Abstract
Halohydrin dehalogenases (HHDHs) have attracted much attention due to their ability to synthesize enantiomerically enriched epoxides and β-haloalcohols. However, most of the HHDHs exhibit low enantioselectivity. Here, a HHDH from the alphaproteobacteria isolate 46_93_T64 (AbHHDH), which shows only poor enantioselectivity in the catalytic resolution of rac-PGE (E = 9.9), has been subjected to protein engineering to enhance its enantioselectivity. Eight mutants (R89K, R89Y, V137I, P178A, N179Q, N179L, F187L, F187A) showed better enantioselectivity than the wild type. The best single mutant N179L (E = 93.0) showed a remarkable 9.4-fold increase in the enantioselectivity. Then, the single mutations were combined to produce the double, triple, quadruple, and quintuple mutants. Among the combinational mutants, the best variant (R89Y/N179L) showed an increased E value of up to 48. The E values of the variants N179L and R89Y/N179L for other epoxides 2–7 were 12.2 to > 200, which showed great improvement compared to 1.2 to 10.5 for the wild type. Using the variant N179L, enantiopure (R)-PGE with > 99% ee could be readily prepared, affording a high yield and a high concentration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enantiomerically pure compounds are important precursors for the synthesis of pharmaceuticals and fine chemicals (Patel 2011; Sun et al. 2018). In recent years, biocatalysts have been extensively used in the production of enantiopure precursors (Savile et al. 2010; Pavlidis et al. 2016; Tentori et al. 2018). Enantioselectivity is one of the prominent catalytic properties of biocatalysts, which makes enzymes a priority for pharmaceutical manufacture (Guo et al. 2014; Ma et al. 2014). However, the poor enantiomeric excess (ee) of products produced by most wild-type enzymes cannot meet the demand for industrial applications, especially when non-natural substrates are used. A number of strategies have been developed and employed to improve the enantioselectivity of enzymes, including medium engineering, substrate engineering, immobilization, and protein engineering (Gao et al. 2014; Xue et al. 2015; de Morais et al. 2018). Strategies for protein engineering, including directed evolution and (semi-) rational design, have achieved remarkable results in enhancing the enantioselectivity of enzymes (van Loo et al. 2004; Reetz et al. 2009; Godinho et al. 2012; Spickermann et al. 2014; Li et al. 2018).
Halohydrin dehalogenases (HHDHs) are multifunctional enzymes that catalyze the dehalogenation of halohydrins to form epoxides. In the reverse direction, they also catalyze the reversible ring opening of various epoxides to form the corresponding β-substituted alcohols (Hasnaoui-Dijoux et al. 2008; Koopmeiners et al. 2017; Zhang et al. 2018). Many HHDHs have been identified in different bacterial strains, and genes encoding some of these have been cloned and heterologously expressed (van Hylckama Vlieg et al. 2001; Koopmeiners et al. 2016; Schallmey and Schallmey 2016). To date, 73 different HHDH enzyme sequences have been discovered that can be classified into seven groups (from A to G) based on the sequence similarity (Schallmey and Schallmey 2016; Koopmeiners et al. 2017; Xue et al. 2018a; Xue et al. 2018b; Xue et al. 2019). However, it seems that HHDH from Agrobacterium radiobacter AD1 (HheC) is the only one with relatively high enantioselectivity among all the identified HHDHs in the transformation of various vicinal halohydrins and epoxides (Schallmey and Schallmey 2016). Thus, it is necessary to increase the enantioselectivity of HHDHs, which so far has been mainly reported for the enzymes from Arthrobacter sp. AD2 (HheA), A. radiobacter AD1 (HheC), and Corynebacterium sp. N-1074 (HheB). In the case of HheC, semi-rational design using iterative saturation mutagenesis resulted in an approximately twofold improvement of R-enantioselectivity in the hydrolytic kinetic resolution of 2-chloro-1-phenylethanol. Notably, the quadruple mutant (P84V/F86P/T134A/N176A) displayed an inversion of enantioselectivity (from ER = 65 to ES = 101) (Guo et al. 2015). In the case of HheA, the double mutant V136Y/L141G also showed an inversion of enantioselectivity (from an ES of 1.7 to an ER of 13) in the kinetic resolution of 2-chloro-1-phenylethanol, while the enzyme activity remained similar to the wild type. Notably, there was a dramatic increase of enantioselectivity toward (S)-2-chloro-1-phenyethanol in the N178A mutant (ES > 200) (Tang et al. 2012). However, this HheA mutant exhibited very low catalytic activity toward vicinal halohydrins. A triple mutant HheB D199H/F71 W/Q125T exhibited the highest R-enantioselectivity in the synthesis of (R)-4-chloro-3-hydroxy-butyronitrile, and the product yield was about twice that of wild-type enzyme. (Watanabe et al. 2016). It is of increasing interest to develop an efficient approach for the generation of HHDH mutants with high enantioselectivity toward halohydrins and epoxides with diverse substituents (Zhang et al. 2019).
PGE and its derivatives are important building blocks for the synthesis of chiral amino alcohols and β-blockers. Up to now, there have been no reported examples of using HHDH to prepare chiral PGE with high ee and yield. Previously, a novel HHDH from the alphaproteobacteria isolate 46_93_T64 has been identified and characterized. The enzyme, named AbHHDH, exhibited moderate enantioselectivity toward PGE (Xue et al. 2019). Therefore, an improvement of the enzyme’s enantioselectivity is necessary before it can be used as efficient biocatalyst for the synthesis of chiral PGE. In this study, homology modeling and molecular docking were performed to identify target amino acids near the bound substrate to enhance the enantioselectivity of AbHHDH. Then, site-saturation mutagenesis and site-directed mutagenesis of positions 89, 137, 178, 179, 180, and 187 were performed. HHDH mutants with enhanced enantioselectivity were screened and characterized. In addition, the application potential of the AbHHDH variant with the highest enantioselectivity was investigated by performing kinetic resolution of rac-PGE.
Materials and methods
Materials
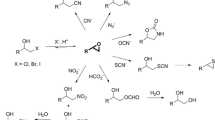
Racemic phenyl glycidyl ether (PGE) (1), (R)-PGE, (S)-PGE, racemic styrene oxide (8), racemic benzyl glycidyl ether (9), racemic naphthyl glycidyl ether (10), isopropyl-β-d-thiogalacto-pyranoside (IPTG), and kanamycin were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Racemic azido alcohols were purchased from Shanghai Nafu Biotechnology Co., Ltd. (China). The racemic epoxides 2–7 (Fig. 1) were synthesized as previously described (Xue et al. 2018b). PrimeSTAR HS DNA polymerase and restriction endonuclease Dpn I were purchased from Takara (Dalian, China). PCR purification and plasmid extraction kits were purchased from Axygen (Hangzhou, China). All primers were synthesized by Synbio Tech (Suzhou, China). All other chemicals used in this study were of analytical grade and commercially available.
Strains, plasmids, and culture conditions
E. coli BL21(DE3) was used as the host cell for the overexpression of wild-type and mutant HHDHs. The pET28a-AbHHDH plasmid constructed in our previous work was used as template (Xue et al. 2019). Recombinant cells were cultured in Luria-Bertani (LB) broth supplemented with kanamycin (50 μg/mL).
Library generation
The template for site-saturation mutagenesis was the plasmid pET28a-AbHHDH, which was extracted from recombinant E. coli. The positions 89, 137, 178, 179, 180, and 187 were selected as target sites. The forward and reverse primers used to generate the mutants are listed in supplemental Table S1. The temperature program of the mutagenic PCR encompassed an initial denaturation step at 98 °C for 3 min, followed by 28 cycles comprising 98 °C for 10 s, 50–55 °C for 15 s and 72 °C for 7 min, and a final extension at 72 °C for 10 min. Then, the samples were digested with Dpn I endonuclease at 37 °C for 3 h, to degrade the parent plasmid (Liu et al. 2017), and the reaction mixtures were transformed into E. coli BL21(DE3) by heat-shock at 42 °C for 90 s. Transformants were spread on LB agar medium (2% agar) containing kanamycin (50 μg/mL) and cultured at 37 °C for 10 h. The positive mutations were further confirmed by DNA sequencing.
Expression and purification
Recombinant E. coli BL21 (DE3) cells carrying either pET28a-AbHHDH or pET28a-AbHHDHmut were pre-cultured at 37 °C in LB medium containing 50 μg/mL kanamycin. When the OD600 of fermentation broth reached 0.6–0.8, the heterologous expression of HHDH was induced at 28 °C for 10 h with 0.1 mM IPTG. After harvesting by centrifugation at 12,000×g for 6 min at 4 °C, the cell pellets were washed twice in Tris-SO4 buffer (20 mM pH 8.0). The wild-type AbHHDH and derivatives were purified to apparent homogeneity by ammonium sulfate precipitation and hydrophobic interaction chromatography, followed by gel chromatography as described before (Xue et al. 2019).
Analytical methods
The conversion ratio and ee of azido alcohols and epoxides were determined by chiral HPLC analysis using Chiralcel AS-H, Chiralcel OD-H, and Chiralpak AD-H columns (250 × 4.6 mm × 5 μm, Daicel, Tokyo, Japan). For details of the chiral analysis methods, see the Supporting Information (Table S2). Absolute configurations of the glycidyl ethers and 1-azido-3-phenoxy-2-propanol were assigned by comparison of their retention time and elution order on the same HPLC column with data from the literature (Ulrich et al. 1993; Zhao et al. 2011; Wu et al. 2013; Kong et al. 2014). The enantiomeric ratio (E) and enantiomeric excess (ee) were calculated using published formulas (Chen et al. 1982).
Enzyme activity assay and measurement of kinetic parameters
The enzyme activities of purified recombinant AbHHDH and variants were assayed using rac-PGE as substrate (Fig. 1). The reaction mixtures (400 μL) consisted of 200 mM Tris-SO4 buffer (pH 7.5), 20 mM epoxide substrate, and the indicated amount of the purified wild-type AbHHDH enzyme or its mutants. The samples were taken at regular time intervals and extracted twice with ethyl acetate. The supernatant was filtered, dried over anhydrous sodium sulfate, and the concentrations of residual substrate and product were measured by HPLC. One unit of HHDH activity was defined as the amount of enzyme required to convert 1 μmol epoxide per min under the described assay conditions.
Kinetic parameters of the wild-type and two selected mutant enzymes toward (R)-PGE and (S)-PGE were determined using purified enzymes at 30 °C in 200 mM Tris-SO4 buffer (pH = 7.5). In the kinetic analysis, the ratios of substrate conversion were controlled within 10% to ensure accuracy. The maximum velocity (Vmax) and Michaelis constants (Km) were determined from plots of the initial velocity (V) versus the substrate concentration ([S]) by non-linear regression analysis. The kcat values were obtained by dividing the Vmax values with the enzyme concentration. The protein concentration was measured according the Bradford method (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as calibration standard.
Homology modeling and molecular docking
The homology sequence analysis was carried out using the BLASTN server at the website of the National Center for Biotechnology Information (NCBI). The crystal structure of Hhe C in the PDB (PDB accession nos. 1PWX), which was 41% identical to AbHHDH, was selected as template. The models of wild-type AbHHDH and its mutants were built using the program Modeller 9.18 with loops refined sufficiently. The quality of the modeled structures were evaluated using the Procheck program. Molecular docking experiments were performed using the program AutoDock 4.2.1, and the docking results were visualized using Pymol (Hu et al. 2017).
Whole-cell biocatalytic conversion
E. coli BL21(DE3) cells harboring the recombinant plasmid pET28a-HHDHmut were cultivated in LB medium. After expression, the cells were harvested by centrifugation at 10,000×g for 6 min and then resuspended in 0.2 M Tris-SO4 buffer (pH 7.5) at a biomass concentration of 50 g/L. The rac-PGE was added to a final concentration of 40 to 400 mM. The reaction was performed at 30 °C and 180 rpm. The reaction was monitored by periodically withdrawing samples. The mixtures were extracted with ethyl acetate and subjected to chiral HPLC analysis.
Statistical analysis
The data represent the means ± standard deviations (SD) from independent triplicates. Statistical analysis was performed using EXCEL (Microsoft Corp., USA) and SPSS (IBM Corp., USA) software (Liu et al. 2017). P < 0.05 was accepted as indicating statistical significance. The graphs were drawn using the Origin 8.0 software.
Accession number of AbHHDH gene
The amino acid sequence of AbHHDH can be accessed through the NCBI Protein database under accession number OUR79898.1 (Xue et al. 2019). Synthetic AbHHDH gene, codon-optimized for heterologous expression in E. coli, has been deposited at GenBank under the accession number MH782148.
Results
Modeling of AbHHDH to identify target residues for mutagenesis
In order to select suitable sites for saturation mutagenesis, we built a molecular model of AbHHDH with the crystal structure of HheC (PDB code: 1PWX, sequence identity 41%) as the template. The Ramachandran plot of the model showed that 91.4% residues fall in the most favored region, 7.7% in the additional allowed region, and 1.0% in the generously allowed region. These results indicated that the AbHHDH model was sufficiently plausible to serve as the basis for targeted mutagenesis. Then, the (S)-PGE was docked into the model of wild-type AbHHDH (Supplemental Fig. S1). Statistical analysis indicated that the residues close to the ligand molecule had a greater influence on the enantioselectivity, and are often chosen as target positions for rational and semi-rational design (Tang et al. 2012; Gu et al. 2015; Xue et al. 2015; Li et al. 2018). Nine residues located within 7 Å of rac-PGE in the modeled structure were chosen for mutations. The catalytic triad, consisting of residues Ser135, Tyr148, and Arg152, was excluded from mutagenesis. As depicted in Fig. 2, the other amino acid residues Arg89, Val 137, Pro178, Asn 179, Phe 180, and Phe187 were selected as the target amino acids for mutation. According to the analysis, the most interesting positions for mutations were Val137, Asn179, and Phe187, which are < 3.5 Å away from the substrate.
Library construction and HHDH variants screening
Saturation mutagenesis provides a method for randomizing the amino acid residues at predetermined positions in an enzyme molecule for building a focused library. Single mutants corresponding to the screened variants were created to determine which mutations contributed to increased selectivity. Six libraries were constructed by saturation mutagenesis, ideally each containing all 20 naturally amino acid substitutions at a specific site. The kinetic resolution of rac-PGE in the presence of azide was chosen as the model reaction. Enzymatic resolution reaction experiments were carried out with all mutants, as well as wild-type AbHHDH (E = 9.9) as a control. The mutant libraries were screened for enzyme activity toward the azidolysis of rac-PGE using ferric chloride, which reacts with azide, leading to a change in the absorbance at 460 nm that can be qualitatively analyzed (Wan et al. 2015). After screening 1000 clones, 400 AbHHDH-positive colonies were selected. Then, the enantioselectivities of 400 selected mutants were screened by HPLC analysis. While no positive mutant with higher than wild-type enantioselectivity was identified at position F180, eight positive mutations (R89K, R89Y, V137I, P178A, N179Q, N179L, F187L, F187A) at five different residual sites were obtained that showed 1.2- to 9.4-fold improvements of enantioselectivity toward PGE compared to wild-type (Table 1). Unfortunately, the mutant V137I, P178A, N179Q, N179L, F187L, and F187A exhibit lower activities with the test substrate than the wild-type enzyme. Thus, an enantioselectivity-activity trade-off was encountered in the saturation mutagenesis of AbHHDH. Changing Arg89 to Tyr raised the E value from 9.9 to 15.5. Similarly, mutating Val 137 to a Ile resulted in an increase of the E value from 9.9 to 12.6. Replacement of Pro178 by Ala gave a more than 1.75-fold increase in the enantioselectivity. Mutation of Phe 187 to Ala raised the E value from 9.9 to 18.3. It was surprising to find that a single-point variant, N179L, showed the highest improvement of enantioselectivity (E = 93.0). N179Q also displayed improved enantioselectivity. Moreover, the variants N179G and N179A exhibited an opposite enantiopreference with E values of 2.5 and 3.3 (Supplemental Table S3). These results indicated that the enantioselectivity of AbHHDH could be fine-tuned by mutations of N179. However, the enantioselectivity of other variants in the position 179 was significantly decreased. Moreover, N179H, N179E, N179D, and N179Y completely lost their enzymatic activity (Supplemental Table S3). These results indicated that although the risk of enzyme inactivation could not be ruled out, these five amino acid residues can be used as excellent targets to efficiently influence the enzyme’s enantioselectivity.
Thus, the six mutations N179L, N179Q, R89Y, V137I, P178A, and F187A were used to construct further combinational mutants, since the effects of mutations are often additive (Gu et al. 2015). The combination of the mutations V137I, P178A, and R89Y, except for the variant R89Y/P178A, resulted in higher E value than that of the corresponding single mutants (Table 1). The V137I/P178A double mutation increased the enantioselectivity for rac-PGE from 9.9 to 25.7. Similarly, R89Y/V137I increased the enantioselectivity from 9.9 to 29. The combinatorial mutant, R89Y/V137I/P178A, gave an approximately 3.5-fold increase in the enantioselectivity as compared with the native enzyme. The combination of the mutations V137I and N179Q also can have an additive effect on enantioselectivity, which increased from 9.9 to 20.8. However, the combination of N179L with other mutations resulted in decreased E values that those of the corresponding single mutants, indicating that the effect of the mutations was subtractive in this case. Further combination into four- and fivefold mutants failed to induce a further improvement.
Kinetic constants of the wild-type AbHHDH and its variants
In the laboratory evolution of enzymes, a trade-off between the targeted characteristic and other basic characteristics is often occured (Guo et al. 2013). In this research, an activity-enantioselectivity trade-off was encountered in the molecular modification of the enantioselectivity of AbHHDH in the resolution of rac-PGE. To analyze the reasons behind the activity-enantioselectivity trade-off, the mutants N179L and R89Y/N179L, as well as wild-type AbHHDH were purified to homogeneity using the method as described before (Fig. 3) (Xue et al. 2019), and their kinetic parameters with two enantiopure substrates were determined (Table 2). The N179L and R89Y/N179L mutations reduced the kcat/Km values considerably. The variant N179L exhibited an obvious increase in Km for both enantiomers. For the substrate rac-PGE, the 9.4-fold improved enantioselectivity of the variant N179L was attributed to a 36.1-fold decrease in catalytic efficiency for (R)-PGE combined with a 3.82-fold reduction in catalytic efficiency for (S)-PGE. A comparison of the kcat/Km values of R89Y/N179L for the azidolysis reaction of the favored (S)-PGE and the disfavored (R)-PGE again showed significant degree of preference. The resulting kcat/Km was decreased less for the faster reacting (S)-PGE than for the disfavored (R)-PGE, resulting in a significant improvement of enantioselectivity, albeit at the expense of overall catalytic efficiency.
Exploring the substrate scope of mutants
The N179L and the R89Y/N179L variants, which displayed a remarkable enhancement in the ring opening of rac-PGE (E = 85.9 and 48) with (S)-enantiopreference, were chosen for more detailed characterization. For this purpose, a small set of structurally diverse aromatic epoxides 2–10 was selected to investigate the enantioselectivity of the variants N179L and N179L/R89Y in the kinetic resolution of epoxides with the azide anion. Enzymatic reactions were performed using the purified recombinant enzymes in Tris-SO4 buffer (pH 7.5) containing NaN3 (50 mM) and epoxide (20 mM) (Fig. S2–S11). Our previous study indicated that the wild-type AbHHDH showed low to moderate enantioselectivity (E = 1.1–20.9) and a certain (S)-preference for most epoxide substrates (Xue et al. 2019). In addition, the change of enantioselectivity showed a close relationship with the structures of the substrate. The results summarized in Table 3 show that mutants N179L and R89Y/N179L can in fact be used as effective biocatalysts in the ring opening of other epoxides, although their enantioselectivity is not always as high as in the catalytic resolution of rac-PGE. A significant increase in enantioselectivity was observed with epoxides 2, 3, 5, and 7 as well as rac-PGE. N179L and R89Y/N179L had enhanced E values (E = 84.3 and 52.79) for rac-3-methylPGE, representing 21- and 13-fold more than that of wild-type AbHHDH. For rac-2-methylPGE the E value increased from 2.0 to 42.5 and 48.2, and for rac-4-methylPGE from 1.4 to 14.7 and 22.3, respectively. As in the case of rac-2-nitroPGE, and different from all the other tested phenyl glycidyl ethers, the (S)-enantiomer remained after the reaction. This means that going from a methyl to a nitro substituent at the phenyl glycidyl ether leads to a reversal of enantioselectivity. Surprisingly, an outstanding E value (> 200) was observed in the case of rac-2-nitroPGE. The mutant showed a more than 20-fold improved enantioselectivity with rac-2-nitroPGE than wild-type AbHHDH. For rac-2-ethylPGE, the variants N179L and N179L/R89Y showed significant increases of enantioselectivity. The enantioselectivity of the mutant N179L (E = 23.2) was increased up to 12.9-fold compared with wild-type AbHHDH (E = 1.8). The enantioselectivity of N179L/R89Y was better, with the E value increasing from 1.8 to 40.9, representing a ~22.7-fold enhancement over the wild type. N179L/R89Y exhibited an enhanced E value (12.8) with rac-naphthyl glycidyl ether, while N179L displayed a slight change of E value compared with the wild-type AbHHDH. For rac-styrene oxide, variants N179L and N179L/R89Y showed almost no obvious change in E value as compared to the wild type. In addition, it was found that the enantioselectivity of the mutant enzymes toward rac-benzyl glycidyl ether decreased.
Enantioselective azidolysis of rac-PGE with resting cells of variant HHDH
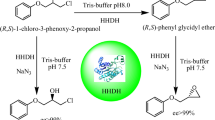
To explore the enzymatic resolution of rac-PGE using the whole cells of recombinant E. coli in the presence of N3−, 40 mM rac-PGE was azidolyzed with recombinant E. coli (N179L) at a biocatalyst loading of 4.5 g cdw/L. As shown in Table 4, the azidolysis rate of the (S)-configuration was much faster than that of the (R)-configuration, leaving 45.4% of the unreacted epoxide (R)-PGE in > 99% ee at 40 min. To further assess variant N179L as a potential biocatalysts, it was tested with higher substrate concentrations. Enantiopure (R)-PGE (> 99% ee) was obtained from 60 to 200 mM rac-PGE with a yield from 44.5 to 43.2% (almost 90% of the theoretical 50%; Table 4). Subsequently, 7.5 g cdw/L of recombinant E. coli was incubated with 300 and 400 mM rac-PGE, respectively. When kinetic resolution of 300 mM rac-PGE was conducted using the recombinant E. coli, enantiopure (R)-PGE with an ee higher than 99% was obtained at a 40.5% yield after 155 min. However, increasing the substrate concentration to 400 mM resulted in an obvious decrease in enantioselectivity. After 5 h, 39.4% of the (R)-PGE epoxide was obtained in > 99% ee, corresponding to an E value of 42.5.
Discussion
Halohydrin dehalogenases (HHDHs) are remarkable enzymes since they represent a useful tool in producing enantiopure epoxides and β-haloalcohols (Koopmeiners et al. 2017). Although the number of newly identified HHDHs has increased significantly in the past 5 years, only few of these novel HHDHs showed good enantioselectivity in the transformation of various substrates (Koopmeiners et al. 2016; Schallmey and Schallmey 2016). There is considerable interest in finding new enantioselective HHDHs (Schallmey et al. 2014; Koopmeiners et al. 2016) or improving the properties of existing enzymes through molecular modification (Schallmey and Schallmey 2016). However, all molecular modification studies of HHDHs have focused on only three different HHDHs, HheA, HheB, and HheC (Schallmey and Schallmey 2016; Watanabe et al. 2016; Wu et al. 2017; Zhang et al. 2018; Zhang et al. 2019). In the present work, the enantioselectivity of HHDH from an alphaproteobacterium as the biocatalyst in the bioresolution of epoxides was improved by protein engineering for the first time.
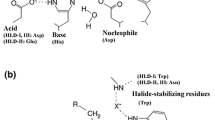
To design rational mutant libraries of HHDH, it was necessary to determine the target location of the mutations. The criteria for identifying target positions were as follows: (1) the residues should be adjacent to the bound substrate; (2) the side chain of enzyme should be oriented toward the bound substrate (Nobili et al. 2013). The docking experiment of rac-PGE into the active site of wild-type AbHHDH was used to identify six positions that might be the promising candidates for engineering the enantioselectivity. Through successive rounds of mutagenesis, thirteen mutants showed higher enantioselectivity than wild-type AbHHDH. In particular, residue 179 seems to play a key role in enzyme enantioselectivity. It was found that for all of the combinatorial mutants, the enantioselectivity was lower than that for mutant N179L. As far as we know, the enantioselectivity of the mutant N179L is the highest among all the known HHDHs for the kinetic resolution of rac-PGE (Mikleusevic et al. 2016; Xue et al. 2018a; Xue et al. 2018b; Xue et al. 2019).In HHDHs, the residues that form the halide-binding site are not strictly conserved, but they certainly include an Asn residue, such as the N179 in AbHHDH (You et al. 2013). The corresponding residues were also studied in HheA (Asn178) and HheC (Asn176), where they also had an obvious impact on the enantioselectivity. The mutant N178A of HheA showed a significant effect on the enzyme’s enantioselectivity toward rac-2-chloro-1-phenylethanol (E > 200) (Tang et al. 2012) and showed inversion of enantioselectivity (from ES = 1.5 to ER = 20) with rac-PGE (Mikleusevic et al. 2016). However, the corresponding N176A mutant of HheC displayed a significant decrease in enantioselectivity for rac-2-chloro-1-phenylethanol (Guo et al. 2015). The mutant N176H exhibited an enhancement of the E value for rac-2-chloro-1-phenylethanol from 65 to 72 (Guo et al. 2015). These results indicated that the Asn residue can be used as excellent targets to efficiently influence the enzyme’s enantioselectivity.
To investigate the structural changes responsible for the improvement of N179L, the HHDH variant with the most significant changes in the azidolysis of rac-PGE, a 3D model was constructed, and substrate docking was used to assess the influence of the mutation on substrate selectivity. In an attempt to understand how the single mutation of the 179 residue could result in such a tremendous change of enantioselectivity for the azidolysis of epoxides, in silico mutagenesis of N179 to L and docking of both enantiomers of PGE into the mutant N179L were performed. The docking results were in agreement with the hypothesis that the proposed catalytic triad indeed mediates the epoxide ring opening reaction catalyzed by AbHHDH. Ser135 appears to bind the PGE and is important for the reduction of the energy barrier by stabilizing to the emerging oxyanion, while Tyr148 acts as a general acid, donating a proton to the PGE (Hopmann and Himo 2008a). It is conducive to stabilizing the position of the PGE in the active site and facilitating the catalytic reaction. In wild-type AbHHDH, the epoxide oxygen atom of (S)-PGE is positioned in such a way that it possible to form two strong hydrogen bonds (d = 2.1 Å and 2.2 Å) with Tyr148 and Ser135, while the (R)-enantiomer forms weaker hydrogen bonds (d = 2.7 Å and 2.8 Å) (Fig. 4a, b). Therefore, the enzyme has high catalytic activity with S-type substrate. The result of the highly enantioselective N179L variant was remarkably different (Fig. 4c, d), and its d value for the preferred (S)-PGE was increased to 2.8 Å. However, in case of the disfavored (R)-PGE, the activated (R)-PGE is positioned farther away (d = 3.9 Å and 4.1 Å), which is not conducive to the ring opening nucleophilic attack. Based on the structure and catalytic mechanism of HheC, it can be inferred that the azide is bound in the halide-binding site of AbHHDH, which is mainly formed by the backbone of residues Ala177-Pro-Asn-Phe180 located in a loop region (de Jong et al. 2003; de Jong et al. 2005; Hopmann and Himo 2008b). The distance between the Cβ atom of the epoxide ring and loop region of the halide-binding site was different for (S)-PGE and (R)-PGE (Fig. 4). In wild-type AbHHDH, the distances were almost the same for both enantiomers (3.9 Å). However, in the highly enantioselective N179L, the △d has increased to 1.2 Å. Moreover, the N179L mutation also represents the substitution of a hydrophobic residue for a polar one, which could affect the microenvironment around the active center. These results allow us to deduce that the high enantioselectivity of N179L in the ring opening reaction of rac-PGE with azide is mainly determined by the great difference in the binding affinity of the two enantiomers. The structural changes in the binding pocket generated by molecular modification are presumed to make the disfavored (R)-PGE more difficult to activate by Ser135 and Tyr148, and at the same time position it farther from the loop region containing the halide-binding site, which slows down the nucleophilic attack.
The influence of the various mutations on enantioselectivity of the enzymes is related to the substrate structure. A range of glycidyl ethers (epoxides 2–10) have been used as substrates for the mutants N179L and R89Y/N179L. We investigated the effect of the methyl-substituent position in the substrate on the enantioselectivity of the mutants and wild-type AbHHDH. For the mutant N179L, mutant R89Y/N179L and wild-type AbHHDH, the highest enantioselectivity was observed in the kinetic resolution of 3-methylPGE and a roughly trend could be observed that the E value decrease as the methyl substituent on the phenyl ring is shifted from the meta-position to the para-position. The mutants N179L and R89Y/N179L showed the highest enantioselectivity (E > 200) toward 2-nitroPGE and the opposite enantiopreference for (R)-epoxide. This phenomenon has also been reported in ring opening reaction of glycidyl ethers catalyzed by epoxide hydrolase from Bacillus megaterium ECU1001. This result may be due to the difference of the charge properties of the two carbon atoms in the epoxide ring caused by the ortho-nitro substitution (Zhao et al. 2011). The E value for the bioresolution of epoxides 2–5, which can be utilized as precursors for the preparation of fine chemicals, were among the highest values reported so far for different HHDHs (Mikleusevic et al. 2016; Xue et al. 2018a; Xue et al. 2018b; Xue et al. 2019). In the epoxide ring opening of substrates 4, 7, and 10, the combination of mutations N179L and R89Y resulted in a higher E value than that of N179L and R89Y, indicating that also in this case the effect of the double mutation was additive.
Furthermore, the practical applicability of the best variant N179L was investigated in the ring opening of rac-PGE using E. coli cells. Enantiopure (R)-PGE were formed in more than 99% ee and a yield from 45.4 to 39.4% with 40 to 400 mM rac-PGE as the substrate. To the best of our knowledge, the HHDH-catalyzed ring opening of rac-PGE at high concentrations by azide has not been described for any other HHDHs. The enantioselectivity and yield for the ring opening reaction were the highest among the known HHDHs (Mikleusevic et al. 2016; Xue et al. 2018a; Xue et al. 2018b; Xue et al. 2019). Although, the activity of the currently available enzyme variants may not be sufficient for practical applications, these variants are a good starting point for additional biocatalyst development. Thus, we will attempt to solve the trade-off between activity and enantioselectivity in future work.
In summary, based on homology modeling and molecular docking, six residues were identified in close vicinity to the bound substrate and their mutants showed altered enantioselectivity. Through semi-rational design, the enantioselectivity of AbHHDH was significantly improved from 9.9 to 94.0 in the azidolysis of rac-PGE. For most tested substrates, the variants showed a good to excellent enantioselectivity. Compared with the wild-type AbHHDH, at least for epoxides 2, 3, 5, and 7 (E = 1.8 to 10.5), the mutants had a dramatically improved enantioselectivity (E = 40.9 to > 200), indicating a greater potential for the biocatalytic enantiomeric resolution of these epoxides. These results indicate that the HHDH variants obtained in this study are promising candidates for the practical synthesis of chiral epoxides and β-substituted alcohols.
References
Chen CS, Fujimoto Y, Girdaukas G, Sih CJ (1982) Quantitative analyses of biochemical kinetic resolutions of enantiomers. J Am Chem Soc 104:7294–7299. https://doi.org/10.1021/ja00389a064
de Jong RM, Tiesinga JJW, Rozeboom HJ, Kalk KH, Tang LX, Janssen DB, Dijkstra BW (2003) Structure and mechanism of a bacterial haloalcohol dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site. EMBO J 22:4933–4944. https://doi.org/10.1093/emboj/cdg479
de Jong RM, Tiesinga JJW, Villa A, Tang LX, Janssen DB, Dijkstra BW (2005) Structural basis for the enantioselectivity of an epoxide ring opening reaction catalyzed by haloalcohol dehalogenase HheC. J Am Chem Soc 127:13338–13343. https://doi.org/10.1021/ja0531733
de Morais JWG, Maia AM, Martins PA, Fernandez-Lorente G, Guisan JM, Pessela BC (2018) Influence of different immobilization techniques to improve the enantioselectivity of lipase from Geotrichum candidum applied on the resolution of mandelic acid. Mol Catal 458:89–96. https://doi.org/10.1016/j.mcat.2018.07.024
Gao PF, Li AT, Lee H, Wang DIC, Li Z (2014) Enhancing enantioselectivity and productivity of P450-catalyzed asymmetric sulfoxidation with an aqueous/ionic liquid biphasic system. ACS Catal 4:3763–3771. https://doi.org/10.1021/cs5010344
Godinho LF, Reis CR, Rozeboom HJ, Dekker FJ, Dijkstra BW, Poelarends GJ, Quax WJ (2012) Enhancement of the enantioselectivity of carboxylesterase a by structure-based mutagenesis. J Biotechnol 158:36–43. https://doi.org/10.1016/j.jbiotec.2011.12.026
Gu JL, Ye LD, Guo F, Lv XM, Lu WQ, Yu HW (2015) Rational design of esterase BioH with enhanced enantioselectivity towards methyl (S)-o-chloromandelate. Appl Microbiol Biotechnol 99:1709–1718. https://doi.org/10.1007/s00253-014-5995-x
Guo F, Xu HM, Xu HN, Yu HW (2013) Compensation of the enantioselectivity-activity trade-off in the directed evolution of an esterase from Rhodobacter sphaeroides by site-directed saturation mutagenesis. Appl Microbiol Biotechnol 97:3355–3362. https://doi.org/10.1007/s00253-012-4516-z
Guo F, Franzen S, Ye LD, Gu JL, Yu HW (2014) Controlling enantioselectivity of esterase in asymmetric hydrolysis of aryl prochiral diesters by introducing aromatic interactions. Biotechnol Bioeng 111:1729–1739. https://doi.org/10.1002/bit.25249
Guo C, Chen YP, Zheng Y, Zhang W, Tao YW, Feng J, Tang LX (2015) Exploring the enantioselective mechanism of halohydrin dehalogenase from Agrobacterium radiobacter AD1 by iterative saturation mutagenesis. Appl Environ Microb 81:2919–2926. https://doi.org/10.1128/aem.04153-14
Hasnaoui-Dijoux G, Elenkov MM, Lutje Spelberg JH, Hauer B, Janssen DB (2008) Catalytic promiscuity of halohydrin dehalogenase and its application in enantioselective epoxide ring opening. ChemBioChem 9:1048–1051. https://doi.org/10.1002/cbic.200890021
Hopmann KH, Himo F (2008a) Quantum chemical modeling of the dehalogenation reaction of haloalcohol dehalogenase. J Chem Theory Comput 4:1129–1137. https://doi.org/10.1021/ct8000443
Hopmann KH, Himo F (2008b) Cyanolysis and azidolysis of epoxides by haloalcohol dehalogenase: theoretical study of the reaction mechanism and origins of regioselectivity. Biochemistry 47:4973–4982. https://doi.org/10.1021/bi800001r
Hu D, Tang CD, Li C, Kan TT, Shi XL, Feng L, Wu MC (2017) Stereoselective hydrolysis of epoxides by reVrEH3, a novel Vigana radiate epoxide hydrolase with high enantioselectivity or high and complementary regioselectivity. J Agric Food Chem 65:9861–9867. https://doi.org/10.1021/acs.jafc.7b03804
Kong XD, Zhou JH, Zeng BB, Xu JH (2014) A smart library of epoxide hydrolase variants and the top hits for synthesis of (S)-β-blocker precursors. Angew Chem Int Ed 53:6641–6644. https://doi.org/10.1002/ange.201402653
Koopmeiners J, Halmschlag B, Schallmey M, Schallmey A (2016) Biochemical and biocatalytic characterization of 17 novel halohydrin dehalogenases. Appl Microbiol Biotechnol 100:7517–7527. https://doi.org/10.1007/s00253-016-7493-9
Koopmeiners J, Diederich C, Solarczek J, Voβ H, Mayer J, Blankenfeldt W, Schallmey A (2017) HheG, a halohydrin dehalogenase with activity on cyclic epoxides. ACS Catal 7:6877–6886. https://doi.org/10.1021/acscatal.7b01854
Li FH, Kong XD, Chen Q, Zheng YC, Xu Q, Chen FF, Fan LQ, Lin GQ, Zhou JH, Yu HL, Xu JH (2018) Regioselectivity engineering of epoxide hydrolase:near-perfect enantioconvergence through a single site mutation. ACS Catal 8:8314–8317. https://doi.org/10.1021/acscatal.8b02622
Liu ZQ, Wu L, Zhang XJ, Xue YP, Zheng YG (2017) Directed evolution of carbonyl reductase from Rhodosporidium toruloides and its application in stereoselective synthesis of tert-butyl (3R,5S)-6-chloro-3,5-dihydroxyhexanoate. J Agric Food Chem 65:3721–3729. https://doi.org/10.1021/acs.jafc.7b00866
Ma HR, Yang X, Lu Z, Liu N, Chen YJ (2014) The “gate keeper” role of Trp222 determines the enantiopreference of diketoreductase toward 2-chloro-1-phenylethanone. PLoS One 9:e103792. https://doi.org/10.1371/journal.pone.0103792
Mikleusevic A, Primozic I, Hrenar T, Salopek-Sondi B, Tang LX, Elenkov MM (2016) Azidolysis of epoxides catalyzed by the halohydrin dehalogenase from Arthrobacter sp. AD2 and a mutant with enhanced enantioselectivity: an (S)-selective HHDH. Tetrahedron:Asymmetr 27:930–935. https://doi.org/10.1016/j.tetasy.2016.08.003
Nobili A, Gall MG, Pavlidis LV, Thompson ML, Schmidt M, Bornscheuer UT (2013) Use of ‘small but smart’ libraries to enhance the enantioselectivity of an esterase from Bacillus stearothermophilus towards tetrahydrofuran-3-yl-acetate. FEBS J 280:3084–3093. https://doi.org/10.1111/febs.12137
Patel RN (2011) Biocatalysis: synthesis of key intermediates for development of pharmaceuticals. ACS Catal 1:1056–1074. https://doi.org/10.1021/cs200219b
Pavlidis IV, Weiβ MS, Genz M, Spurr P, Hanlon SP, Wirz B, Iding H, Bornscheuer UT (2016) Identification of (S)-selective transaminases for the asymmetric synthesis of bulky chiral amines. Nat Chem 8:1076–1082. https://doi.org/10.1038/nchem.2578
Reetz MT, Bocola M, Wang LW, Sanchis J, Cronin A, Arand M, Zou JY, Archelas A, Bottalla AL, Naworyta A, Mowbray S (2009) Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage. J Am Chem Soc 131:7334–7343. https://doi.org/10.1021/ja809673d
Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, Devine PN, Huisman GW, Hughes GJ (2010) Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329:305–309. https://doi.org/10.1126/science.1188934
Schallmey A, Schallmey M (2016) Recent advances on halohydrin dehalogenases-from enzyme identification to novel biocatalytic applications. Appl Microbiol Biotechnol 100:7827–7839. https://doi.org/10.1007/s00253-016-7750-y
Schallmey M, Koopmeiners J, Wells E, Wardenga R, Schallmey A (2014) Expanding the halohydrin dehalogenase enzyme family: identification of novel enzymes by database mining. Appl Environ Microbiol 80:7303–7315. https://doi.org/10.1128/AEM.01985-14
Spickermann D, Hausmann S, Degering C, Schwaneberg U, Leggewie C (2014) Engineering of highly selective variants of Parvibaculum lavamentivorans alcohol dehydrogenase. ChemBioChem 15:2050–2052. https://doi.org/10.1002/cbic.201402216
Sun HH, Zhang HF, Ang EH, Zhao HM (2018) Biocatalysis for the synthesis of pharmaceuticals and pharmaceutical intermediates. Bioorgan Med Chem 26:1275–1284. https://doi.org/10.1016/j.bmc.2017.06.043
Tang LX, Zhu XC, Zheng HR, Jiang RX, Elenkov MM (2012) Key residues for controlling enantioselectivity of halohydrin dehalogenase from Arthrobacter sp. AD2 revealed by structure-guided directed evolution. Appl Environ Microb 78:2631–2637. https://doi.org/10.1128/AEM.06586-11
Tentori F, Brenna E, Colombo D, Crotti M, Gatti FG, Ghezzi MC, Pedrocchi-Fantoni G (2018) Biocatalytic approach to chiral β-nitroalcohols by enantioselective alcohol dehydrogenase-mediated reduction of α-nitroketones. Catalysts 8:308–318. https://doi.org/10.3390/catal8080308
Ulrich A, Volker M, Manfred PS (1993) Chromatographic resolution of chiral intermediates in β-adrenergic blocker synthesis on chiral stationary phases. Chirality 5:554–559. https://doi.org/10.1002/chir.530050712
van Hylckama Vlieg JET, Tang LX, Lutje Spelberg JH, Smilda T, Poelarends GJ, Bosma T, Merode AEJ, Fraaije MW, Janssen DB (2001) Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J Bacteriol 183:5058–5066. https://doi.org/10.1128/JB.183.17.5058-5066.2001
van Loo B, Lutje Spelberg JH, Kingma J, Sonke T, Wubbolts MG, Janssen DB (2004) Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem Biol 11:981–990. https://doi.org/10.1016/j.chembiol.2004.04.019
Wan NW, Liu ZQ, Xue F, Huang K, Tang LJ, Zheng YG (2015) An efficient high-throughput screening assay for rapid directed evolution of halohydrin dehalogenase for preparation of β-substituted alcohols. Appl Microbiol Biotechnol 99:4019–4029. https://doi.org/10.1007/s00253-015-6527-z
Watanabe F, Yu F, Ohtaki A, Yamanaka Y, Noguchi K, Odaka M, Yohda M (2016) Improvement of enantioselectivity of the B-type halohydrin hydrogen-halidelyase from Corynebacterium sp. N-1074. J Biosci Bioeng 122:270–275. https://doi.org/10.1016/j.jbiosc.2016.02.003
Wu SK, Li AT, Chin YS, Li Z (2013) Enantioselctive hydrolysis of racemic and meso-epoxides with recombinant Escherichia coli expressing epoxide hydrolase from Sphinggomonas sp. HXN-200: preparation of epoxides and vicinal diols in high ee and high concentration. ACS Catal 3:752–759. https://doi.org/10.1021/cs300804v
Wu ZY, Deng WF, Tong YP, Liao Q, Xin DM, Yu HS, Feng J, Tang LX (2017) Exploring the thermostable properties of halohydrin dehalogenase from Agrobacterium radiobacter AD1 by a combinatorial directed evolution strategy. Appl Microbiol Biotechnol 101:3201–3211. https://doi.org/10.1007/s00253-017-8090-2
Xue YP, Shi CC, Xu Z, Jiao B, Liu ZQ, Huang JF, Zheng YG, Shen YC (2015) Design of nitrilases with superior activity and enantioselectivity towards sterically hindered nitrile by protein engineering. Adv Synth Catal 357:1741–1750. https://doi.org/10.1002/adsc.201500039
Xue F, Gao J, Zhang L, Li H, Huang H (2018a) Identification and characterization of a novel halohydrin dehalogenase from Bradyrhizobium erythrophlei and its performance in preparation of both enantiomers of epichlorohydrin. Catal Lett 148:1181–1189. https://doi.org/10.1007/s10562-017-2292-1
Xue F, Ya XJ, Tong Q, Xiu YS, Huang H (2018b) Heterologous overexpression of Pseudomonas umsongensis halohydrin dehalogenase in Escherichia coli and its application in epoxide asymmetric ring opening reactions. Process Biochem 75:139–145. https://doi.org/10.1016/j.procbio.2018.09.018
Xue F, Ya XJ, Xiu YS, Tong Q, Wang YQ, Zhu XH, Huang H (2019) Exploring the biocatalytic scope of a novel enantioselective halohydrin dehalogenase from an alphaproteobacterium. Catal Lett 149:629–637. https://doi.org/10.1007/s10562-019-02659-0
You ZY, Liu ZQ, Zheng YG (2013) Properties and biotechnological applications of halohydrin dehalogenases: current state and future perspectives. Appl Microbiol Biotechnol 97:9–21. https://doi.org/10.1007/s00253-012-4523-0
Zhang XJ, Shi PX, Deng HZ, Wang XX, Liu ZQ, Zheng YG (2018) Biosynthesis of chiral epichlorohydrin using an immobilized halohydrin dehalogenase in aqueous and non-aqueous phase. Bioresour Technol 263:483–490. https://doi.org/10.1016/j.biortech.2018.05.027
Zhang XJ, Deng HZ, Liu N, Gong YC, Liu ZQ, Zheng YG (2019) Molecular modification of a halohydrin dehalogenase for kinetic regulation to synthesize optically pure (S)-epichlorohydrin. Bioresour Technol 276:154–160. https://doi.org/10.1016/j.biortech.2018.12.103
Zhao J, Chu YY, Li AT, Ju X, Kong XD, Pan J, Tang Y, Xu JH (2011) An unusual (R)-selective epoxide hydrolase with high activity for facile preparation of enantiopure glycidyl ethers. Adv Synth Catal 353:1510–1518. https://doi.org/10.1002/adsc.201100031
Funding
This project was financially supported by the National Natural Science Foundation of China (No. 21606192 and No. 21878154), the National Key Research and Development Program of China (No. 2018YFC1604101), the China Postdoctoral Science Foundation (No. 2016M601795), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (No. 16KJB180029), and the Nature Science Foundation of Jiangsu Province (No. BK20170988).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1246 kb)
Rights and permissions
About this article
Cite this article
Xue, F., Zhang, LH. & Xu, Q. Significant improvement of the enantioselectivity of a halohydrin dehalogenase for asymmetric epoxide ring opening reactions by protein engineering. Appl Microbiol Biotechnol 104, 2067–2077 (2020). https://doi.org/10.1007/s00253-020-10356-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10356-x