Abstract
Ophiocordyceps sinensis, one of the well-known and precious fungal species in the world, parasitizes soil-dwelling larvae of ghost moths on the Tibetan Plateau. The genetic intractability of this extremely psychrophilic and slow-growing O. sinensis fungus is a major limitation on the molecular study. In this study, an Agrobacterium tumefaciens–mediated genetic transformation (ATMT) system for this fungus was established. ATMT procedure was optimized based on the fungal recipient, Agrobacterium strains, and different co-cultivation conditions. Blastospores were ideal recipients for this system. Acetosyringone (AS) was not essential for the transformation of O. sinensis. Sixty to 100 hygromycin B–resistant transformants per 1 × 106 blastospores were obtained. Southern blot analysis indicated the presence of a random single-copy integration of T-DNA into the O. sinensis genome. The insertional transformants with altered growth characters such as colony, blastospore, and fruiting body production were selected to identify the T-DNA flanking sequences by modified hiTAIL-PCR and FPNI-PCR techniques. Eight genes, including genes for an MFS transporter, a 2-oxoglutarate dehydrogenase, a DNA-directed RNA polymerase III complex subunit Rpc37, a cytochrome oxidase subunit I, a mitochondrial import inner membrane translocase subunit tim54, a cytidine deaminase, a phosphoribosylaminoimidazole carboxylase, and a histone H3-like centromeric protein cse-4 were identified. This ATMT system provides a useful tool for gene discovery and characterization of O. sinensis and contributes to the better understanding of the mysterious life cycle of O. sinensis and the molecular interaction between this fungus and its host insects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ophiocordyceps sinensis (Berk.) G. H. Sung et al. is one of the well-known and precious fungal species in the world. O. sinensis parasitizes soil-dwelling larvae of ghost moths on the Tibetan Plateau with an altitude between 3600 and 5000 m (Sung et al. 2007; Tao et al. 2016). The sclerotium and fruiting body after the parasitism (also called Chinese cordyceps, Zhang et al. 2012) have been used for medicinal treatment and health food since the fifteenth century and its value is demonstrated by modern pharmacological science (Holliday and Cleaver 2008; Hu et al. 2013; Wu et al. 2006; Zhang et al. 2006). The trade of Chinese cordyceps becomes a primary source of income for local herdsmen (Stone 2008). O. sinensis is extremely psychrophilic and slow-growing and needs 50 days for hyphal growth and more than 90 days for the induction of stromata in a rice-based medium (Cao et al. 2015). The Thitarodes/Hepialus host insects of this fungus need 1 to 30 years in laboratory and 3 to 5 years in nature, to complete a developmental life cycle (Tao et al. 2016). Due to the long growth period of O. sinensis, over-exploitation, habitat degradation, and climate warming, natural production of the Chinese cordyceps has greatly decreased (Hopping et al. 2018; Qiu et al. 2016). It was listed as an “endangered species for protection” by the Chinese government in 1999.
Mass cultivation of the Chinese cordyceps in commercial mode has been established (Li et al. 2016; Li et al. 2019a; Qiu et al. 2016). However, the degeneration of the fungus, high larval mortality by pathogens, and low infection and mummification rate constrained the efficient production of the Chinese cordyceps (Lu et al. 2015; Qin et al. 2018; Qiu et al. 2016; Zhou et al. 2014). In nature, the fungal reproductive cycle appears to be highly synchronized with the life cycle of its host insect (Zhang et al. 2014). However, the molecular basis of this life cycle, the induction of primordium, and sexual development remain unclear. Compared with other entomopathogenic fungi such as Beauveria bassiana and Metarhizium anisopliae causing the death of host larvae within a few days (Roberts and St Leger 2004; Thomas and Read 2007; Wang et al. 2004), this fungus takes 5 to more than 12 months to kill the host larvae, which provides a valuable system to study the interaction between the pathogenic fungi and the host insects (Liu et al. 2019). Although recent advances have provided some insights into the adaptation to highland environment, sexual reproduction and fungal pathogenicity of O. sinensis based on high-throughput sequencing (Li et al. 2019b; Meng et al. 2015; Rao et al. 2019; Xia et al. 2016; Xiang et al. 2014; Zhong et al. 2016), establishment of an efficient and stable genetic transformation system is prerequisite for characterizing the fungal gene functions.
Agrobacterium tumefaciens–mediated transformation (ATMT) is an efficient genetic transformation method for insertional mutagenesis and gene transfer to a wide variety of plant and fungal species (Bundock et al. 1995; de Groot et al. 1998; Ishida et al. 1996; Michielse et al. 2005; Mullins et al. 2001; Zhong et al. 2007). ATMT is also regarded as an efficient alternative to other DNA transfer methods for its flexibility and efficiency of the protocol, predominant single-copy insertion, and high homologous recombination frequencies (Jiang et al. 2013; Kozák et al. 2018; Michielse et al. 2008). T-DNA random insertion and integration are predominantly dependent on host factors, recipient, acetosyringone, pH of the co-cultivation medium, Agrobacterium strains, and cell densities (Bundock et al. 1995; Martínez-Cruz et al. 2017; Shao et al. 2015; Zheng et al. 2011). However, no report on tools for DNA transfer in the medicinal fungus O. sinensis is found. In this study, ATMT system of O. sinensis KD1223 was constructed, a T-DNA insertion mutant library was screened for mutants with altered growth phenotype, and the flanking sequences of T-DNA insertion sites of eight mutants were obtained by TAIL-PCR and FPNI-PCR. The establishment of this transformation system would provide an efficient tool for genetic studies of O. sinensis, especially on better understanding of fungal biology and interaction with host insects.
Materials and methods
Strain, culture media, and culture conditions
Three A. tumefaciens strains AGL-1, GV3101, and LBA4404 were kind gifts of Prof. Zhongkang Wang from Chongqing University, China, and used for the transformation on Luria-Bertani (LB) medium at 28 °C. Escherichia coli strain Trans1-T1 (TransGen Biotech, Beijing, China) was used as a host for the propagation of plasmid DNA and for the amplification of the binary vector on LB medium at 37 °C.
O. sinensis strain KD1223 was a subculture of strain KD11-2 (GDMCC 60594), which was isolated from fruiting bodies of O. sinensis in Sichuan, China, in May 2012, and maintained at − 80 °C in 40% glycerol in Guangdong Institute of Applied Biological Resources, Guangzhou, China. The stock culture was transferred to liquid PPDA medium (Liu et al. 2018) and incubated at 13 °C on a 100-rpm rotary shaker for seed culture. The resulting fungal cultures after 45 days of inoculation was harvested with three layers of sterile lens papers to remove hyphae and large particles. The filtered solutions were centrifuged at 5000 rpm for 10 min at 10 °C and the supernatant was discarded. Harvested conidia or blastospores (spindle cells) (Supplemental Fig. S1) were re-suspended in 5-mL sterile 1 × phosphate-buffered saline (PBS, pH 7.2) solution. Pure conidia were harvested from liquid PPDA medium where cut artificial fruiting bodies (about 1 cm3) were incubated for 14 days. Both conidia and blastospores were used as a recipient in this study. The numbers of conidia and blastospores were counted using a hemocytometer and a light microscope (Eclipse 80i; Nikon, Tokyo, Japan) at × 400 magnifications.
ATMT of O. sinensis
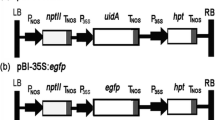
A binary T-DNA vector pEX4-hpt-eGFP carrying the optimized hygromycin B phosphotransferase gene (hpt) under the control of the Aspergillus nidulans gpdA promoter was used to construct the mutant library of O. sinensis. This vector, a kind gift of Prof. Zide Jiang from South China Agricultural University, China, was reconstructed based on pPZP200 by inserting the oligonucleotides LoxP1 and LoxP2 (Sun et al. 2014), and it was then introduced into A. tumefaciens AGL-1, GV3101, and LBA440 cells by the freeze-thaw method, respectively. ATMT of O. sinensis was carried out according to the method as previously described (Mullins et al. 2001; Zheng et al. 2011). To test hygromycin B sensitivity of O. sinensis, 107 blastospores were spread on PPDA plates supplemented with different hygromycin B concentrations (50, 100, 150, 200, 300, and 400 μg mL−1).
A. tumefaciens AGL-1, GV3101, and LBA4404 carrying pEX4-hpt-eGFP were grown respectively on LB supplemented with 100 μg mL−1 rifampicin and 100 μg mL−1 spectinomycin at 28 °C until OD600 value was 2.0. Then, induction liquid medium (IM) in the presence (IM + AS) and absence (IM-AS) of 200 μM acetosyringone (AS, Sigma-Aldrich, MO, USA) (Michielse et al. 2008; Zheng et al. 2011) was used to dilute above cultures until OD600 value to 0.15–0.2 and re-incubated at 28 °C until OD600 value equal to 0.6. One hundred microliters of pre-induced A. tumefaciens culture was mixed in a sterile Eppendorf tube with 100 μL blastospores suspension at three concentrations (1 × 106 blastospores mL−1, 1 × 107 blastospores mL−1 and 1 × 108 blastospores mL−1, respectively) and spread evenly onto cellophane sheets (Guangzhou Probe Instrument, Guangzhou, China) on induction medium plates at different pH values (5.0–8.0) in the presence (IM + AS) and absence (IM-AS) of 200 μM AS. Co-cultivation was performed at 16 °C and 22 °C in the dark for 48–96 h. Subsequently, the cellophane sheets were transferred onto PPDA plates supplemented with 300 μg mL−1 cefotaxime, 100 μg mL−1 ampicillin, and 300 μg mL−1 hygromycin B to select the transformants at 13 °C. Both IM and PPDA plates contained 2.5% (w/v) agar and were air-dried before use. All the experiments were carried out at least twice in triplicate.
The resulting O. sinensis transformants were cultured on PPDA at 13 °C for 60 days. The mycelia were transferred to a 250-mL flask containing 50 mL of liquid PPDA medium. The flasks were incubated at 13 °C on a 100-rpm shaker for 45 days. For the fruiting body production, the transformants cultured on PPDA at 13 °C for 60 days were transferred to 4 °C for 120 days to stimulate the development of stromata and fruiting body (Cao et al. 2015). The phenotypic characters of transformants including shape and color of the forming colonies and the fruiting body formation in PPDA, together with the formation of hyphae and blastospores in liquid PPDA medium, were observed at × 400 magnification using an optical microscope.
Genomic DNA extraction, PCR, and Southern blot analyses
Total DNA of the transformants was extracted as previously described (Liu et al. 2018). Mycelia of a single colony was inoculated into 3-mL liquid PPDA medium supplemented with 300 μg mL−1 hygromycin B and 300 μg mL−1 cefotaxime and cultured at 13 °C for 60 days. The resulting mycelia and spores were collected by centrifugation at 8000 rpm for 10 min at 10 °C and washed twice with sterile distilled water. The genomic DNA was extracted using DNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany). The primers used in this study were listed in Supplemental Table S1. Specific primers for promoter gpdA and the hpt gene of T-DNA (PgpdA-F and Hpt-R) were used for PCR characterization and Southern blot analysis. Specific primers for eGFP gene of T-DNA were also used for PCR characterization to detect the integration of eGFP gene into the genomic DNA of O. sinensis transformants. PCR parameters included an initial denaturing step of 5 min at 94 °C, followed by 35 cycles consisting each of 30 s at 94 °C, 30 s at 60 °C, and 45 s at 72 °C and a final elongation for 10 min at 72 °C.
For further Southern blot analysis of the transformants, the 977-bp fragment for promoter gpdA and the hpt gene of T-DNA amplified with the primers PgpdA-F and Hpt-R was digoxigenin (DIG)-labeled as a specific probe for signal detection. The genomic DNA samples (10 mg) were digested with EcoRI and separated by electrophoresis in 0.8% agarose gels, blotted to Hybond™-N+ membranes (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Standard procedures for probe labeling, hybridization, and immunological detection were used according to the manufacturer’s protocol (Roche Diagnostics GmbH, Mannheim, Germany). O. sinensis transformants were successively cultured on PPDA plates in the presence or absence of hygromycin B selection pressure. The T-DNA integrations were further confirmed by PCR and Southern blot analysis.
Identification of T-DNA flanking sequences by hiTAIL-PCR and FPNI-PCR
Both high-efficiency TAIL-PCR (hiTAIL-PCR) (Liu and Chen 2007), and fusion primer and nested integrated PCR (FPNI-PCR) (Wang et al. 2011) were employed to clone the T-DNA left- and right-border flanking sequences from the selected transformants (all the primers were listed in Supplemental Table S1). Four arbitrary degenerate primers (LAD1–4), AC1, and nested genomic walking specific primers (LB1–3, RB1–3) were used for hiTAIL-PCR. Nine arbitrary degenerate primers (FP1–9) in combination with nested FP-specific primers (FSP1 and FSP2) and nested genomic walking specific primers (FLB1–3, FRB1–3) were used for FPNI-PCR. The tertiary hiTAIL-PCR and FPNI-PCR products were cloned into pEASY-T1 vector (TransGen Biotech, Beijing, China) and sequenced by the Sangon Biotech. (Guangzhou, China). A diagnostic PCR fragment from T-DNA with primers in Supplemental Table S1 was used to verify the read-through of the LB border and RB border. T-DNA flanking sequences were used as queries to search for nucleotide similarities by the Blastn and Blastx algorithm at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). hiTAIL-PCR was also applied to obtain the upstream and downstream sequences of the T-DNA flanking sequences. Total RNA was extracted according to the instruction of the RNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany). The integrity of the total RNA was checked by agarose gel electrophoresis. Full-length genes were then verified by RT-PCR. The resulting homology was analyzed by multiple sequence alignment using the MEGA 5.1 software (Tamura et al. 2011). Classification of sequences was performed using SMART (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de) for the annotation of protein domains and the Gene Ontology resource (GO; http://geneontology.org) for structured, computable knowledge regarding the functions of genes and gene products.
Data analysis
All the data were expressed as means ± SE. One-way ANOVA analysis of variance was used and the significance between treatments was analyzed by Tukey’s honestly significant different test (SPSS17.0, SPSS Inc., Chicago, IL, USA) or t tests for experiments with two treatments (P < 0.05).
Results
Validation of O. sinensis transformants
O. sinensis was completely inhibited on PPDA medium supplemented with 150 μg mL−1 hygromycin B (Supplemental Fig. S2). A higher 300 μg mL−1 hygromycin B was used for the selection of resistant transformants. From the putative transformants, a 977-bp PCR product was amplified to confirm the presence of promoter gpdA and the hpt gene of T-DNA (Fig. 1a). Similarly, a 437-bp fragment amplified from the tested transformants demonstrated the integration of eGFP gene of T-DNA (Fig. 1b). For further confirmation of the T-DNA integration into O. sinensis and the copy numbers of the transformants, Southern blot analysis was performed. Genomic DNA of each transformant digested with EcoRI was probed with a 977-bp fragment for promoter gpdA and the hpt gene of T-DNA (Fig. 2a). The variety of hybridization patterns suggested that integration of T-DNA into the host genome occurred randomly and 83.33% of the transformants appeared to possess a single insertion of the PgpdA-hpt cassette (Fig. 2b). PCR and Southern blot analysis showed that T-DNA maintained under hygromycin B selective pressure while lost in almost 50% transformants in the third generations without the selection pressure.
Polymerase chain reaction characterization of hygromycin-resistant transformants of Ophiocordyceps sinensis. a Polymerase chain reaction characterization of the hpt gene integrated into the transformants (a 977-bp PgpdA-hpt fragment). b Polymerase chain reaction characterization of the eGFP gene integrated into the transformants (a 437-bp eGFP fragment). Lanes 1–15, gDNAs of O. sinensis randomly selected hygromycin-resistant transformants; WT, gDNA of wild type; CK, positive control with plasmid DNA of pEX4-hpt-eGFP. M, DNA molecular marker.
Southern blot analysis of Ophiocordyceps sinensis representative hygromycin-resistant transformants. a Probe with a 977-bp DIG-labeled PgpdA-hpt fragment used for the Southern blot analysis of transformant gDNAs. bEcoRI digested genomic DNA of 14 transformants. Lanes 1–14, gDNA of hygromycin-resistant transformants; CK, positive control with plasmid DNA of pEX4-hpt-eGFP. M, DIG-labeled marker.
Evaluation of factors influencing the transformation efficiency
Transformation of O. sinensis was successful when blastospores were used as the recipient, while it failed when conidia as the recipient. In several transformations, there was a watery layer of blastospores when co-cultivated with A. tumefaciens AGL-1, which resulted in the failure of the transformation. It seemed that the plates should be dry enough for co-cultivation. Co-cultivation of AGL-1 carrying pEX4-hpt-eGFP with blastospores for 72 h resulted in the appearance of putative hygromycin B–resistant transformants after approximately 55 days on the selective PPDA plates (Supplemental Fig. S3).
The ATMT procedure of O. sinensis was also optimized by the evaluation of several factors on transformation efficiency. The results showed that transformation efficiency of O. sinensis correlated strongly with A. tumefaciens strains, pH of the co-cultivation plate, and co-cultivation time and temperature (Fig. 3). AGL-1 and GV3101 showed the highest transformation efficiency whereas LBA4404 was the lowest (Fig. 3a). The numbers of putative hygromycin B–resistant transformants were significantly higher on co-cultivation medium at pH 6.0 and 6.5 than those at other pH values (Fig. 3b). The transformation efficiency increased with the prolongation of co-cultivation time to 4 days (Fig. 3c). Co-cultivation at 16 °C obtained more transformants than those at 22 °C (Fig. 3d). Therefore, co-cultivation of 107 blastospores with AGL-1 strain carrying pEX4-hpt-eGFP on PPDA medium with a pH value at 6.5 at 16 °C for 72 h was used in the following transformation.
The effect of AS on the transformation efficiency was also determined. After pre-cultivation of the AGL-1 strain carrying pEX4-hpt-eGFP in the presence or absence of 200 μM AS, 100 μL pre-induced culture equally mixed with 100 μl 1 × 107 blastospores was spread evenly onto CO-IM plates with or without 200 mM AS and cultivated for 72 h. The numbers of putative hygromycin B-resistant transformants after transfer onto selective PPDA plates for 55 days were counted. The results showed that the supplement of AS into the pre-cultivation medium and co-cultivation medium was not necessary for the transformation of O. sinensis. However, the numbers of transformants in IM+AS/CO-IM + AS, IM+AS/CO-IM-AS, and IM-AS/CO-IM+AS conditions were significantly higher than those in IM-AS/CO-IM-AS condition (F3,23 = 12.043, P < 0.001; Table 1). The effects of the supplement of AS during pre-cultivation and co-cultivation on the percentages of positive “transformants” were determined by PCR amplification using specific primers for promoter gpdA and hpt gene of T-DNA (PgpdA-F and Hpt-R). The percentages of positive transformants were 43.75%, 40.00%, and 41.25% in conditions IM+AS/CO-IM+AS, IM+AS/CO-IM-AS, and IM-AS/CO-IM+AS, respectively, while 37.5% in IM-AS/CO-IM-AS (Table 1).
Screening for mutant phenotypes
The transformants with different phenotypic characteristics on solid or liquid PPDA medium were selected when compared with the wild type (i) (Fig. 4), based on the colonies with fewer aerial hyphae (a); with more villous hyphae (b); with slower growth rate without conidial sporulation and no blastopore produced in PPDA medium 60 days after inoculation (c); with more aerial hyphae (d, f, h); and with a delayed hyphal development (e, g). After the induction to stimulate the development of fruiting bodies as previously described (Cao et al. 2015), fruiting bodies were grown only from the colony of the wild type (j).
Identification of the sequences flanking the inserted T-DNA in transformants
To identify the genomic DNA sequences flanking the inserted T-DNA, both hiTAIL-PCR and FPNI-PCR techniques were employed. Just one arbitrary primer LAD1 or the combination of arbitrary primer LAD1 and LAD2 could result in the successful amplification of desired products from the transformants. For FPNI-PCR, one arbitrary primer FP6 was suitable for the amplification of the transformants. The amplified PCR products ranged from 0.5 to 1.5 kb. Among 8 selected transformants, the promoter gpdA and Hyg B–resistance gene hpt of the T-DNA were intact. Alignment was used to compare 8 T-DNA flanking sequences with O. sinensis genomic sequences for T-DNA insertion site identification (Fig. 5). Truncation sequences ranging from 1 to 17 bp in size of the inserted T-DNA occurred on both LB and RB borders. In 2 of 8 selected transformants, a read-through of the LB border and a subsequent 626-bp fragment including the streptomycin resistance gene sequence were inserted into the O. sinensis genome. T-DNA insertion sites were gene coding subregions of the predicted genes for mutant phenotypes A, C, D, F, and G; intron subregions of the predicted gene for mutant phenotypes B and H; and a promoter subregion of the predicted gene for mutant phenotype E.
Blastn and Blastx analyses showed that the 8 flanking genomic sequences from the transformants A–H were homologous to known genes or their encoded proteins MFS transporter, 2-oxoglutarate dehydrogenase (E1), DNA-directed RNA polymerase III complex subunit Rpc37, cytochrome oxidase subunit I, mitochondrial import inner membrane translocase subunit tim54, phosphoribosylaminoimidazole carboxylase, cytidine deaminase, and histone H3-like centromeric protein cse-4, respectively, as found in the GenBank database (Table 2). SMART software analysis also indicated that the MFS transporter of O. sinensis is homologous to MFS_1 and contains a transmembrane region (residues 492–514), that the putative 2-oxoglutarate dehydrogenase (E1) is homologous to E1_dh and contains a C-terminal catalytic domain (residues 624–992) and an N-terminal peptide (residues 21–61), that the putative DNA-directed RNA polymerase III complex subunit Rpc37 has a typical structural feature of Sin_N, that the putative cytochrome oxidase subunit I has a typical structural feature of cox1, that the mitochondrial import inner membrane translocase subunit tim54 has a typical structural feature of tim54, that the phosphoribosylaminoimidazole carboxylase contains a C-terminal catalytic domain (residues 122–309) and an N-terminal AIRC domain (residues 449–598), that the cytidine deaminase contains a cytidine and deoxycytidylate deaminase zinc-binding region required for its catalytic activity (residues 221–342), and that histone H3-like centromeric protein cse-4 is homologous to a H3 protein that has typical structural features (residues 28–133).
Discussion
In this study, ATMT was first introduced to construct insertional mutants of O. sinensis, which is an extremely psychrophilic and slow-growing fungus. Due to the low germination rate of conidia, blastospores were demonstrated to be ideal recipients for the transformation of O. sinensis.
Factors influencing ATMT frequency include the fungal recipient, pH, Agrobacterium strain, and the co-cultivation conditions. Optimal conditions for each fungus or isolate to obtain the maximum transformants are required (Combier et al. 2003; Michielse et al. 2004; Mullins et al. 2001; Zheng et al. 2011). O. sinensis grows in a low and narrow temperature range, usually at 12 to 18 °C and the hyphae grow faster at 22 °C but stop to grow above 25 °C (Qiu et al. 2016; Wang et al. 2003). The pH values have significant effects on the growth of O. sinensis, and the normal growth could be obtained only at pH 6.0–7.0 (Supplemental Fig. S4). The co-cultivation time and different strains of A. tumefaciens are also important factors influencing the transformation of A. tumefaciens. Therefore, in the present study, O. sinensis was transformed with three Agrobacterium strains, AGL-1, GV3101, and LBA4404, and the co-cultivation of O. sinensis and A. tumefaciens was performed at 16 °C and 22 °C for different co-cultivation times. The result indicated that high and reproducible transformation efficiency of this fungus was obtained on media at pH 6.5 when co-cultivation was performed with AGL-1 at 16 °C for 72 h.
Acetosyringone (AS), a phenolic compound used to induce the expression of vir genes for the T-DNA transfer of A. tumefaciens, has been found to be essential for the ATMT of the entomopathogenic fungi B. bassiana (Leclerque et al. 2004) and Metarhizium (Nomuraea) rileyi (Shao et al. 2015). However, AS was not essential for the successful transformation of O. sinensis. Transformants with hygromycin B resistance were detected at the absence of AS during either pre-cultivation or co-cultivation without the dramatical reduction of the positive colonies, like the transformation of the plant pathogen Fusarium oxysporum (Mullins et al. 2001) and the entomopathogenic fungus Cordyceps militaris (Zheng et al. 2011).
The results of Southern blot showed that the rate of a single-copy insertion of T-DNA accounted for about 83%. Single-copy insertion into a genome is usually expected in order to obtain the gene that is responsible for the derived phenotype. Some reports implied that the rate of T-DNA single-copy insertion was negatively related to co-cultivation time (Mullins et al. 2001). However, the co-cultivation time should be long enough to guarantee the transformation efficiency in O. sinensis. Now the high-throughput sequencing technology can replace the Southern blot for molecular identification of O. sinensis mutants.
The stability of the transformants was checked and almost 50% transformants were lost in the third generations without selection pressure. This phenomenon was also found with variable frequency in other filamentous fungi, such as Mucor miehei (Martínez-Cruz et al. 2017), Podosphaera xanthii (Monfort et al. 2003), Aspergillus sojae (Mora-Lugo et al. 2014), and Backusella lamprospora (Nyilasi et al. 2008). The presence of a high number of transposons in their genomes might be responsible for the reduction of the stability of T-DNA (Martínez-Cruz et al. 2017; Spanu et al. 2010). The presence of a single-copy insertion of T-DNA into the O. sinensis genome was confirmed in the positive transformants, which is consistent with other reports (Mullins et al. 2001; Zheng et al. 2011).
The altered morphological phenotypes in colonies, blastospores, and fruiting body production of O. sinensis were detected from the insertional transformants obtained by ATMT. TAIL-PCR and FPNI-PCR techniques were established for O. sinensis to obtain the T-DNA flanking regions. Eight genes, namely genes for an MFS transporter, a 2-oxoglutarate dehydrogenase, a DNA-directed RNA polymerase III complex subunit Rpc37, a cytochrome oxidase subunit I, a mitochondrial import inner membrane translocase subunit tim54, a phosphoribosylaminoimidazole carboxylase, a cytidine deaminase, and a histone H3-like centromeric protein cse-4, were identified to be involved in the altered morphology of colonies and in the degenerated fruiting body production.
The molecular function of the genes or proteins was predicted by the Gene Ontology resource. The MFS transporter is mainly predicted to be involved in transporter activity (GO:0005215). The MFS transporters, single-polypeptide secondary carriers, are capable only of transporting small solutes including simple sugars, inositol, oligosaccharides, amino acids, nucleoside, drugs, organophosphate esters, Krebs cycle metabolites, and many organic and inorganic anions and cations (Nelissen et al. 1997; Pao et al. 1998). The disruption of an MFS transporter might affect the nutrient absorption and metabolism for the growth of O. sinensis mutant. Phosphoribosylaminoimidazole carboxylase is predicted to be involved in phosphoribosylaminoimidazole carboxylase activity (GO:0004638). It is involved in the subpathway of inosine monophosphate (IMP) biosynthesis, which is itself part of the purine metabolism. Disruption of IMP biosynthesis genes such as ade16 and ade17 resulted in adenine auxotrophy, double disruption of ade16 and ade17 led to adenine/histidine auxotrophy like in the ade3 mutant yeast strains (Tibbetts and Appling 2000). Cytidine deaminase is predicted to function in ATP:3′-cytidine-cytidine-tRNA adenylyltransferase activity (GO:0052929). It scavenges exogenous and endogenous cytidine and 2′-deoxycytidine for uridine monophosphate (UMP) biosynthesis. Disruption of cytidine deaminases would abolish cytidine deaminase activity (Kurtz et al. 1999). DNA-directed RNA polymerase III complex subunit Rpc37 is predicted to function in DNA-directed DNA polymerase activity (GO:0003887), RNA-directed DNA polymerase activity (GO:0003964), and DNA-directed 5′-3′ RNA polymerase activity (GO:0003899). Histone H3, together with other histones, forms the core of the nucleosome, which forms the octameric structure to wrap DNA in a left-handed manner. Histones can undergo several different types of post-translational modifications that affect transcription, DNA repair, DNA replication, and chromosomal stability (Thatcher and Gorovsky 1994).
2-Oxoglutarate dehydrogenase (E1), cytochrome oxidase subunit I, and mitochondrial import inner membrane translocase subunit tim54 are located in the mitochondria. Mitochondria have a translocation machinery for the import and assembly of nuclear-encoded proteins (Koehler 2004; Truscott et al. 2003). E1 is a component of the 2-oxoglutarate dehydrogenase complex, which catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO2 (Sato et al. 2002). Cytochrome oxidase subunit I is involved in the pathway oxidative phosphorylation and transfers electrons from reduced cytochrome c to molecular oxygen (Zheng et al. 2015). Tim54, a component of the inner membrane tim22 complex, is involved in protein import, assembly, and turnover pathways in the mitochondrion (Hwang et al. 2007). Fungal degeneration was found closely related to mitochondria, including oxidative stress, mtDNA alterations, mtDNA glycation, and mitochondrial dysfunctions. The features of cytochrome c release, calcium overload, decreasing activation of dehydrogenase, and upregulation of apoptosis inducing factors were evident in mitochondria of degenerated strains (Li et al. 2014; Li et al. 2008; Lin et al. 2010; Wang et al. 2005).
In conclusion, an optimal protocol for ATMT of O. sinensis was established. The genes involved in the altered growth characters of the transformants were identified. This provides an important genetic tool for studying the mysterious biology and genetics of O. sinensis.
References
Bundock P, den Dulk-Ras A, Beijersbergen A, Hooykaas PJ (1995) Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J 14:3206–3214. https://doi.org/10.1002/j.1460-2075.1995.tb07323.x
Cao L, Ye YS, Han RC (2015) Fruiting body production of the medicinal Chinese caterpillar mushroom, Ophiocordyceps sinensis (Ascomycetes), in artificial medium. Int J Med Mushrooms 17:1107–1112. https://doi.org/10.1615/IntJMedMushrooms.v17.i11.110
Combier JP, Melayah D, Raffier C, Gay G, Marmeisse R (2003) Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Microbiol Lett 220:141–148. https://doi.org/10.1016/s0378-1097(03)00089-2
de Groot MJA, Bundock P, Hooykaas PJJ, Beijersbergen AGM (1998) Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16:839–842. https://doi.org/10.1038/nbt0998-839
Holliday J, Cleaver M (2008) Medicinal value of the caterpillar fungi species of the genus Cordyceps (Fr.) link (Ascomycetes). Int J Med Mushrooms 10:219–234. https://doi.org/10.1615/IntJMedMushr.v10.i3.30
Hopping KA, Chignell SM, Lambin EF (2018) The demise of caterpillar fungus in the Himalayan region due to climate change and overharvesting. Proc Natl Acad Sci 115:11489–11494. https://doi.org/10.1073/pnas.1811591115
Hu X, Zhang Y, Xiao G, Zheng P, Xia Y, Zhang X, St Leger RJ, Liu X, Wang C (2013) Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin Sci Bull 58:2846–2854. https://doi.org/10.1007/s11434-013-5929-5
Hwang DK, Claypool SM, Leuenberger D, Tienson HL, Koehler CM (2007) Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J Cell Biol 178:1161–1175. https://doi.org/10.1083/jcb.200706195
Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T (1996) High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat Biotechnol 14:745–750. https://doi.org/10.1038/nbt0696-745
Jiang D, Zhu W, Wang Y, Sun C, Zhang KQ, Yang J (2013) Molecular tools for functional genomics in filamentous fungi: recent advances and new strategies. Biotechnol Advances 31:1562–1574. https://doi.org/10.1016/j.biotechadv.2013.08.005
Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20:309–335. https://doi.org/10.1146/annurev.cellbio.20.010403.105057
Kozák L, Szilágyi Z, Vágó B, Kakuk A, Tóth L, Molnár I, Pócsi I (2018) Inactivation of the indole-diterpene biosynthetic gene cluster of Claviceps paspali by Agrobacterium-mediated gene replacement. Appl Microbiol Biotechnol 102:3255–3266. https://doi.org/10.1007/s00253-018-8807-x
Kurtz JE, Exinger F, Erbs P, Jund R (1999) New insights into the pyrimidine salvage pathway of Saccharomyces cerevisiae: requirement of six genes for cytidine metabolism. Curr Genet 36:130–136. https://doi.org/10.1007/s002940050482
Leclerque A, Wan H, Abschütz A, Chen S, Mitina GV, Zimmermann G, Schairer HU (2004) Agrobacterium-mediated insertional mutagenesis (AIM) of the entomopathogenic fungus Beauveria bassiana. Curr Genet 45:111–119. https://doi.org/10.1007/s00294-003-0468-2
Li L, Pischetsrieder M, St Leger RJ, Wang C (2008) Associated links among mtDNA glycation, oxidative stress and colony sectorization in Metarhizium anisopliae. Fungal genet biol 45:1300–1306. https://doi.org/10.1016/j.fgb.2008.06.003
Li L, Hu X, Xia Y, Xiao G, Zheng P, Wang C (2014) Linkage of oxidative stress and mitochondrial dysfunctions to spontaneous culture degeneration in Aspergillus nidulans. Mol Cell Proteomics 3:449–461. https://doi.org/10.1074/mcp.M113.028480
Li WJ, Dong CH, Liu XZ, Li QP, Xia JM, Liang F (2016) Research advances in artificial cultivation of Chinese cordyceps. Mycosystema 35:375–387. https://doi.org/10.13346/j.mycosystema.160003
Li X, Liu Q, Li W, Li Q, Qian Z, Liu X, Dong C (2019a) A breakthrough in the artificial cultivation of Chinese cordyceps on a large-scale and its impact on science, the economy, and industry. Crit Rev Biotechnol 39:181–191. https://doi.org/10.1080/07388551.2018.1531820
Li X, Wang F, Liu Q, Li Q, Qian Z, Zhang X, Li K, Li W, Dong C (2019b) Developmental transcriptomics of Chinese cordyceps reveals gene regulatory network and expression profiles of sexual development-related genes. BMC Genomics 20:337. https://doi.org/10.1186/s12864-019-5708-z
Lin QQ, Qiu XH, Zheng ZL, Xie CH, Xu ZF, Han RC (2010) Characteristics of the degenerate strains of Cordyceps militaris. Mycosystema 29:670–677. https://doi.org/10.13346/j.mycosystema.2010.05.019
Liu YG, Chen Y (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43:649–656. https://doi.org/10.2144/000112601
Liu GQ, Qiu XH, Cao L, Han RC (2018) Scratching stimuli of mycelia influence fruiting body production and ROS-scavenging gene expression of Cordyceps militaris. Mycobiology 46:382–387. https://doi.org/10.1080/12298093.2018.1544769
Liu GQ, Han RC, Cao L (2019) Artificial cultivation of the Chinese cordyceps from injected ghost moth larvae. Environ Entomol 48:1088–1094. https://doi.org/10.1093/ee/nvz099
Lu Z, Shi P, He Y, Zhang D, He Z, Chen S, Tu Y, Li L, Liu F, Zeng W (2015) Review on natural enemies and diseases in the artificial cultivation of Chinese caterpillar mushroom, Ophiocordyceps sinensis (Ascomycetes). Int J Med Mushrooms 17:693–700. https://doi.org/10.1615/IntJMedMushrooms.v17.i7.90
Martínez-Cruz J, Romero D, de Vicente A, Pérez-García A (2017) Transformation of the cucurbit powdery mildew pathogen Podosphaera xanthii by Agrobacterium tumefaciens. New Phytol 213:1961–1973. https://doi.org/10.1111/nph.14297
Meng Q, Yu HY, Zhang H, Zhu W, Wang ML, Zhang JH, Zhou GL, Li X, Qin QL, Hu SN, Zou Z (2015) Transcriptomic insight into the immune defenses in the ghost moth, Hepialus xiaojinensis, during an Ophiocordyceps sinensis fungal infection. Insect Biochem Mol Biol 64:1–15. https://doi.org/10.1016/j.ibmb.2015.06.014
Michielse CB, Ram AFJ, Hooykaas PJJ, van den Hondel CAMJJ (2004) Role of bacterial virulence proteins in Agrobacterium-mediated transformation of Aspergillus awamori. Fungal Genet Biol 41:571–578. https://doi.org/10.1016/j.fgb.2004.01.004
Michielse CB, Hooykaas PJ, van den Hondel CA, Ram AF (2005) Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr Genet 48:1–17. https://doi.org/10.1007/s00294-005-0578-0
Michielse CB, Hooykaas PJJ, van den Hondel CAMJJ, Ram AFJ (2008) Agrobacterium-mediated transformation of the filamentous fungus Aspergillus awamori. Nat Protoc 3:1671. https://doi.org/10.1038/nprot.2008.154
Monfort A, Cordero L, Maicas S, Polaina J (2003) Transformation of Mucor miehei results in plasmid deletion and phenotypic instability. FEMS Microbiol Lett 224:101–106. https://doi.org/10.1016/s0378-1097(03)00421-x
Mora-Lugo R, Zimmermann J, Rizk AM, Fernandez-Lahore M (2014) Development of a transformation system for Aspergillus sojae based on the Agrobacterium tumefaciens-mediated approach. BMC Microbiol 14:247–248. https://doi.org/10.1186/s12866-014-0247-x
Mullins ED, Chen X, Romaine P, Raina R, Geiser D, Kang S (2001) Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91:173–180. https://doi.org/10.1094/phyto.2001.91.2.173
Nelissen B, Wachter RD, Goffeau A (1997) Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol Rev 21:113–134. https://doi.org/10.1016/s0168-6445(97)00053-3
Nyilasi I, Papp T, Csernetics A, Vagvolgyi C (2008) Agrobacterium tumefaciens -mediated transformation of the zygomycete fungus Backusella lamprospora. J Basic Microbiol 48:59–64. https://doi.org/10.1002/jobm.200700221
Pao SS, Paulsen IT, Saier MH (1998) Major Facilitator Superfamily. Microbiol Mol Biol Rev 62:1-34. PMID: 9529885
Qin QL, Zhou GL, Zhang H, Meng Q, Zhang JH, Wang HT, Miao L, Li X (2018) Obstacles and approaches in artificial cultivation of Chinese cordyceps. Mycology 9:7–9. https://doi.org/10.1080/21501203.2018.1442132
Qiu XH, Cao L, Han RC (2016) The progress, issues and perspectives in the research of Ophiocordyceps sinensis. J Environ Entomol 38:1–23. https://doi.org/10.3969/j.issn.1674-0858.2016.01.1
Rao ZC, Cao L, Qiu XH, Han RC (2019) Comparative transcriptome analysis reveals molecular strategies of ghost moth Thitarodes armoricanus in response to hypoxia and anoxia. J Insect Physiol 112:23–34. https://doi.org/10.1016/j.jinsphys.2018.11.001
Roberts DW, St Leger RJ (2004) Metarhizium spp., cosmopolitan insect-pathogenic fungi: mycological aspects. Adv Appl Microbiol 54:1–70. https://doi.org/10.1016/s0065-2164(04)54001-7
Sato H, Tachifuji A, Tamura M, Miyakawa I (2002) Identification of the YMN-1 antigen protein and biochemical analyses of protein components in the mitochondrial nucleoid fraction of the yeast Saccharomyces cerevisiae. Protoplasma 219:51–58. https://doi.org/10.1007/s007090200005
Shao C, Yin Y, Qi Z, Li R, Song Z, Li Y, Wang Z (2015) Agrobacterium tumefaciens-mediated transformation of the entomopathogenic fungus Nomuraea rileyi. Fungal Genet Biol 83:19–25. https://doi.org/10.1016/j.fgb.2015.08.002
Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Loren van Themaat EV, Brown JKM, Butcher SA, Gurr SJ, Lebrun M-H, Ridout CJ, Schulze-Lefert P, Talbot NJ, Ahmadinejad N, Ametz C, Barton GR, Benjdia M, Bidzinski P, Bindschedler LV, Both M, Brewer MT, Cadle-Davidson L, Cadle-Davidson MM, Collemare J, Cramer R, Frenkel O, Godfrey D, Harriman J, Hoede C, King BC, Klages S, Kleemann J, Knoll D, Koti PS, Kreplak J, López-Ruiz FJ, Lu X, Maekawa T, Mahanil S, Micali C, Milgroom MG, Montana G, Noir S, O’Connell RJ, Oberhaensli S, Parlange F, Pedersen C, Quesneville H, Reinhardt R, Rott M, Sacristán S, Schmidt SM, Schön M, Skamnioti P, Sommer H, Stephens A, Takahara H, Thordal-Christensen H, Vigouroux M, Weßling R, Wicker T, Panstruga R (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330:1543–1546. https://doi.org/10.1126/science.1194573
Stone R (2008) Last stand for the body snatcher of the Himalayas? Science 322:1182. https://doi.org/10.1126/science.322.5905.1182
Sun L, Yan M, Ding Z, Liu Y, Du M, Xi P, Liao J, Ji L, Jiang Z (2014) Improved dominant selection markers and co-culturing conditions for efficient Agrobacterium tumefaciens-mediated transformation of Ustilago scitaminea. Biotechnol Lett 36:1309–1314. https://doi.org/10.1007/s10529-014-1486-5
Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol 57:5–59. https://doi.org/10.3114/sim.2007.57.01
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Tao Z, Cao L, Zhang Y, Ye YS, Han RC (2016) Laboratory rearing of Thitarodes armoricanus and Thitarodes jianchuanensis (Lepidoptera: Hepialidae), hosts of the Chinese medicinal mungus Ophiocordyceps sinensis (Hypocreales: Ophiocordycipitaceae). J Econ Entomol 109:176–181. https://doi.org/10.1093/jee/tov319
Thatcher TH, Gorovsky MA (1994) Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res 22:174–179. https://doi.org/10.1093/nar/22.2.174
Thomas MB, Read AF (2007) Can fungal biopesticides control malaria? Nat Rev Microbiol 5:377–383. https://doi.org/10.1038/nrmicro1638
Tibbetts AS, Appling DR (2000) Characterization of two 5-aminoimidazole-4-carboxamide ribonucleotide transformylase/inosine monophosphate cyclohydrolase isozymes from Saccharomyces cerevisiae. J Biol Chem 275:20920–20927. https://doi.org/10.1074/jbc.M909851199
Truscott KN, Brandner K, Pfanner N (2003) Mechanisms of protein import into mitochondria. Curr Biol 13:R326–R337. https://doi.org/10.1016/S0960-9822(03)00239-2
Wang SF, Zhu HX, Zhu P (2003) The preliminary cultivation and observation on the vitality of Hirsutella sinensis. Edible Fungi China 22:4–6. https://doi.org/10.13629/j.cnki.53-1054.2003.06.002
Wang CS, Fan M, Li Z, Butt TM (2004) Molecular monitoring and evaluation of the application of the insect-pathogenic fungus Beauveria bassiana in Southeast China. J Appl Microbiol 96:861–870. https://doi.org/10.1111/j.1365-2672.2004.02215.x
Wang CS, Butt TM, St Leger RJ (2005) Colony sectorization of Metarhizium anisopliae is a sign of ageing. Microbiology 151:3223–3236. https://doi.org/10.1099/mic.0.28148-0
Wang Z, Ye SF, Li JJ, Zheng B, Bao MZ, Ning GG (2011) Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol 11:109–120. https://doi.org/10.1186/1472-6750-11-109
Wu YL, Sun HX, Qin F, Pan YJ, Sun CR (2006) Effect of various extracts and a polysaccharide from the edible mycelia of Cordyceps sinensis on cellular and humoral immune response against ovalbumin in mice. Phytother Res 20:646–652. https://doi.org/10.1002/ptr.1921
Xia F, Chen X, Guo MY, Bai XH, Liu Y, Shen GR, Li YL, Lin J, Zhou XW (2016) High-throughput sequencing-based analysis of endogenetic fungal communities inhabiting the Chinese Cordyceps reveals unexpectedly high fungal diversity. Sci Rep 6:33437–33437. https://doi.org/10.1038/srep33437
Xiang L, Li Y, Zhu Y, Luo H, Li C, Xu X, Sun C, Song J, Shi L, He L, Sun W, Chen S (2014) Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics 103:154–159. https://doi.org/10.1016/j.ygeno.2014.01.002
Zhang G, Huang Y, Bian Y, Wong JH, Ng TB, Wang H (2006) Hypoglycemic activity of the fungi Cordyceps militaris, Cordyceps sinensis, Tricholoma mongolicum, and Omphalia lapidescens in streptozotocin-induced diabetic rats. Appl Microbiol Biotechnol 72:1152–1156. https://doi.org/10.1007/s00253-006-0411-9
Zhang YJ, Li EW, Wang CS, Li YL, Liu XZ (2012) Ophiocordyceps sinensis, the flagship fungus of China: terminology, life strategy and ecology. Mycology 3:2–10. https://doi.org/10.1080/21501203.2011.654354
Zhang YJ, Zhang S, Li YL, Ma SL, Wang CS, Xiang MC, Liu X, An Z, Xu J, Liu X (2014) Phylogeography and evolution of a fungal-insect association on the Tibetan Plateau. Mol Ecol 23:5337–5355. https://doi.org/10.1111/mec.12940
Zheng Z, Huang CH, Cao L, Xie CH, Han RC (2011) Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in medicinal fungus Cordyceps militaris. Fungal Biol 115:265–274. https://doi.org/10.1016/j.funbio.2010.12.011
Zheng ZL, Qiu XH, Han RC (2015) Identification of the genes involved in the fruiting body production and cordycepin formation of Cordyceps militaris fungus. Mycobiology 43:37–42. https://doi.org/10.5941/MYCO.2015.43.1.37
Zhong YH, Wang XL, Wang TH, Jiang Q (2007) Agrobacterium-mediated transformation (AMT) of Trichoderma reesei as an efficient tool for random insertional mutagenesis. Appl Microbiol Biotechnol 73:1348–1354. https://doi.org/10.1007/s00253-006-0603-3
Zhong X, Gu L, Li S, Kan X, Zhang G, Liu X (2016) Transcriptome analysis of Ophiocordyceps sinensis before and after infection of Thitarodes larvae. Fungal Biol 120:819–826. https://doi.org/10.1016/j.funbio.2016.02.003
Zhou XW, Li LJ, Tian EW (2014) Advances in research of the artificial cultivation of Ophiocordyceps sinensis in China. Crit Rev Biotechnol 34:233–243. https://doi.org/10.3109/07388551.2013.791245
Funding
This study was supported by the National Nature Science Foundation of China (31900368), Natural Science Foundation of Guangdong Province (2018A030310489), Guangzhou Science and Technology Projects (201604020030; 201803010087), and GDAS Special Project of Science and Technology Development (2018GDASCX-0107).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 428 kb)
Rights and permissions
About this article
Cite this article
Liu, G., Cao, L., Rao, Z. et al. Identification of the genes involved in growth characters of medicinal fungus Ophiocordyceps sinensis based on Agrobacterium tumefaciens–mediated transformation. Appl Microbiol Biotechnol 104, 2663–2674 (2020). https://doi.org/10.1007/s00253-020-10417-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10417-1