Abstract
Biological nitrogen fixation (BNF) is an important natural biochemical process converting the inert dinitrogen gas (N2) in the atmosphere to ammonia (NH3) in the N cycle. In this study, the nifH gene was chosen to detect the diazotrophic microorganisms with high-throughput sequencing from five acidic forest soils, including three natural forests and two re-vegetated forests. Soil samples were taken in two seasons (summer and winter) at two depth layers (surface and lower depths). A dataset of 179,600 reads obtained from 20 samples were analyzed to provide the microbial community structure, diversity, abundance, and relationship with physiochemical parameters. Both archaea and bacteria were detected in these samples and diazotrophic bacteria were the dominant members contributing to the biological dinitrogen fixation in the acidic forest soils. Cyanobacteria, Firmicutes, Proteobacteria, Spirocheates, and Verrucomicrobia were observed, especially the Proteobacteria as the most abundant phylum. The core genera were Bradyrhizobium and Methylobacterium from α-Proteobacteia, and Desulfovibrio from δ-Proteobacteia in the phylum of Proteobacteia of these samples. The diversity indices and the gene abundances of all samples were higher in the surface layer than the lower layer. Diversity was apparently higher in re-vegetated forests than the natural forests. Significant positive correlation to the organic matter and nitrogen-related parameters was observed, but there was no significant seasonal variation on the community structure and diversity in these samples between the summer and winter. The application of high-throughput sequencing method provides a better understanding and more comprehensive information of diazotrophs in acidic forest soils than conventional and PCR-based ones.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Biological nitrogen fixation (BNF), the biological reduction of dinitrogen gas (N2) to biologically available ammonium, contributes to about half of annual nitrogen inputs into the biosphere by natural processes (Galloway et al. 2004; Vitousek et al. 1997). The process is associated with various microorganisms with a wide variety of physiologies from obligate aerobes to obligate anaerobes (Boyd et al. 2011; Dos Santos et al. 2012; Raymond et al. 2004). The taxonomic distribution of diazotrophs is restricted to bacteria and archaea without specific known genes encoding for this process in the Eukarya (Boyd et al. 2011; Dos Santos et al. 2012; Raymond et al. 2004).
Diazotrophic microorganisms of both bacteria and archaea are globally significant in their ability to provide the sole natural biological source of fixed nitrogen from the biosphere, a significant process of nitrogen transformation in the ecosystems (Cleveland et al. 1999). BNF is catalyzed by nitrogenase, an ATP-hydrolyzing, redox-active complex of two evolutionarily conserved proteins, the dinitrogenase α2β2 heterotetramer (NifD and NifK proteins) and the dinitrogenase reductase γ2 homodimer (NifH protein) (Homer et al. 1995; Raymond et al. 2004). There are three known types of nitrogenase, the molybdenum (Mo)-nitrogenase composed of two components namely Fe protein and MoFe protein; the vanadium (V)-nitrogenase comprised of Fe protein and VFe protein; and the iron (Fe)-only nitrogenase (Fe protein and Fe protein), which are sometimes referred to as the alternative nitrogenase (Bishop et al. 1986; Chisnell et al. 1988). The nifH gene, which encodes Fe protein, is highly conserved across the bacterial and archaeal domains and has been successfully used in culture-independent investigations of microbial N2 fixing communities in various environments including terrestrial (Chowdhury et al. 2009; Poly et al. 2001), marine (Moisander et al. 2006; Steppe and Paerl 2005), extremely hydrothermal vent (Mehta and Baross 2006), termite gut (Yamada et al. 2007), anthropogenic (Moseman-Valtierra et al. 2016), and other niches (Liao and Inglett 2014; Lincoln and Vitousek 2016). As a result, non-culturable microorganisms yield a large database based on nitrogenase genes and become one of the largest non-ribosomal gene datasets (Zehr et al. 2003). Therefore, this makes nifH gene a reference marker for studying diazotrophs in the different environments with most of the members remaining to be cultured.
Up to date, four groups of diazotrophs have been described based on the phylogeny of 16S rRNA gene and nifH gene (Raymond et al. 2004; Zehr et al. 2003). Cluster I consists of aerobic and facultative anaerobic nitrogen fixers including Actinobacteria, Cyanobacteria, Protebacteria and Firmicutes. Cluster II includes anfH, paralog of nifH gene, which is an alternative nitrogenase. Cluster III contains predominately obligate anaerobes, including sulfate-reducing bacteria of Delta-proteobacteria, Spirochaetes, and methanogenic archaea. Cluster IV contains of yet to be characterized nitrogenase paralogs and pigment biosynthesis genes of all phototrophs (Fujita et al. 1992; Nomata et al. 2006; Raymond et al. 2004; Souillard et al. 1988; Staples et al. 2007).
Forest soils are a crucial factor in terrestrial ecosystems especially in terms of energy and nutrient flow, and forest soil microbial communities play an essential role to forest productivity and climatic changes. Both symbiotic BNF via a root-nodule association and non-symbiotic BNF through free-living N2 fixers occur in forest ecosystem through autotrophic and heterotrophic process. In this study, forest soil samples from Nanling Nature Reserve located in Guangdong province in southern China, the heart of subtropical evergreen broadleaf forest, were taken from natural forest and re-vegetated forest. To better understand the significance of nitrogen fixing by microbial contributors to the global nitrogen equilibrium in this study, high-throughput sequencing technique was applied to reveal the distribution and abundance of nitrogen-fixing microbial community in the acidic forest soils of natural and re-vegetated forests.
Materials and methods
Sample collection and soil characterization
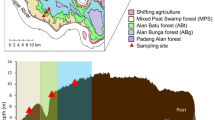
Soil samples were collected from five representative forest types and locations in the Nanling National Nature Reserve in Guangdong province, China. As previously described, three natural forests (HLS/HLD, PS/PD, EBLS/EBLD) and two re-vegetated forests (CLYS/CLYD, CLMS/CLMD) were defined based on the forest types (Gan et al. 2016; Meng et al. 2016; Meng et al. 2017). Sampling was performed in January of 2015 (winter) and August of 2015 (summer) from two soil depths at each site: the surface (0–10 cm) and the lower one (20–40 cm) of bulk soil profile. Each site was sampled randomly with three replicates. These samples were immediately separated into two portions for physiochemical characteristic measurement and molecular analysis after field collection. They were kept and then transported in ice-cold box to the laboratory immediately.
The following soil characteristics were used in the subsequent statistical analyses: pH values were measured in a soil slurry with soil to water ratio of 1:1 using a pH meter (Starter 3C, OHAUS, Pine Brook, NJ, USA), soil moisture content (%) measured through determining the constant weight of samples after drying in oven at 105 °C for 24 h, total organic matter measured by K2Cr2O7 method (g/kg), total nitrogen obtained by the Kjeldahl method (TN, g/kg), hydrolyzed nitrogen determined by KOH diffusion method, total phosphorous, and available phosphorous digested by HClO4+H2SO4 and NaHCO3, respectively and measured with the colorimetry method, exchangeable Al, and total potassium (TK, g/kg) detected by KF and HF with HClO4 methods (Lu 2000; Sparks 1996).
DNA extraction
Genomic DNA was extracted from approximately 0.25 g of soil sample using the PowerSoil® DNA isolation kit (MO BIO Laboratories, Inc. USA) in accordance with the manufacturer’s instructions. And the concentration was measured using NanoDrop™ ND-2000 (Thermo Scientific, Inc. USA).
Construction of barcode nifH gene libraries
nifH gene as a molecular biomarker was amplified to assess the diversity and composition of diazotrophic communities in these soil samples. IGK-DVV functional PCR primers were used to amplify the target fragment of the nifH gene (Gaby and Buckley 2012). A 12 nt unique barcode was fused to the 5′ end of the forward primer. Amplification was performed using a thermal cycler (Takara, Japan) with the following steps of 95 °C for 5 min and 35 cycles consisting of 94 °C for 30 s, 58 °C for 45 s, and 72 °C for 45 s; and elongation at 72 °C for 7 min at last. The PCR reactions were conducted in triplicate independently for each sample to minimize random PCR bias. Amplicons were purified by using GFX™ PCR DNA and Gel Band Purification Kit by following the manufacturer’s protocol (Amersham Biosciences, UK) and qualities measured by a Nanodrop spectrophotometer. Finally, a total of 20 libraries were labeled with a unique oligonucleotide barcode and high-throughput sequencing of Illumia MiSeq technology was used to obtain nucleotide sequences.
MiSeq sequencing data processing
The original data were processed by the following steps: firstly, merged two pair ends into one read by using FLASH-1.2.8 (Magoč and Salzberg 2011); secondly, split assembled data into individual samples according to the unique barcode via fastx-toolkit software (http://hannonlab.cshl.edu/fastx_toolkit); thirdly, filtered out low quality components of combined reads through the command of “split_libraries” under QIIME software and assigned to each library (Caporaso et al. 2012), the filtering criterion was set as “-s 15 -k -a 6 -r -l 150 -b 12 -M 5 -e 0”. Finally, raw reads were obtained after these steps.
Raw nucleotide sequences from each library were processed using the FrameBot function under Ribosomal Database Project (RDP) functional gene (Fungene) (http://fungene.cme.msu.edu) for further conduct dereplication, chimera checking, detecting, and frameshift correcting (Fish et al. 2013). Finally, 8980 sequences for per clone library were obtained after a series of tests, and combined into a single file as input to the QIIME for clustering and OTU picking based on 90% similarity criterion. The representative OTUs were analyzed by MG-RAST (Glass et al. 2010) and local blast for taxonomy information.
Raw reads were deposited in the European Nucleotide Short Read Archive (SRA) under accession PRJEB12432, sequence accession numbers ERS1118546–ERS1118565.
Quantification real-time PCR for the nifH gene
To quantify the abundance of nifH gene in each acidic forest soil, the qPCR assays were performed on an ABI 7000 Sequence detection system (Applied Biosystems, CA) using SYBR green method. The standard plasmid with a known copy number was made by a positive sequenced clone with primer set of IGK and DVV. Standard curves were generated by a 10 serial of dilution of the standard plasmid. Triplicate reactions (20 μl volume per reaction) were performed independently, including 1 μl of DNA template (10~50 ng DNA as templates), 1 μl of each primer (10 μM), and 10 μl of SYBR-Green PCR Master Mix (Applied Biosystems, CA). The reaction cycling conditions were in accordance with the PCR procedures described above. Gene copy numbers of standard plasmid dilution were calculated by original plasmid concentration measured by Nanodrop, and applied into the equation: quantities in gene copies/μl = (concentration ng/μl × 6.022 × 1023) / (length×1 × 109 × 660). The R2 value and amplification efficiency were 0.996 and 88.59%, respectively, suggesting the results were reliable. The gene abundance was calculated to gene copies/g dry soil based by taking into the water contents in these soil samples.
Statistical data analyses
Stack bar charts for delineating taxonomic profiles of nifH community of all libraries were originated from QIIME-based files, and visualized by Origin8 and Adobe illustrator software. Mantel Test analysis implemented in QIIME was conducted to analyze the correlation of physicochemical parameters with UniFrac and Bray-Curtis distance matrices from beta diversity analysis. Principal Coordinate Analysis (PCoA) on depicting the dissimilar relationship of samples was conducted in four categories of forest type, depth, site, and season based on both phylogenetic and non-phylogenetic distance matrix methods. Pearson correlation analysis between physicochemical parameters and diversity, abundance properties was conducted in R software. Statistical analysis of alpha diversity was conducted in R software by ANOVA.
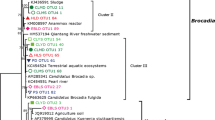
In order to display the community clearly, the phylogenetic tree of the 50 most abundant OTUs were constructed by MEGA 6 with references from GenBank, and visualized by iTOL online (Letunic and Bork 2007) (http://itol.embl.de/).
Results
Characteristics of the forest soils
Soil properties of 11 physicochemical characteristics including pH, organic matter, total N, total K, ammonium-N, nitrate-N, hydrolyzed N, exchangeable Al3+, SO42−, moisture content, and available P were described previously (Meng et al. 2016). Briefly, the pH values of these soils were very acidic ranging from 3.56 to 4.66 and they can be defined as strongly acidic soils (USDA 1999). The organic matter, total nitrogen, ammonium-N, nitrate-N, and moisture contents were significantly higher in natural forest than re-vegetated forest at Nanling Nature Reserve (p < 0.05). Soil properties also varied between the two depth layers. In natural forest, lower soil layer had significantly lower total N, hydrolyzed N, available P, NH4+-N, NO3−-N, organic matter, and soil moisture content, but relatively higher pH value (p < 0.05) comparing with the surface soil layer. However, soil characteristics did not show significant difference between the two layers in re-vegetated forest. These parameters were higher in winter than in summer. The detailed information is shown in Fig. S1.
Sequencing statistics and alpha diversity estimates
A total of 179,600 reads remained after a series of tests and 8980 sequences on average for each sample were retrieved and used for further statistical analysis (Table 1). A total of 2907 OTUs were obtained based on 90% similarity. The coverage value for all samples ranged from 0.97 to 0.99 (Table 1), suggesting that a deep sequencing was achieved in this study, and the results verified by rarefaction curves for all samples showed saturation, indicating an adequate assessment of the microbial community composition in each sample (Fig. S2). The alpha diversity index of Chao1 ranged from 606.33 to 1062.75 of surface layer was significantly higher than that of lower layer, which ranged from 210.77 to 756.7. The observed OTU value of 388–640 in surface layer was higher than 124–485 in the lower layer. Also, the Shannon value in the surface (5.54–6.52) was higher than in the lower layer (2.37–5.59). The other diversity indexes include PD_whole_tree and Simpson values also showed a similar trend. On the contrary, no significant difference was observed between winter and summer.
Community composition and phylogeny of diazotrophs
Taxonomic composition on the phylum level in each sample is shown (Fig. 1), including Methanomicrobia (< 0.14%), Cyanobacteria (< 3.16%), Proteobacteria (α-, β-, γ-, and δ-Proteobacteria) (87.62%–97.96%), Firmicutes (< 7.7%), Spirochaetes (< 0.16%), Verrucomicrobia (< 3.46%), and other minor phyla (1.51–5.15%, sequences < 1%). Among the group of Proteobacteria, the α-Proteobacteria and δ-Proteobacteria were the most predominant classes, while the β-Proteobacteria and γ-Proteobacteria were the minor components. Viewing the community of different forest types, natural forest was dominated by α-Proteobacteria especially the Bradyrhizobium, while re-vegetated forest was mainly by δ-Proteobacteria. In natural forests, the community structure showed a slight difference specifically at the highest elevation (HLS and HLD) with more γ-Proteobacteria than the other two sites. While in re-vegetated forests, there were apparent differences in the ratio of each phylum between the young (7 years) and matured forests (27 years). No significant difference could be detected in community composition between winter and summer, but significant difference was observed between the two depth layers at any of the same site.
In order to reduce the excessive complexity in analysis at the genus level, the first 50 most dominant core OTUs with coverage from 57.7 to 99.1% in these samples were used for further analysis (Fig. S3). A phylogenetic tree was constructed based on deduced amino acid sequences of the core OTUs and related known species sequences downloaded from GenBank using NJ method to form three major clusters (Fig. 2). Cluster I includes α-, β- and γ-Proteobacteria related to nitrogen fixers with Mo-dependent nitrogenase, closely affiliated with sequences from Bradyrhizobium, Rhodopseudomonas, Bacillus, etc. Cluster III was more related to sulfate-reducing bacteria of Desulfovibrio of δ-Proteobacteria. Sequences in cluster IV were closely related to species of protochlorophy II or chlorophy II biosynthesis. Overall, well-known nitrogen fixers from various clusters were detected and recovered in the samples of this study. Based on the taxonomy composition on genus level (Fig. 3), Bradyrhizobium was the most dominant genus of the core community, accounting for 45.96% of the community; Methylobacterium, 12.77% of the core community; Desulfovibrio, 25.66% of dominant community; and the other genera including Rhodopseudomonas, Acidiphilium, Asaia, Rhodovulm, Gluconacetobacter, Thiocapsa, Cynechococcus, Panibacillus, Bacillus, Diplosphaera and Pelomonas for < 2% of the main communities.
Beta diversity of diazotrophs in acidic forest soils
Three different methods, phylogenetic unweighted UniFrac and weighted UniFrac and non-phylogenetic Bray-Curtis distance matrices, were used to evaluate the beta diversity, but similar results of the Principal Coordinate Analysis (PCoA) were obtained, yielding a confirmation on the diversity trends in forest soils. There were four categories: forest type, soil depth, site, and season for further analysis. In details, the first principal (PC1) plus the second principal (PC2) accounted for only 29.14% of the total contribution based on Unweighted UniFrac method (Fig. 4), but PC1 added PC2 made a 77.82% contribution in weighted UniFrac method (Fig. S4), and 46.56% contribution of PC1 and PC2 were made by Bray-Curtis method (Fig. S5). A similar trend was obtained from different methods. More specifically, the natural forest soil samples and the re-vegetated forest soil samples were clustered into separate groups based on forest types (Fig. 4a) and soil depths (Fig. 4b); but no obvious differentiation among the five sites (Fig. 4c); and no seasonal differences were detected in our samples based on all three methods (Figs. 4d, S4 and S5). Correspondingly, the PCoA analysis results of total taxonomic community, the impacting factors on beta diversity of four categories were similar to the distribution of total community.
Abundance of diazotrophs
The abundance of diazotrophs quantified with nifH gene in these forest soils is shown (Fig. 5). In details, the gene copy numbers were from 1.03 × 107 to 7.20 × 107 per gram of dry soil in the surface layer and from 8.84 × 105 to 1.82 × 107 per gram of dry soil in the lower layer with significant difference between the two layers (t test, p < 0.05) (Table S2). For forest type, the quantities of gene abundance ranged from 105 to 107 in natural forest and 106 to 107 gene copies per gram of dry soil in re-vegetated forest. There was no significant difference observed between the two types of forests, the abundance in the surface layer showed an increase with the elevation while an opposite trend in the lower depth layer was detected in natural forests. The gene copies in young forest (7 years) were lower than those in matured forest (27 years) for re-vegetated forests. According to seasons, the gene abundance ranged from 8.84 × 105 to 4.75 × 107 gene copies per gram of dry soil in winter were slightly higher than that in summer during natural forests, while had a reverse trend in re-vegetated forests.
Discussion
Biological dinitrogen fixation is carried out exclusively by N2-fixing bacteria and archaea, a very important driver of primary productivity. Tracking the biomarker of nifH gene directly offers a better insight into N2 fixation capacity in forest soils and under different management conditions. In our study, high-throughput sequencing techniques were applied instead of classical cultivation approaches and clone libraries to detect the deeper community structure and abundance of N2 fixers in subtropical forest soils at Nanling National Nature Reserve, a subtropical forest ecosystem. The PCR primer set of IGK and DVV was used to detect the diazotrophs community and diversity with good coverage (97–99%) and high diversity obtained, indicating the effectiveness and efficiency of these primers for the soil samples of this study. The next generation sequencing method was used successfully to provide reliable results in analysis of community composition and diversity of diazotrophs compared to traditional clone library method.
Community structures of diazotrophic microorganisms
According to the study, Proteobacteria was the most dominant phylum in the N2 fixation community of acidic forest soils, including α-Proteobacteria, β-Proteobacteria, γ-Proteobacteria, and δ-Proteobacteria. This was not surprising, especially for the class of α-Proteobacteria with occurrence from aquatic to terrestrial ecosystems (Collavino et al. 2014; Flores-Mireles et al. 2007; Jing et al. 2015; Turk-Kubo et al. 2014; Wang et al. 2013). The most abundant genera of diazotrophic bacteria were Bradyrhizobium, Methylobacterium, and Desulfovibrio. In this study, Bradyrhizobium were found to be the most dominant type of N2 fixers, contributing up to 45% of the core community. Bradyrhizobium are very common slow-growing bacteria, and known to have a better adaption and tolerance to acidic conditions compared to the other genera of rhizobia (Graham 1992; Koponen et al. 2003). In addition, many Bradyrhizobium are able to tolerate alternating cycles of dry and wet as well as anoxic conditions during the season transitions (Koponen et al. 2003), playing an important role in tropical and subtropical forests. Interestingly, in one of our previous studies at this site on denitrification, Bradyrhizobium were also found to be one of the most abundant denitrifiers in samples based on both nirK gene and nosZ gene (Meng et al. 2017), indicating that Bradyrhizobium plays an essential role in the nitrogen cycle in forest system. In other studies, the involvement of Bradyrhizobium in both N2 fixation and denitrification was also reported (Bedmar et al. 2005; Sánchez et al. 2011). In our study, the dominance of Bradyrhizobium in both N2 fixing and denitrification may imply its importance for both processes. Methylobacterium spp. are important methylotrophs in soil and on surface of leaves and other plant parts. This is probably a reason for the relative higher abundance of Methylobacterium in surface layer than lower layer of these samples. Methylobacterium are facultative methylotrophs with ability to grow by reducing carbon compounds containing one or more carbon atoms (Chistoserdova et al. 2003), e.g., methylamine; methanol; and C2, C3, and C4 compounds; including the methanol derived from the leaves of plants (Fedorov et al. 2011; Mizuno et al. 2012). While Desulfovibrio, as a member of δ-Proteobacteria, use hydrogen, organic acids or alcohols as electron donors for sulfate reduction as a typical anaerobic sulfate-reducing bacteria (Heidelberg et al. 2004). In our study, the genus Desulfovibrio was mostly observed in re-vegetated soils and had a relative high abundance in lower layer regardless of the forest ages. This is in agreement with the reports that Desulfovibrio sp. possesses unique alternative non-heme iron proteins with superoxide reductase, playing important role in the nitrogen cycle (Radajewski et al. 2002; Stacheter et al. 2013).
There was no sequence detected in Cluster II of the core community. This is not surprising because diazotrophs of Cluster II contain anfH gene and nitrogenase from some archaea, which are below detection or have a very low presence in soils specifically nature environment (Zehr et al. 2003). Diazotrophs from Cluster IV were recovered from our study, involving in bacteriachlorophy II and chlorophy II biosysthesis genes (nifH-like) to all phototrophs, confirming the pair of PCR primers had a good coverage (Gaby and Buckley 2012) and previous studies showed that Cluster IV are associated with plant roots (Flores-Mireles et al. 2007).
Diversity of diazotrophic microorganisms
A significant difference of α-diversity was observed in diazotrophs between the surface layer and lower layer, with higher diversity and richness in the surface layer than the lower depth layer (Table S1). These results were positively correlated with organic matters and nitrogen-related parameters after statistical variation test (p < 0.05). On one hand, the nutrients in surface layer accumulated by litter deposition to provide dissolved nutrients and enough oxygen for diazotrophic microorganism growth, on the other hand, root-nodule symbiosis diazotrophs and non-symbiotic free-living N2 fixers require different habitats. According to the forest types, the diversity indexes containing PD_whole_tree, Chao1, and observed_species were slightly higher in re-vegetated forest than the natural forest (Table S1). This observation fits with the report that the diversity of Bradyrhizobium decreased after deforestation of the secondary forests (Ormeño-Orrillo et al. 2012). The diversity positively related to the parameters of organic matter, hydrolyzed N, NH4+-N, NO3−-N, and moisture content in natural forest (p < 0.05) in both summer and winter, but no significant correlation between the diversity and physiochemical parameters in re-vegetated forest soils (Chan et al. 2008; Nüsslein and Tiedje 1999; Ormeño-Orrillo et al. 2012). In light of locations, the genetic diversity of nifH was reduced along the elevation (HLS/HLD > PS/PD > EBLS/EBLD) in natural forest, nutrients (nitrogen-related) with vegetation type affect the diversity. Moreover, in re-vegetated forest, the diversity value was higher in matured forest than young forest, high level of moisture content decreased the diversity, probably due to anthropogenic activities (Jing et al. 2015). In contrast to other studies, there was no significant correlation detected between summer and winter (Table S1), keeping consistent of community composition.
For β-diversity, the clear differences of distribution and principal clustering in PCoA graph showed the variation between different depths, forest types, and sites (Fig. 5, Fig. S4, Fig. S5). It is also consistent with previous studies that the availability of O2, which changes the bacterial community of anaerobic bacteria and aerobic bacteria with depth (Andersen et al. 2013). Vegetation types were also the impact factors to the soil community structure (Li et al. 2014). The pH rather than temperature, plays an important role in shaping community structure like other studies (Fierer & Jackson 2006; Griffiths et al. 2011).
Environmental parameters to the abundance of diazotrophs
As mentioned above, environmental parameters were correlated to community diversity, but no significant correlation could be observed between environmental parameters and abundance of N2 fixers overall. The reasons for this situation probably are because: (1) the low pH as described in forest soil was strongly acidic to inhibit N2 fixation and reduced the abundance (Lauber et al. 2009; Li et al. 2014); (2) vegetation type of plant species is important in influencing the rate of N2 fixation and abundance (Nüsslein and Tiedje 1999; Ormeño-Orrillo et al. 2012); (3) anthropogenic factor of human impact on the abundance of N2 fixers; and (4) different soil types in natural forest (yellow soil) and re-vegetate forest (red soil) lead to different abundances. Overall, several factors mixed together may cause the abundance indirectly. Noticeably, the gene copy numbers at the surface were significantly higher than that at deep layer (p = 0.007), implying that nutrients and oxygen concentration strongly associated with depths and are consistent with other studies (Mergel et al. 2001). Specifically, Bradyrhizobium was dominant in most of the samples at the surface and it was known to have close interaction with plant’s root through nodule formation (Gully et al. 2013; McDermoti and Graham 1990). Interestingly, the abundance of diazotrophs was higher in winter than summer in natural forest but had an opposite trend in re-vegetated forest. Besides the different dominant types of vegetation favoring different types of N2 fixer, soil texture (yellow soil above 700 m and red soil below 700 m) probably was another one important factor that caused different results (e Silva et al. 2011; Gan et al. 2016). Considering the community and abundance together, forest type, depth, and vegetation species together may be cofactors on diazotrophs, no single factor was found to directly correlate with the abundance.
In summary, this study investigated the community structure, diversity, and abundance on the diazotrophs of the forest soils through the application of high-throughput sequencing on the functional nifH gene. In the acidic forest soils, the N2-fixation communities included Methanomicrobia, Cyanobacteria, Firmicutes, Proteobacteria, Spirochaetes, Verrucomicrobia, and other minor phylum. Specifically, α-Proteobacteria was the dominant class including orders of Rhizobiales, Rhodobacterales, and Sphingomonadales. The genus of Desulfovibrio belonging to δ-Proteobacteria was dominant in re-vegetated forest, not natural forest. The community structure was very stable and no significant differences between seasons. Significant variations were detected between depths, forest type, and also different locations. The abundance was higher in surface than deep layer, positively correlated to physical-chemical parameters of organic matter, total N, NH4+-N, NO3−-N, and moisture content. This study provides valuable reference for future diazotrophs in subtropical forest soil through the application of high-throughput sequencing.
References
Andersen R, Chapman SJ, Artz RRE (2013) Microbial communities in natural and disturbed peatlands: a review. Soil Biol Biochem 57:979–994. https://doi.org/10.1016/j.soilbio.2012.10.003
Bedmar E, Robles E, Delgado M (2005) The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem Soc Trans 33:141–144. https://doi.org/10.1042/BST0330141
Bishop PE, Hawkins ME, Eady R (1986) Nitrogen fixation in molybdenum-deficient continuous culture by a strain of Azotobacter vinelandii carrying a deletion of the structural genes for nitrogenase (nifHDK). Biochem J 238:437–442. https://doi.org/10.1042/bj2380437
Boyd ES, Hamilton TL, Peters JW (2011) An alternative path for the evolution of biological nitrogen fixation. Front Microbiol 2:205. https://doi.org/10.3389/fmicb.2011.00205
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. https://doi.org/10.1038/ismej.2012.8
Chan OC, Casper P, Sha LQ, Feng ZL, Fu Y, Yang XD, Ulrich A, Zou XM (2008) Vegetation cover of forest, shrub and pasture strongly influences soil bacterial community structure as revealed by 16S rRNA gene T-RFLP analysis. FEMS Microbiol Ecol 64:449–458. https://doi.org/10.1111/j.1574-6941.2008.00488.x
Chisnell JR, Premakumar R, Bishop PE (1988) Purification of a second alternative nitrogenase from a nifHDK deletion strain of Azotobacter vinelandii. J Bacteriol 170:27–33
Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME (2003) Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol 185:2980–2987. https://doi.org/10.1128/JB.185.10.2980-2987.2003
Chowdhury SP, Schmid M, Hartmann A, Tripathi AK (2009) Diversity of 16S-rRNA and nifH genes derived from rhizosphere soil and roots of an endemic drought tolerant grass, Lasiurus sindicus. Eur J Soil Biol 45:114–122. https://doi.org/10.1016/j.ejsobi.2008.06.005
Cleveland CC, Townsend AR, Schimel DS, Fisher H, Howarth RW, Hedin LO, Perakis SS, Latty EF, Von Fischer JC, Elseroad A, Wasson MF (1999) Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Glob Biogeochem Cycles 13:623–645. https://doi.org/10.1029/1999GB900014
Collavino MM, Tripp HJ, Frank IE, Vidoz ML, Calderoli PA, Donato M, Zehr JP, Aguilar OM (2014) nifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2-fixing community dynamics. Environ Microbiol 16:3211–3223. https://doi.org/10.1111/1462-2920.12423
Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R (2012). Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13:162. https://doi.org/10.1186/1471-2164-13-162
e Silva MCP, Semenov AV, van Elsas JD, Salles JF (2011) Seasonal variations in the diversity and abundance of diazotrophic communities across soils. FEMS Microbiol Ecol 77:57–68. https://doi.org/10.1111/j.1574-6941.2011.01081.x
Fedorov DN, Doronina NV, Trotsenko YA (2011) Phytosymbiosis of aerobic methylobacteria: new facts and views. Microbiology 80:443–454. https://doi.org/10.1134/S0026261711040047
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103:626–631. https://doi.org/10.1073/pnas.0507535103
Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4:291. https://doi.org/10.3389/fmicb.2013.00291
Flores-Mireles AL, Winans SC, Holguin G (2007) Molecular characterization of diazotrophic and denitrifying bacteria associated with mangrove roots. Appl Environ Microbiol 73:7308–7321. https://doi.org/10.1128/AEM.01892-06
Fujita Y, Takahashi Y, Chuganji M, Matsubara H (1992) The nifH-like (frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol 33:81–92
Gaby JC, Buckley DH (2012) A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS One 7:e42149. https://doi.org/10.1371/journal.pone.0042149
Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP, Asner GP, Cleveland CC, Green PA, Holland EA, Karl DM, Michaels AF, Porter HJ, Vöosmarty CJ (2004) Nitrogen cycles: past, present, and future. Biogeochemistry 70:153–226. https://doi.org/10.1007/s10533-004-0370-0
Gan XH, Zhang FQ, Gu JD, Guo YD, Li ZQ, Zhang WQ, Xu XY, Zhou Y, Wen XY, Xie GG (2016) Differential distribution patterns of ammonia-oxidizing archaea and bacteria in acidic soils of Nanling National Nature Reserve forests in subtropical China. Antonie Van Leeuwenhoek 109:237–251. https://doi.org/10.1007/s10482-015-0627-8
Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F (2010) Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc 2010(1):pdb. prot5368. https://doi.org/10.1101/pdb.prot5368
Graham PH (1992) Stress tolerance in Rhizobium and Bradyrhizobium, and nodulation under adverse soil conditions. Can J Microbiol 38:475–484. https://doi.org/10.1139/m92-079
Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS (2011) The bacterial biogeography of British soils. Environ Microbiol 13:1642–1654. https://doi.org/10.1111/j.1462-2920.2011.02480.x
Gully D, Sadowsky MJ, Giraud E, Xu L, Chaintreuil C, Gargani D (2013) Photosynthetic Bradyrhizobium sp. strain ORS285 is capable of forming nitrogen-fixing root nodules on soybeans (Glycine max). Appl Environ Microbiol 79:2459–2462. https://doi.org/10.1128/AEM.03735-12
Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM (2004) The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nature Biotechnol 22:554–559. https://doi.org/10.1038/nbt959
Homer MJ, Dean DR, Roberts GP (1995) Characterization of the γ protein and its involvement in the metallocluster assembly and maturation of dinitrogenase from Azotobacter vinelandii. J Biol Chem 270:24745–24752. https://doi.org/10.1074/jbc.270.42.24745
Jing HM, Xia XM, Liu HB, Zhou Z, Wu C, Nagarajan S (2015) Anthropogenic impact on diazotrophic diversity in the mangrove rhizosphere revealed by nifH pyrosequencing. Front Microbiol 6:1172. https://doi.org/10.3389/fmicb.2015.01172
Koponen P, Nygren P, Domenach AM, Le Roux C, Saur E, Roggy JC (2003) Nodulation and dinitrogen fixation of legume trees in a tropical freshwater swamp forest in French Guiana. J Trop Ecol 19:655–666. https://doi.org/10.1017/S0266467403006059
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. https://doi.org/10.1128/AEM.00335-09
Letunic I, Bork P (2007) Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. https://doi.org/10.1093/nar/gkw290
Li H, Ye D, Wang X, Settles ML, Wang J, Hao Z, Zhou L, Dong P, Jiang Y, Ma ZS (2014) Soil bacterial communities of different natural forest types in Northeast China. Plant Soil 383:203–216. https://doi.org/10.1007/s11104-014-2165-y
Liao X, Inglett PW (2014) Dynamics of periphyton nitrogen fixation in short-hydroperiod wetlands revealed by high-resolution seasonal sampling. Hydrobiologia 722:263–277. https://doi.org/10.1007/s10750-013-1709-0
Lincoln NK, Vitousek P (2016) Nitrogen fixation during decomposition of sugarcane (Saccharum officinarum) is an important contribution to nutrient supply in traditional dryland agricultural systems of Hawai'i. Int J Agric Sustain 14:214–230. https://doi.org/10.1080/14735903.2015.1071547
Lu RK (2000) Methods of agriculture chemical analysis. China Agriculture Scientech Press, Beijing
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
McDermoti TR, Graham PH (1990) Competitive ability and efficiency in nodule formation of strains of Bradyrhizobium japonicum. Appl Environ Microbiol 56:3035–3039
Mehta MP, Baross JA (2006) Nitrogen fixation at 92 °C by a hydrothermal vent archaeon. Science 314:1783–1786. https://doi.org/10.1126/science.1134772
Meng H, Wang YF, Chan HW, Wu RN, Gu J-D (2016) Co-occurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in subtropical acidic forest soils. Appl Microbiol Biotechnol 5:1–13. https://doi.org/10.1007/s00253-016-7585-6
Meng H, Wu RN, Wang YF, Gu J-D (2017) A comparison of denitrifying bacterial community structures and abundance in acidic soils between natural forest and re-vegetated forest of Nanling nature reserve in southern China. J Environ Manag 198:41–49. https://doi.org/10.1371/journal.pone.0042149.t002
Mergel A, Schmitz O, Mallmann T, Bothe H (2001) Relative abundance of denitrifying and dinitrogen-fixing bacteria in layers of a forest soil. FEMS Microbiol Ecol 36:33–42. https://doi.org/10.1111/j.1574-6941.2001.tb00823.x
Mizuno M, Yurimoto H, Yoshida N, Iguchi H, Sakai Y (2012) Distribution of pink-pigmented facultative methylotrophs on leaves of vegetables. Biosci Biotechnol Biochem 76:578–580. https://doi.org/10.1271/bbb.110737
Moisander PH, Shiue L, Steward GF, Jenkins BD, Bebout BM, Zehr JP (2006) Application of a nifH oligonucleotide microarray for profiling diversity of N2-fixing microorganisms in marine microbial mats. Environ Microbiol 8:1721–1735. https://doi.org/10.1111/j.1462-2920.2006.01108.x
Moseman-Valtierra S, Levin LA, Martin RM (2016) Anthropogenic impacts on nitrogen fixation rates between restored and natural Mediterranean salt marshes. Mar Ecol 37:370–379. https://doi.org/10.1111/maec.12289
Nomata J, Mizoguchi T, Tamiaki H, Fujita Y (2006) A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis reconstitution of chlorophyllide a reductase with purified x-protein (bchX) and yz-protein (bchY-bchZ) from Rhodobacter capsulatus. J Biol Chem 281:15021–15028. https://doi.org/10.1074/jbc.M601750200
Nüsslein K, Tiedje JM (1999) Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl Environ Microbiol 65:3622–3626
Ormeño-Orrillo E, Rogel-Hernández MA, Lloret L, López-López A, Martínez J, Barois I, Martínez-Romero E (2012) Change in land use alters the diversity and composition of Bradyrhizobium communities and led to the introduction of Rhizobium etli into the tropical rain forest of Los Tuxtlas (Mexico). Microb Ecol 63:822–834. https://doi.org/10.1007/s00248-011-9974-9
Poly F, Ranjard L, Nazaret S, Gourbière F, Monrozier LJ (2001) Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl Environ Microbiol 67:2255–2262. https://doi.org/10.1128/AEM.67.5.2255-2262.2001
Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, Nedwell DB, Prosser JI, Murrell JC (2002) Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331–2342. https://doi.org/10.1099/00221287-148-8-2331
Raymond J, Siefert JL, Staples CR, Blankenship RE (2004) The natural history of nitrogen fixation. Mol Biol Evol 21:541–554. https://doi.org/10.1093/molbev/msh047
Sánchez C, Tortosa G, Granados A, Delgado A, Bedmar EJ, Delgado MJ (2011) Involvement of Bradyrhizobium japonicum denitrification in symbiotic nitrogen fixation by soybean plants subjected to flooding. Soil Biol Biochem 43:212–217. https://doi.org/10.1016/j.soilbio.2010.09.020
Souillard N, Magot M, Possot O, Sibold L (1988) Nucleotide sequence of regions homologous to nifH (nitrogenase Fe protein) from the nitrogen-fixing archaebacteria Methanococcus thermolithotrophicus and Methanobacterium ivanovii: evolutionary implications. Mol Evol J 27:65–76. https://doi.org/10.1007/BF02099731
Sparks DL, Page AL, Helmke PA, Loeppert RH, Soltanpour PN, Tabatabai MA, Johnston CT, Sumner ME (1996). Methods of soil analysis. part 3 - chemical methods. Wisconsin, USA.
Stacheter A, Noll M, Lee CK, Selzer M, Glowik B, Ebertsch L, Mertel R, Schulz D, Lampert N, Drake HL (2013) Methanol oxidation by temperate soils and environmental determinants of associated methylotrophs. ISME J 7:1051–1064. https://doi.org/10.1038/ismej.2012.167
Staples CR, Lahiri S, Raymond J, Von Herbulis L, Mukhophadhyay B, Blankenship RE (2007) Expression and association of group IV nitrogenase nifD and nifH homologs in the non-nitrogen-fixing archaeon Methanocaldococcus jannaschii. Bacteriol J 189:7392–7398
Steppe T, Paerl HW (2005) Nitrogenase activity and nifH expression in a marine intertidal microbial mat. Microb Ecol 49:315–324. https://doi.org/10.1007/s00248-004-0245-x
Turk-Kubo KA, Karamchandani M, Capone DG, Zehr JP (2014) The paradox of marine heterotrophic nitrogen fixation: abundances of heterotrophic diazotrophs do not account for nitrogen fixation rates in the eastern tropical South Pacific. Environ Microbiol 16:3095–3114. https://doi.org/10.1111/1462-2920.12346
US Department of Agriculture (1999) Soil taxonomy—a basic system of soil classification for making and interpreting soil surveys. USDA, Washington.
Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, Schindler DW, Schlesinger WH, Tilman DG (1997) Human alteration of the global nitrogen cycle: sources and consequences. Ecol Appl 7:737–750. https://doi.org/10.1890/1051-0761(1997)007[0737:HAOTGN]2.0.CO;2
Wang Q, Quensen JF, Fish JA, Lee TK, Sun Y, Tiedje JM, Cole JR (2013) Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. MBio 4:e00592–e00513. https://doi.org/10.1128/mBio.00592-13
Yamada A, Inoue T, Noda S, Hongoh Y, Ohkuma M (2007) Evolutionary trend of phylogenetic diversity of nitrogen fixation genes in the gut community of wood-feeding termites. Mol Ecol 16:3768–3777. https://doi.org/10.1111/j.1365-294X.2007.03326.x
Zehr JP, Jenkins BD, Short SM, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5:539–554. https://doi.org/10.1046/j.1462-2920.2003.00451.x
Funding
This study was funded by the National Natural Science Foundation of China (grant no. 31470562 to YFW), a Hong Kong PhD Fellowship (HM), and RGC GRF grant no. 701913 (J-DG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1199 kb)
Rights and permissions
About this article
Cite this article
Meng, H., Zhou, Z., Wu, R. et al. Diazotrophic microbial community and abundance in acidic subtropical natural and re-vegetated forest soils revealed by high-throughput sequencing of nifH gene. Appl Microbiol Biotechnol 103, 995–1005 (2019). https://doi.org/10.1007/s00253-018-9466-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9466-7