Rahmati-Holasoo H (2016) Boule gene expression underpins the meiotic arrest in spermatogenesis in male rainbow trout (Oncorhynchus mykiss) exposed to DEHP and butachlor. Gen Comp Endocrinol 225:235–241
Anbumani S, Mohankumar MN (2014) Cytogenotoxicity assessment of monocrotophos and butachlor at single and combined chronic exposures in the fish Catla catla (Hamilton). Environ Sci Pollut Res 22:4964–4976
Anees S, Suhail S, Pathak N, Zeeshan M (2014) Potential use of rice field cyanobacterium Nostoc muscorum in the evaluation of butachlor induced toxicity and their degradation. Bioinformation 10:365–370
Bhatt P, Gangola S, Bhandari G, Zhang W, Maithani D, Mishra S, Chen S (2021) New insights into the degradation of synthetic pollutants in contaminated environments. Chemosphere 259:128827
Bhatt P, Rene ER, Kumar AG, Zhang W, Chen S (2020) Binding interaction of allethrin with esterase: bioremediation potential and mechanism. Bioresour Technol 315:123845
Bhatti AA, Haq S, Bhat RA (2017) Actinomycetes benefaction role in soil and plant health. Microb Pathog 111:458–467
Birolli WG, Vacondio B, Alvarenga N, Seleghim MHR, Porto ALM (2018) Enantioselective biodegradation of the pyrethroid (+/-)-lambda-cyhalothrin by marine-derived fungi. Chemosphere 197:651–660
Chakraborty SK, Bhattacharyya A (1991) Degradation of butachlor by two soil fungi. Chemosphere 23:99–105
Chau ND, Sebesvari Z, Amelung W, Renaud FG (2015) Pesticide pollution of multiple drinking water sources in the Mekong Delta, Vietnam: evidence from two provinces. Environ Sci Pollut Res Int 22:9042–9058
Chen C, Zou WB, Cui GL, Tian JC, Wang YC, Ma LM (2020) Ecological risk assessment of current-use pesticides in an aquatic system of Shanghai, China. Chemosphere 257:127222
Chen K, Chen Q, Wang GX, Ni HY, He J, Yan X, Gu JG, Li SP (2015) Sphingomonas chloroacetimidivorans sp. nov., a chloroacetamide herbicide-degrading bacterium isolated from activated sludge. Antonie van Leeuwenhoek 108:703–710
Chen Q, Wang CH, Deng SK, Wu YD, Li Y, Yao L, Jiang JD, Yan X, He J, Li SP (2014) Novel three-component Rieske non-heme iron oxygenase system catalyzing the N-dealkylation of chloroacetanilide herbicides in Sphingomonads DC-6 and DC-2. Appl Environ Microbiol 80:5078–5085
Chu CW, Chen Q, Wang CH, Wang H, Sun ZG, He Q, He J, Gu JG (2016) Roseomonas chloroacetimidivoranssp. nov., a chloroacetamide herbicide-degrading bacterium isolated from activated sludge. Antonie Van Leeuwenhoek 109:611–618
Coleman S, Linderman R, Hodgson E, Rose HL (2001) Comparative metabolism of chloroacetamide herbicides and selected metabolites in human and rat liver microsomes. Environ Health Perspect 108:1151–1157
Cycoń M, Mrozik A, Piotrowska-Seget Z (2017) Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: a review. Chemosphere 172:52–71
Deknock A, De Troyer N, Houbraken M, Dominguez-Granda L, Nolivos I, Van Echelpoel W, Forio MAE, Spanoghe P, Goethals P (2019) Distribution of agricultural pesticides in the freshwater environment of the Guayas river basin (Ecuador). Sci Total Environ 646:996–1008
Duc HD, Thuy NTD, Truc HTT, Nhu NTH, Oanh NT (2020) Degradation of butachlor and propanil by Pseudomonas sp. strain But2 and Acinetobacter baumannii strain DT. FEMS Microbiol Lett 367:18
Dwivedi S, Singh BR, Al-Khedhairy AA, Alarifi S, Musarrat J (2010) Isolation and characterization of butachlor-catabolizing bacterial strain Stenotrophomonas acidaminiphila JS-1 from soil and assessment of its biodegradation potential. Lett Appl Microbiol 51:54–60
Dwivedi S, Saquib Q, Al-Khedhairy AA, Musarrat J (2012) Butachlor induced dissipation of mitochondrial membrane potential, oxidative DNA damage and necrosis in human peripheral blood mononuclear cells. Toxicology 302:77–87
Farombi EO, Ajimoko YR, Adelowo OA (2008) Effect of butachlor on antioxidant enzyme status and lipid peroxidation in fresh water African catfish, (Clarias gariepinus). Int J Environ Res Public Health 5:423–427
French KE, Zhou ZR, Terry N (2020) Horizontal ‘gene drives’ harness indigenous bacteria for bioremediation. Sci Rep 10:15091–15102
Friedman CL, Lemley AT, Hay A (2006) Degradation of chloroacetanilide herbicides by Anodic Fenton. J Agric Food Chem 54:2640–2651
Gao Y, Jin L, Shi H, Chu Z (2015) Characterization of a novel butachlor biodegradation pathway and cloning of the debutoxylase (Dbo) gene responsible for debutoxylation of butachlor in Bacillus sp. hys-1. J Agric Food Chem 63:8381–8390
Gautam S, Sood NK, Gupta K, Joshi C, Gill KK, Kaur R, Chauhan I (2020) Bioaccumulation of pesticide contaminants in tissue matrices of dogs suffering from malignant canine mammary tumors in Punjab, India. Heliyon 6:e05274
Griffith CM, Morgan MA, Dinges MM, Mathon C, Larive CK (2018) Metabolic profiling of chloroacetanilide herbicides in earthworm coelomic fluid using 1H NMR and GC–MS. J Proteome Res 17:2611–2622
Guo HR, Yin LC, Zhang SC, Feng WR (2010) The toxic mechanism of high lethality of herbicide butachlor in marine flatfish flounder, Paralichthys olivaceus. J Ocean U China 9:257–264
Hosseini N, Toosi MR (2019) Removal of 2,4-D, glyphosate, trifluralin, and butachlor herbicides from water by polysulfone membranes mixed by graphene oxide/TiO2 nanocomposite: study of filtration and batch adsorption. J Environ Health Sci Eng 17:247–258
Hou Y, Dong W, Wang F, Li J, Shen W, Li Y, Cui Z (2014) Degradation of acetochlor by a bacterial consortium of Rhodococcus sp. T3-1, Delftia sp.T3-6 and Sphingobium sp. MEA3-1. Lett Appl Microbiol 59:35–42
Hussain R, Khan A, Mahmood F, Rehan S, Ali F (2014) Clinico-hematological and tissue changes induced by butachlor in male Japanese quail (Coturnix japonica). Pestic Biochem Physiol 109:58–63
Jolodara NR, Karimi S, Bouteh E, Balist J, Prosser R (2021) Human health and ecological risk assessment of pesticides from rice production in the Babol Roud River in Northern Iran. Sci Total Environ 772:144729
Karami A, Karbalaei S, Zad BF, Ismail A, Simpson SL, Courtenay SC (2016) Alterations in juvenile diploid and triploid African catfish skin gelatin yield and amino acid composition: effects of chlorpyrifos and butachlor exposures. Environ Pollut 215:170–177
Kaur G, Dogra N, Singh S (2018) Health risk assessment of occupationally pesticide-exposed population of cancer prone area of Punjab. Toxicol Sci 165:157–169
Kaur P, Randhawa SK, Duhan A, Bhullar MS (2017) Influence of long term application of butachlor on its dissipation and harvest residues in soil and rice. B Environ Contam Toxicol 98:874–880
Kaur R, Goyal D (2020) Biodegradation of butachlor by Bacillus altitudinis and identification of metabolites. Curr Microbiol 77:2602–2612
Kim H, Min J, Park J, Lee S, Lee J (2011) Erythema multiforme major due to occupational exposure to the herbicides alachlor and butachlor. Emerg Med Australas 23:103–105
Kim NH, Kim DU, Kim I, Ka JO (2013) Syntrophic biodegradation of butachlor by Mycobacterium sp. J7A and Sphingobium sp. J7B isolated from rice paddy soil. FEMS Microbiol Lett 344:114–120
Kitamoto HK, Shinozaki Y, Cao XH, Morita T, Konishi M, Tago K, Kajiwara H (2011) Phyllosphere yeasts rapidly break down biodegradable plastics. AMB Express 1:44
Kumari N, Narayan OP, Rai LC (2012) Cyanobacterial diversity shifts induced by butachlor in selected Indian rice fields in Eastern Uttar Pradesh and Western Bihar analyzed with PCR and DGGE. J Microbiol Biotechnol 2:1–12
Li XJ, Li Y, Zhao LX, Sun Y, Zhang XL, Chen XD, Weng LP, Li YT (2019) Efficient removal of butachlor and change in microbial community structure in single-chamber microbial fuel cells. Int J Environ Res Public Health 16:3897
Lin Z, PangS ZhangW, MishraS BhattP, Chen S (2020) Degradation of acephate and its intermediate methamidophos: mechanisms and biochemical pathways. Front Microbiol 11:2045
Liu H, Cao L, Lu P, Ni H, Li YX, Yan X, Hong Q, Li SP (2012) Biodegradation of butachlor by Rhodococcus sp. strain B1 and purification of its hydrolase (ChlH) responsible for N-dealkylation of chloroacetamide herbicides. J Agric Food Chem 60:12238–12244
Mahmoodi NM, Arami M, Limaee NY, Gharanjig K, Nourmohammadian F (2007) Nanophotocatalysis using immobilized titanium dioxide nanoparticle. Mater Res Bull 42:797–806
Matthes B, Böger P (2002) Chloroacetamides affect the plasma membrane. Z Naturforsch C J Biosci 57:843–852
Mishra S, Zhang W, Lin Z, Pang S, Huang Y, Bhatt P, Chen S (2020) Carbofuran toxicity and its microbial degradation in contaminated environments. Chemosphere 259:127419
Mohanty SS, Jena HM (2018a) Evaluation of butachlor biodegradation efficacy of Serratia ureilytica strain AS1: a statistical optimization approach. Int J Environ Sci Technol 16:5807–5816
Mohanty SS, Jena HM (2018b) Process optimization of butachlor bioremediation by Enterobacter cloacae using Plackett Burman design and response surface methodology. Process Saf Environ 119:198–206
Mohanty SS, Jena HM (2019) A systemic assessment of the environmental impacts and remediation strategies for chloroacetanilide herbicides. J Water Process Eng 31:100860
Mukherjee I, Das SK, Kumar A (2018) Atmospheric CO2 level and temperature affect degradation of pretilachlor and butachlor in indian soil. B Environ Contam Toxicol 100:856–861
Ni YY, Zheng JW, Zhang J, Wang BZ, He J, Li SP (2011) Isolation of chloracetanilide herbicides-degrading Bacterium Y3B-1 and its degradability to chloracetanilide herbicides. Chin J Appl Environ Biol 17:711–716
Oanh NT, Duc HD, Ngoc DTH, Thuy NTD, Hiep NH, Van HN (2020) Biodegradation of propanil by Acinetobacter baumannii DT in a biofilm-batch reactor and effects of butachlor on the degradation process. FEMS Microbiol Lett 367:5
Ou YH, Lin JK (1992) Biotransformation of butachlor through mercapturic acid pathway in rat tissue homogenates. J Toxicol Environ Health 35:19–28
Pang S, Lin Z, Zhang W, Mishra S, Bhatt P, Chen S (2020) Insights into the microbial degradation and biochemical mechanisms of neonicotinoids. Front Microbiol 11:868
Raut AK, Kulshrestha G (2008) Biotransformation of butachlor by soil fungi. Toxicol Environ Chem 61:109–116
Satish GP, Ashokrao DM, Arun SK (2017) Microbial degradation of pesticide: a review. Afr J Microbiol Res 11:992–1012
Seok SJ, Choi SC, Gil HW, Yang JO, Lee EY, Song HY, Hong SY (2012) Acute oral poisoning due to chloracetanilide herbicides. J Korean Med Sci 27:111–114
Shuman GME, Singleton GR, Propper CR (2017) Competition and pesticide exposure affect development of invasive (Rhinella marina) and native (Fejervarya vittigera) rice paddy amphibian larvae. Ecotoxicology 26:1293–1304
Shuman GME, Singleton GR, Forsman AM, Hines S, Christodoulides N, Daniels KD, Propper CR (2020) Developmental assays using invasive cane toads, Rhinella marina, reveal safety concerns of a common formulation of the rice herbicide, butachlor. Environ Pollut 272:115955
Singh J, Kadapakkam NY (2018) Prospecting Ammoniphilus sp. JF isolated from agricultural fields for butachlor degradation. 3 Biotech 8:164
Singh S, Singh PP (2013) In-silico analysis suggests alterations in the function of XisA protein as a possible mechanism of butachlor toxicity in the nitrogen fixing cyanobacterium Anabaena sp. PCC 7120. Bioinformation 9:701–706
Takahashi G, Yokofuji H, Terayama M, Yoshinao D, Fujino Y, Inoue Y, Endo S (2020) Course of matrix metalloproteinase-1 and pulmonary oxygenation in acute respiratory distress syndrome caused by oral ingestion of large doses of oxadiazon/butachlor emulsion: a case report. Acute Med Surg 7:e552
Toan PV, Sebesvari Z, Bläsing M, Rosendahl I, Renaud FG (2013) Pesticide management and their residues in sediments and surface and drinking water in the Mekong Delta, Vietnam. Sci Total Environ 452-453:28–39
Torabi E, Wiegert C, Guyot B, Vuilleumier S, Imfeld G (2020) Dissipation of S-metolachlor and butachlor in agricultural soils and responses of bacterial communities: insights from compound-specific isotope and biomolecular analyses. J Environ Sci 92:163–175
Trenkamp S, Martin W, Tietjen K (2004) Specific and differential inhibition of very-long-chain fatty acid elongases from Arabidopsis thaliana by different herbicides. Proc Natl Acad Sci U S A 101:11903–11908
Tripthi DK, Varma RK, Singh S, Sachan M, Guerriero G, Kushwaha BK, Bhardwaj S, Ramawat S, Sharma S, Singh VP, Prasad SM, Devendra KC, DubeyNK SS (2020) Silicon tackles butachlor toxicity in rice seedlings by regulating anatomical characteristics, ascorbate-glutathione cycle, proline metabolism and levels of nutrients. Sci Rep 10:14078
Wang JH, Lu YT, Chen YY (2007) Comparative proteome analysis of butachlor-degrading bacteria. Environ Geol 53:1339–1344
Yang CM, Yulai W, Wang MM, Chen HY (2013) Plant species influence microbial metabolic activity and butachlor biodegradation in a riparian soil from Chongming Island, China. Geoderma 193-194:165–171
Yu YL, Chen YX, Luo YM, Pan XD, He YF, Wong MH (2003) Rapid degradation of butachlor in wheat rhizosphere soil. Chemosphere 50:771–774
Zhang J, Zheng JW, Liang B, Wang CH, Cai S, Ni YY, He J, Li SP (2011) Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro. J Agric Food Chem 59:4614–4621
Zheng JW, Li R, Zhu JC, Zhang J, He J, Li SP, Jiang JD (2012) Degradation of the chloroacetamide herbicide butachlor by Catellibacterium caeni sp. nov DCA-1T. Inter Biodeter Biodegr 73:16–22
Acknowledgments
This study was partially funded by grants from the Key-Area Research and Development Program of Guangdong Province (2018B020206001, 2020B0202090001), the Natural Science Foundation of Guangdong Province (2021A1515010889), the National Natural Science Foundation of China (31401763) and the Guangdong Special Branch Plan for Young Talent with Scientific and Technological Innovation (2017TQ04N026).
Author information
Authors and Affiliations
Contributions
SC conceived of the presented idea. ZL and SP contributed to the writing and prepared the figures and tables. ZZ, XW, PB, and SC participated in revising the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lin, Z., Pang, S., Zhou, Z. et al. Current insights into the microbial degradation for butachlor: strains, metabolic pathways, and molecular mechanisms. Appl Microbiol Biotechnol 105, 4369–4381 (2021). https://doi.org/10.1007/s00253-021-11346-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11346-3