Abstract
The d-xylose oxidative pathway (XOP) has recently been employed in several recombinant microorganisms for growth or for the production of several valuable compounds. The XOP is initiated by d-xylose oxidation to d-xylonolactone, which is then hydrolyzed into d-xylonic acid. d-Xylonic acid is then dehydrated to form 2-keto-3-deoxy-d-xylonic acid, which may be further dehydrated then oxidized into α-ketoglutarate or undergo aldol cleavage to form pyruvate and glycolaldehyde. This review introduces a brief discussion about XOP and its discovery in bacteria and archaea, such as Caulobacter crescentus and Haloferax volcanii. Furthermore, the current advances in the metabolic engineering of recombinant strains employing the XOP are discussed. This includes utilization of XOP for the production of diols, triols, and short-chain organic acids in Escherichia coli, Saccharomyces cerevisiae, and Corynebacterium glutamicum. Improving the d-xylose uptake, growth yields, and product titer through several metabolic engineering techniques bring some of these recombinant strains close to industrial viability. However, more developments are still needed to optimize the XOP pathway in the host strains, particularly in the minimization of by-product formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cellulosic, lignocellulosic, and aquatic biomass are attractive biorenewable resources for the production of valuable compounds (Naik et al. 2010; Singh et al. 2011). Among which, lignocellulosic biomass was regarded as the most promising alternative feedstock due to its abundance and lower carbon footprint (Blaschek et al. 2010; Naik et al. 2010). Regardless of saccharification or hydrolysis method, lignocellulosic biomass hydrolysates contain fermentable sugars with d-glucose as the most abundant followed by d-xylose (Cheng and Timilsina 2011; Kobayashi and Fukuoka 2013). Conversion of d-glucose to biofuels or high-value chemicals has been employed in recombinant microorganisms, e.g., biosynthesis of 1,4-butanediol, isobutanol, and 1,3-propanediol (Nakamura and Whited 2003; Atsumi et al. 2010; Yim et al. 2011). This subject has been thoroughly discussed in another review by Rabinovitch-Deere et al. (2013). A more viable and sustainable route of biomass conversion would be the simultaneous utilization of d-glucose and d-xylose. This is a key factor in the technological development of xylose utilization in recombinant microorganisms (Dumon et al. 2012).
The metabolism of d-xylose has been well studied in bacteria and fungi (Karhumaa et al. 2007; Bettiga et al. 2008; Dumon et al. 2012). There are two distinct and well-known pathways for d-xylose degradation: the isomerase (XIP) and oxo-reductive pathways (ORP). The XIP is typically found in bacteria wherein d-xylose is isomerized to d-xylulose, phosphorylated into d-xylulose-5-phosphate, which then enters the pentose phosphate pathway (PPP) for the synthesis of cellular metabolites (Fig. 1). On the other hand, the ORP is more common in fungi, wherein d-xylose is reduced to d-xylitol, then oxidized to d-xylulose. Similar to the XIP, d-xylulose is phosphorylated before it enters the PPP (Fig. 1).
Several studies have focused on enhancing the efficiency of d-xylose utilization by engineering the XIP or ORP in recombinant microorganisms. Despite the success, there are limitations that need to be addressed. One major issue in the ORP of engineered yeast is xylitol accumulation, which is believed to be a consequence of co-factor imbalance in the host cell (Matsushika et al. 2009). This can be attributed to the co-factor preference of xylose reductase towards NADPH and xylitol dehydrogenase towards NAD (Fig. 1). On the other hand, the XIP forms xylulose through a one-step isomerization reaction which does not require redox co-factors. However, the key challenge in the XIP is the slow xylose uptake/assimilation rate. In an earlier study, the performances of the XIP from Piromyces and the ORP from Scheffersomyces stipitis were compared by recruiting the appropriate genes in an isogenic yeast strain (Karhumaa et al. 2005, 2007; Bettiga et al. 2008). The host cell carries the following genetic modifications: overexpression of xylulokinase, deletion of native non-specific aldose reductase, and overexpression of oxidative PPP enzymes. All of these gene alterations were predetermined to be essential in achieving an industrially relevant rate of xylose uptake. The comparison showed that the ORP has a higher specific ethanol productivity as a result of a faster d-xylose consumption rate. However, XIP afforded higher product yield despite its incomplete d-xylose utilization.
d-xylose consumption in microorganisms through the XIP and ORP is continuously being optimized by various research groups and laboratories. Interestingly, recent studies have employed an alternative pathway for d-xylose metabolism called the xylose oxidative pathway (XOP). This review introduces the XOP, the essential genetic elements, and the natural regulatory mechanisms from its source microorganism. Advancements in the use of XOP in metabolic engineering and synthetic biology are also discussed, as well as its future prospects and impacts on the field of industrial biotechnology.
Xylose oxidative pathway in bacteria and archaea
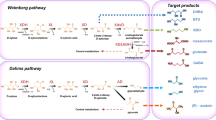
The XOP is found in most archaea and some bacterial and fungal strains. Its distinguishing feature from XIP and ORP is its non-phosphorylated intermediates (Fig. 1). In XOP, d-xylose is initially oxidized to d-xylonolactone in the presence of NAD(P)-dependent xylose dehydrogenase. d-Xylonolactone is subsequently hydrolyzed either spontaneously or by lactonases to form d-xylonic acid, which is further converted to 2-keto-3-deoxy-d-xylonic acid (KDX) by xylonate dehydratase. KDX may go through two separate routes: (a) further dehydration then oxidation to form α-ketoglutarate, which enters the tricarboxylic acid cycle, or by (b) aldol cleavage between carbon 3 and 4 to form pyruvate and glycolaldehyde, wherein both compounds can be assimilated for cellular metabolism (Fig. 1). The former route is known as the Weimberg pathway, while the latter describes the Dahms pathway (Weimberg 1961; Dahms 1974).
The XOP has several essential attributes which may be considered advantageous over the traditional XIP and ORP. One of its salient features is its relatively fewer enzymatic reaction steps involved in forming key glycolytic intermediates such as α-ketoglutarate for the Weimberg pathway and pyruvate and glycolate for the Dahms pathway (Fig. 1). In contrast, XIP and ORP require twice the number of reactions to convert d-xylose to pyruvate. Another key feature of XOP is its independence from other carbohydrate assimilative pathways, relatively simplifying the metabolic network when other carbon sources are present. On the other hand, XIP and ORP are interconnected to several metabolic/catabolic pathways such as the PPP, gluconeogenesis, and ribonucleotide synthesis. The complexity of such metabolic networks could pose as a huge challenge in improving the carbon yield. Finally, XOP reactions are more energetically favorable compared to XIP and ORP (Fig. 2). Thermodynamic favorability is indicated by how low (preferably negative value) the change in Gibbs energy (ΔrG) is for each enzymatic reaction (Flamholz et al. 2012). In the case of the Weimberg and Dahms pathways in XOP, each reaction step has a highly negative ΔrG values indicating favorable energetics.
Thermodynamic analysis of xylose metabolic pathway reaction steps towards ethanol production. Isomerase pathway step no. 1, isomerase; 2, xylulose kinase; 3, aldol reaction; 4, glyceraldehyde 3-phosphate dehydrogenase; 5, phosphoglycerate kinase; 6, phosphoglycerate mutase; 7, enolase; 8, pyruvate kinase; 9, pyruvate decarboxylase; 10, alcohol dehydrogenase. Oxo-reductive pathway step no. 1, reductase; 2, dehydrogenase; 3, xylulose kinase; 4, aldol reaction; 5, glyceraldehyde 3-phosphate dehydrogenase; 6, phosphoglycerate kinase; 7, phosphoglycerate mutase; 8, enolase; 9, pyruvate kinase; 10, pyruvate decarboxylase; 11, alcohol dehydrogenase. Weimberg pathway step no. 1, xylose dehydrogenase; 2, lactonase; 3, xylonate dehydratase; 4, 2-keto-3-deoxy-d-xylonate dehydratase; 5, semialdehyde dehydrogenase; 6, isocitrate dehydrogenase; 7, isocitrate lyase; 8, malate synthase; 9, malate dehydrogenase; 10, pyruvate decarboxylase; 11, alcohol dehydrogenase. Dahms pathway step no. 1, xylose dehydrogenase; 2, lactonase; 3, xylonate dehydratase; 4, aldolase; 5, pyruvate decarboxylase; 6, alcohol dehydrogenase
Xylose dehydrogenase activity was first reported in Pseudomonas (Lockwood and Nelson 1946). Members of this genus were capable of secreting d-xylonic acid into the culture media and displayed negative growth phenotypes on d-xylonic acid. Aside from Pseudomonas, xylose oxidation was also observed in other bacteria such as Gluconobacter oxydans (Buchert and Viikari 1988; Zhang et al. 2013) and in fungi Starmera quercuum (formerly Pichia quercuum) (Suzuki and Onishi 1973) and Trichoderma reesei (formerly Hypocrea jecorina) (Berghäll et al. 2007). Further exploration of this distinctive d-xylose metabolic pathway led to the discovery of the Weimberg and Dahms pathways (Weimberg 1961; Dahms 1974).
In the case of archaeal strains, the first observed xylose dehydrogenase activity was from Haloarcula marismortui (Johnsen and Schönheit 2004). Genome sequence analysis of H. marismortui did not show homologs for genes that catalyze d-xylose metabolism through XIP or ORP (Brouns et al. 2006). Furthermore, H. marismortui did not exhibit d-xylose isomerase or oxo-reductive enzyme activities. However, putative genes (homologs) that encode for the necessary enzymes of a functional Weimberg pathway were identified in its genome (Brouns et al. 2006).
The first detailed genetic analysis on the Weimberg pathway genes or gene clusters in bacteria was carried out in the genome sequences of Azospirillum brasilense and Caulobacter crescentus (also known as Caulobacter vibrioides) (Watanabe et al. 2006a, b, c; Stephens et al. 2007a). Since then, more archaeal and bacterial strains were discovered to metabolize d-xylose through the XOP including Haloferax volcanii (Johnsen et al. 2009), Sulfolobus solfataricus (Nunn et al. 2010), Sulfolobus acidocaldarius (Nunn et al. 2010), Arthrobacter nicotinovorans (Mihasan et al. 2013), and Pseudomonas taiwanensis (Köhler et al. 2015).
C. crescentus
Gram-negative C. crescentus is an oligotrophic bacterium that is widely found in freshwater environments. It has been used as a model microorganism for studies on prokaryotic cell cycle due to its ability to form two types of daughter cells during cell division: a flagellated “swarmer” cell and a “stalked” cell that has a protruding stem with an adhesive holdfast. The genome of C. crescentus has been sequenced and was found to contain several gene clusters that ensure survival during nutrient starvation or drastic changes in environmental conditions (Nierman et al. 2001). One notable finding during the annotation of the genome sequence is the absence of gene homologs for XIP and ORP. This indicated that d-xylose assimilation in C. crescentus operates through a different route. Transcriptomics analysis of C. crescentus when grown on d-xylose as sole carbon source revealed expression of genes coding for hydrolytic exoenzymes (xylosidases and hydrolases) and a gene cluster that was initially predicted to encode enzymes for the Weimberg pathway (Hottes et al. 2004). These genes were assigned to the locus tag CC_0823 to CC_0819 and were later confirmed as a d-xylose-inducible xylXABCD operon, encoding for the enzymes of a functional Weimberg pathway (Fig. 3) (Stephens et al. 2007a, b).
The xylB gene (locus tag CC_0821) from the xyl operon of C. crescentus was initially annotated as an oxidoreductase (Nierman et al. 2001), which carries conserved domains for short-chain dehydrogenase/reductase (SDR) family. Several studies have confirmed the physiological function of XylB for the catalysis of d-xylose oxidation with preference for NAD as its co-factor (Stephens et al. 2007a; Liu et al. 2012). The oxidation product (xylonolactone) then undergoes spontaneous lactone ring opening. Alternatively, enzyme catalyzed hydrolysis of xylonolactone is facilitated by a xylonolactonase encoded by the xylC gene. The activity of this enzyme was confirmed for d-xylonic acid production in Saccharomyces cerevisiae (Toivari et al. 2012; Nygård et al. 2014). As for other enzyme products of the xyl operon, the activities were initially predicted based on homology matches from available bioinformatics databases. For example, XylD has a high sequence similarity with the 6-phosphogluconate dehydratase involved in the Entner-Doudoroff pathway in E. coli (Carter et al. 1993; Stephens et al. 2007a) while XylA has homology with the α-ketoglutarate semialdehyde dehydrogenase from A. brasilense (Watanabe et al. 2006a, b; Stephens et al. 2007a). Additionally, XylX was hypothesized to catalyze the second dehydration step in the pathway. It has conserved domains which are characteristic of the fumarylacetoacetate hydrolase family (Stephens et al. 2007a). In the case of XylD, recent studies have confirmed that it belongs to the IlvD/EDD dehydratase family of enzymes which have Fe-S clusters (Andberg et al. 2016). It prefers d-xylonic acid as its substrate, but it also has considerable activity towards d-gluconic acid. Furthermore, crystallographic analysis showed that XylD has a homotetrameric quaternary structure (Rahman et al. 2016; Rahman et al. 2018). Despite the limited biochemical data for XylX, XylA, and XylD, these enzymes have been successfully employed (along with XylB and XylC) in the metabolic engineering of recombinant strains, as discussed in the succeeding sections (Meijnen et al. 2009; Radek et al. 2014; Tai et al. 2016; Zhao et al. 2017; Rossoni et al. 2018; Wasserstrom et al. 2018).
The regulation of the xyl operon was first described to be under the control of a xylose-inducible promoter Pxyl located upstream of the xylX gene (Meisenzahl et al. 1997). Expression of XylX was found to be induced by d-xylose and is not repressed by d-glucose, thereby showing no carbon catabolite repression in C. crescentus. The presence of the Pxyl sequence (or motif) was found upstream of the genes displaying increased expressions during growth on d-xylose (Hottes et al. 2004). It was later established that the xyl operon is controlled by the transcription factor XylR coded by the xylR gene (locus tag CC_3065) located more than 2.4 Mbp upstream of xylX (Meisenzahl et al. 1997; Stephens et al. 2007b). XylR, a member of the LacI family of transcription factors, binds to a predicted operator sequence located in the middle of the Pxyl sequence. Without d-xylose, XylR binds to the operator sequence and blocks the transcription of xyl genes. But in the presence of d-xylose, XylR interacts with d-xylose or any d-xylose-derivative molecule resulting in a decreased binding affinity of XylR to the operator sequence. This allows RNA polymerase to access the promoter site, leading to the transcription of the xyl genes (Stephens et al. 2007b).
H. marismortui
As mentioned above, XIP and ORP for xylose assimilation are predominant in bacteria and fungi. These two pathways are rarely found in microbes belonging to the Archaea domain, which suggests that archaeal organisms may employ a different pathway for d-xylose assimilation such as the XOP. The first report supporting this assumption describes the discovery of a novel xylose dehydrogenase in the halophilic archaeon H. marismortui (Johnsen and Schönheit 2004). It was initially observed that H. marismortui was capable of utilizing d-xylose for growth. However, neither d-xylose isomerase nor xylulokinase activity was detected (Johnsen and Schönheit 2004). Instead, xylose dehydrogenase activity was found which utilizes NAD and NADP as electron acceptors, with preference to the latter. Comparison of the amino acid sequences of the H. marismortui xylose dehydrogenase and C. crescentus XylB reveals two very different enzymes (Johnsen and Schönheit 2004; Stephens et al. 2007a). While the xylose dehydrogenase of C. crescentus belongs to the SDR family, that of H. marismortui (encoded by gene with locus tag of rrnAC3034) contains conserved domains for the glucose-fructose oxidoreductase (GFOR) family (Johnsen and Schönheit 2004; Baliga et al. 2004). Further genetic analysis of the H. marismortui genome revealed that ORFs near rrnAC3034 are genes encoding putative enzymes for a functional Weimberg pathway (Fig. 3) (Brouns et al. 2006). Upstream of rrnAC3034 are two ORFs: rrnAC3033 annotated as a xylonolactonase-encoding gene and dgoA2 annotated as a xylonate dehydratase-encoding gene. Somewhere downstream of rrnAC3034 are the genes aldY3 and hpcEb, which are predicted to encode ketoglutarate semialdehyde dehydrogenase and 2-keto-3-deoxy-d-xylonate dehydratase, respectively. In addition to dgoA2, another putative dehydratase encoded by dgoA4 is located at 0.51 MB of the H. marismortui chromosome. This gene has approximately 80% nucleotide sequence identity to dgoA2 while its gene product has 75% sequence match to dgoA2. The genome of H. marismortui also contains a homolog of a secondary xylose dehydrogenase HVO_B0029 from H. volcanii (see next section). This gene has a locus tag rrnAC2929 and is annotated as gfo1.
H. volcanii
Although the first and partial evidence for the existence of the Weimberg pathway in Archaea was from H. marismortui (Johnsen and Schönheit 2004), its first detailed characterization was from another halophilic archaeon, H. volcanii (Johnsen et al. 2009). Complete characterization of the Weimberg pathway in H. volcanii was carried out through 13C-labeling experiments, enzyme characterization and assays, shotgun DNA microarray, and in-frame gene deletion experiments (Johnsen et al. 2009). It was shown that the d-xylose-inducible genes in H. volcanii are those with the locus tags HVO_B0028 for xylose dehydrogenase, HVO_B0030 for xylonolactonase, HVO_B0038A for xylonate dehydratase, HVO_B0027 for KDX dehydratase, and HVO_B0039 for α-ketoglutarate semialdehyde dehydrogenase (Fig. 3). Unlike the C. crescentus xylose dehydrogenase, the gene product of HVO_B0028 shows NADP-dependent xylose dehydrogenase activity and contains conserved domains belonging to the GFOR family. Interestingly, the gene product of HVO_0029 also shows NADP-dependent xylose dehydrogenase activity (Johnsen et al. 2009). In-frame deletion of either HVO_B0028 or HVO_B0029 revealed that HVO_B0028 is essential for d-xylose assimilation and is therefore the physiologically relevant xylose dehydrogenase-coding gene (Johnsen et al. 2009). Another interesting observation on the arrangement of the aforementioned structural genes is that HVO_B0032 encodes for the enzyme responsible for the initial step of oxidative l-arabinose assimilation in H. volcanii (Johnsen et al. 2013). Noting that this gene is clustered with the genes involved in the Weimberg pathway, it was proposed that the gene cluster has a common transcriptional regulator. This was later validated through experimental evidence showing XacR, the gene product of HVO_B0040, as the transcriptional activator for both d-xylose and l-arabinose assimilation (Johnsen et al. 2015). Unlike the XylR of C. crescentus, XacR belongs to the IcIR family of transcriptional regulators and acts as a repressor for its own transcription in H. volcanii.
Besides having a common transcriptional regulator, it was also proposed that H. volcanii shares common structural genes for d-xylonic acid and l-arabonic acid conversion to α-ketoglutarate (Johnsen et al. 2013). However, it remains unclear whether the two pathways converge at the formation of α-ketoglutarate intermediate or at the C4 epimerization of either sugar acids. The former relies on the promiscuity of the subsequent dehydration and oxidation steps of the sugar acids, while the latter requires the presence of an unknown and uncharacterized C4-epimerase (Johnsen et al. 2013).
Sulfolobus sp.
Another well-studied archaeal strain that employs the XOP belongs to the Sulfolobus genus. The oxidative d-arabinose metabolism (an analog of Weimberg pathway) was initially described in S. solfataricus (Brouns et al. 2006). The necessary enzymes for the conversion of d-arabinose to 2-ketoglutarate were detected and confirmed through proteomics and enzyme assays. d-arabinose is oxidized to d-arabonic acid by arabinose dehydrogenase encoded by aradH (locus tag SSO1300). d-arabonic acid is then dehydrated to 2-keto-3-deoxy-d-arabonate by arabonate dehydratase encoded by araD (locus tag SSO3124). 2-Keto-3-deoxy-d-arabonate is further dehydrated to 2-ketoglutarate semialdehyde by 2-keto-3-deoxy-d-arabonate dehydratase encoded by kdaD (locus tag SSO3118). Finally, 2-ketoglutarate semialdehyde is oxidized to 2-ketoglutarate by ketoglutarate semialdehyde dehydrogenase encoded by dopDH (locus tag SSO3117). Interestingly, these structural genes are located in a different locus in the S. solfataricus genome.
The existence of the abovementioned d-arabinose oxidative metabolism in S. solfataricus implies that d-xylose metabolism may follow the same route. Interestingly, some of the enzymes of the d-arabinose degradation pathway have shown certain degrees of promiscuity towards d-xylose (Archer et al. 2013). This kind of promiscuity was also observed in enzymes involved in glycolysis. S. solfataricus metabolizes d-glucose through the Entner-Doudoroff pathway, which may be referred to as the C6 counterpart of the XOP. It is initiated by d-glucose oxidation to d-gluconic acid by d-glucose dehydrogenase encoded by dhg-1. This enzyme has a broad spectrum of substrates that include d-galactose, d-xylose, l-arabinose, 6-deoxy-d-glucose, and d-fucose, all of which have rates > 50% relative to d-glucose and NADP as a co-factor (Lamble et al. 2003). Furthermore, the 2-keto-3-deoxy-d-gluconate aldolase is also capable of catalyzing the condensation of glycolaldehyde and pyruvate, the products of the aldol cleavage of 2-keto-3-deoxy-d-xylonate (Danson et al. 2007). These observations led to the establishment of a functional Dahms pathway in S. solfataricus (Nunn et al. 2010). The reactions in the pathway are catalyzed by a promiscuous glucose dehydrogenase dhg-1, xylonate dehydratase encoded by SS02665, and KDX aldolase encoded by dapA-3 (Fig. 3) (Nunn et al. 2010). Interestingly, S. acidocaldarius assimilates d-xylose through the Weimberg pathway based on preliminary experimental evidence (Nunn et al. 2010).
Metabolic engineering of the XOPs
During the past 5 years, the Weimberg and Dahms pathways have been gaining momentum in their application in the field of metabolic engineering and microbial cell factories. The following section discusses the recent advances in the development of different microbial strains for d-xylose assimilation through the XOP.
Pseudomonas
Successful heterologous expression of XOP enzymes in a recombinant host was first demonstrated in Pseudomonas putida S12 (Meijnen et al. 2009). This effort was motivated by the fact that P. putida naturally oxidizes d-xylose to d-xylonic acid by the promiscuous glucose dehydrogenase (Gcd) and that the accumulated d-xylonic acid must be further utilized to prevent carbon loss. It was observed that the expression of xylonate dehydratase (XylD) was enough to establish a functional Weimberg pathway in P. putida (Meijnen et al. 2009). Co-expression of other enzyme components of the pathway, specifically XylX and XylA (Fig. 4), improved d-xylose assimilation. However, growing P. putida on d-xylose through the XOP extended its lag phase (Meijnen et al. 2009). Carbon catabolite effect was also observed when it was grown on both d-glucose and d-xylose, which is most likely caused by the preference of Gcd for d-glucose. Transcriptome analysis revealed that the Entner-Doudoroff pathway genes, eda and edd, were downregulated when d-xylose assimilation operates through the XOP (Meijnen et al. 2012). Another notable change in the transcriptome is the increased expression level of crp, which encodes for the Crp protein. Considering that Crp is known as a global regulatory protein, it can be inferred that d-xylose utilization through the XOP in P. putida triggered a broad response which affects Crp-regulated genes (Meijnen et al. 2012). These genes may not necessarily be involved in carbon utilization since the exact biological role of Crp in P. putida remains unknown (Milanesio et al. 2011).
Escherichia coli
1,2,4-Butanetriol production
One of the earliest metabolic engineering studies in E. coli through the XOP was for the production of 1,2,4-butanetriol (BTO), a four-carbon compound used as a precursor for the production of plasticizers in explosives (Niu et al. 2003). The pathway involves the first three reactions of the XOP wherein d-xylose is converted to KDX. KDX is then converted to BTO through sequential decarboxylation and reduction reactions (Fig. 5). The strategy involved the use of two different strains: Pseudomonas fragi that converts d-xylose into d-xylonic acid and a recombinant E. coli that converts d-xylonic acid to BTO (Niu et al. 2003). It is noteworthy that E. coli strains from the K12 lineage showed capability to assimilate d-xylonic acid (Liu et al. 2013). This catabolic pathway is an analog of the Dahms pathway but lacks xylose dehydrogenase activity. The genes encoding for the enzymes were initially categorized as cryptic prophage-associated elements: yjhG and yagF as dehydratases and yjhH and yagE as aldolases (Fig. 5) (Shimada et al. 2017; Shimada et al. 2018). However, recent biochemical studies have demonstrated the possible biological roles of these genes. For example, the d-xylonate dehydratase YjhG was shown to be highly active towards KDX, with optimum activity at 30 °C and pH 8.0 (Jiang et al. 2015). Bioinformatic studies also revealed that YjhG belongs to the IlvD/EDD dehydratase family of enzymes. Meanwhile, the crystal structure of the KDX aldolase enzyme YagE (Manicka et al. 2008) suggested that it functions as an aldolase for both five- and six-carbon substrates. Aside from the reported biochemical data, several studies have also presented genetic evidence suggesting that YjhG and YagF have xylonate dehydratase activities, whereas YjhH and YagE have KDX aldolase activities (Liu et al. 2013; Valdehuesa et al. 2014; Cao et al. 2015; Cabulong et al. 2017; Wang et al. 2018; Cabulong et al. 2018). These works are further discussed in the following paragraphs.
Xylose oxidative pathway assembled in E. coli for the production of different target compounds. Gene names are enclosed in parenthesis. Blue, native to the host strain; red, recruited from C. crescentus or P. putida (in the case of mdlC gene). No genes were assigned to the reductase and dehydrogenase reactions since the E. coli host carries a variety of these promiscuous enzymes
Based on a previous study by Niu et al. (2003), BTO production was integrated into a single E. coli W3110 strain (a K12 strain derivative) by recruiting XylB from C. crescentus and MdlC (a decarboxylase) from P. putida (Valdehuesa et al. 2014; Valdehuesa et al. 2015). The lactonase, dehydratase, and alcohol dehydrogenase activities required for the pathway were inherent to the E. coli host. The native XIP of E. coli was blocked by disruption of the xylAB gene cluster, and the KDX aldolase expression was prevented thru the deletion of yjhH and yagE genes (Valdehuesa et al. 2014). Furthermore, the main aldehyde reductase which catalyzes the last step of the BTO pathway was identified to be YqhD (Valdehuesa et al. 2015). Finally, a yield of 12.82% was achieved from the prototype strain (Valdehuesa et al. 2014). Further optimization of the BTO pathway in E. coli identified dehydratase and decarboxylase reactions as the rate-liming steps (Sun et al. 2016). One study addressed this issue by co-overexpression of chaperone proteins GroES-GroEL and Tig (trigger factor) (Lu et al. 2016). Another study demonstrated that overexpression of ADH2 from S. cerevisiae (with or without YqhD overexpression) increased BTO production (Zhang et al. 2016). Furthermore, competing reactions that consume KDX were inactivated by deleting the yiaE gene in the host genome. The optimized strain was capable of producing BTO up to 5 g/L which is equivalent to a molar yield of around 36% (Zhang et al. 2016).
For E. coli strains that belong to the B lineage, recruitment of the full set of enzymes was required since these strains lack the yjh and yag gene clusters. Enzymes from C. crescentus (XylB and XylC), E. coli K12 (YjhG), and P. putida (MdlC) were introduced to E. coli BL21 Star, along with the overexpression of a native alcohol dehydrogenase (AdhP) (Cao et al. 2015). The resulting strain was capable of producing BTO with a molar yield of 27.7%. In a more recent work by Wang et al. (2018), BTO production was improved by enzyme homolog screening. Results showed that the best enzymes for building the BTO pathway in E. coli BL21 were XylD (xylonate dehydratase) from C. crescentus, KdcA (decarboxylase) from Lactococcus lactis, and the native AdhP (aldehyde reductase) from E. coli (Wang et al. 2018). Using the optimized strain, around 5.1 g/L BTO (molar yield of 9%) was produced from d-xylose and 3.4 g/L BTO (molar yield of 5.4%) from corncob hydrolysates.
3,4-Dihydroxybutyric acid production
3,4-Dihydroxybutyric acid (DHBA) can be synthesized by modifying the last step of the BTO production pathway. Instead of a reduction reaction, the 3,4-dihydroxybutanal is oxidized to form DHBA (Fig. 5). To enable DHBA production in E. coli, overexpression of succinate semialdehyde dehydrogenase gene gabD from P. putida or yneI from E. coli was implemented (Wang et al. 2017b). The best strain was capable of producing 1.27 g/L DHBA in shake flask experiments. One way to improve DHBA production from the prototype strain is through the inactivation of any native aldehyde reductase in E. coli that catalyzes the formation of BTO. Based on earlier works, AdhP and YqhD are the best candidates for gene deletion experiments (Valdehuesa et al. 2015; Wang et al. 2018). However, E. coli possesses several aldehyde reductases that have different cellular functions, which makes this task difficult to perform without inducing adverse effects to the host phenotype (Wang et al. 2017b). Other strategies to improve DHBA production are inactivation of competing pathways, improvement of cellular redox balance, and enhancement of cell growth by using dual-substrate systems (Gao et al. 2017).
Ethylene glycol and glycolic acid
The first report on the biosynthesis of ethylene glycol (EG) employed the Dahms pathway (Fig. 5) in a recombinant E. coli (Liu et al. 2013). Genetic evidence demonstrating the catalytic activities of the enzymes from the yag and yjh operon in E. coli was presented through enzyme assays and gene deletion–complementation experiments. The best EG-producing strain has an inactive XIP and an overexpressed XylB from C. crescentus and the native YqhD from E. coli (Liu et al. 2013). The strain was capable of accumulating 11.7 g/L EG which is equivalent to a molar yield of 70.8%.
One of the issues in the microbial EG production through the Dahms pathway is the accumulation of d-xylonic acid at the onset of cultivation (Liu et al. 2013). Although the secreted d-xylonic acid in the medium is re-assimilated later, the whole process lowered the productivity and the pH (albeit temporarily) of the medium. This problem was addressed by expression of XylB under the control of a weak and constitutive promoter (Cabulong et al. 2017). Furthermore, EG production was enhanced upon overexpression of the native YjgB from E. coli, which was identified as the most suitable aldehyde reductase. Rational engineering of the host for higher EG production required inactivation of the chromosomal yjgB and aldA genes (AldA catalyzes conversion of EG to glycolic acid). The final strain was capable of producing EG up to a molar yield of 98% (Cabulong et al. 2017).
Similar to the BTO and DHBA biosynthetic pathways, glycolic acid (GA) can be produced from the Dahms pathway by shifting from glycolaldehyde reduction to oxidation (Fig. 5). This was done by simple overexpression of the native E. coli lactate dehydrogenase aldA and inactivation of the GA consuming step catalyzed by the native E. coli glycolate dehydrogenase glcD (Cabulong et al. 2018). Moreover, xylonate dehydratase reaction was identified as the bottleneck in the GA pathway (i.e., the Dahms pathway in general), which is consistent with the findings from the BTO biosynthesis studies. Increasing the GA titer further required overexpression of yagF (xylonate dehydratase), yagE (KDX aldolase), ycdW (glyoxylate reductase), and aceAK (glyoxylate shunt enzymes) (Cabulong et al. 2018). On the other hand, construction of the GA production pathway in E. coli B strain requires recruitment of all the enzymes in the Dahms pathway (Liu et al. 2018). Due to the variation in host genetic background compared to the K12-derivative strain, optimization of GA production in B strains requires a slightly different set of gene overexpression and deletion, particularly when the XIP is working in tandem with the Dahms pathway. For GA biosynthesis, the optimized E. coli B strain with a functional XIP requires overexpression of xylB, xylC, yjhG, yjhH, and aldA genes and the deletion of ackA (acetate kinase) (Liu et al. 2018).
1,4-Butanediol
The first report featuring the production of 1,4-butanediol (BDO) using the XOP demonstrated the synergy of synthetic biology and metabolic engineering techniques (Liu and Lu 2015). Initially, the conversion of d-xylose to BDO was established in E. coli through heterologous expression of xylB, xylX, and mdlC (Fig. 5). The host strain has xylA, yjhH, and yagE gene knockouts, which rendered it incapable of consuming d-xylose through the XIP and the aldol cleavage of KDX. Genetic controllers were then constructed for the BDO pathway. The synthetic genetic circuit was designed to achieve autonomous regulation of the heterologous gene expressions, thus eliminating the need for artificial and expensive inducer molecules such as IPTG (Liu and Lu 2015). This genetic controller exploits the quorum sensing mechanism from Vibrio cholerae, known as the lux system.
Alternatively, BDO production was also achieved by extending the BTO pathway (Wang et al. 2017a). The critical step in this pathway is the dehydration of BTO (Fig. 5). To date, there are no known enzymes that naturally catalyze the conversion of BTO to 4-hydroxybutanal. Hence, rational engineering was employed to enable naturally existing dehydratases to convert the non-natural substrate BTO. A dehydratase enzyme that utilizes similar molecules as BTO, for example, the 1,2-propanediol dehydratase from Klebsiella oxytoca encoded by ppdACB gene cluster, was selected as candidate. Certain point mutations around the active site of this enzyme rendered it active towards BTO, which resulted in BDO production up to 0.2 g/L (Wang et al. 2017a).
Yeast
The XOP pathway was first utilized in yeast for the production of d-xylonic acid (Toivari et al. 2010). Compared to E. coli, d-xylonic acid production in S. cerevisiae was demonstrated by expression of xylose dehydrogenase gene, xyd1, from T. reesei (Berghäll et al. 2007). This enzyme prefers NADP as co-factor compared to XylB. Genetic modifications of the host strain included inactivation of the xylose reductase gene GRE3 to reduce xylitol accumulation. On the other hand, expression of xylB from C. crescentus increased the d-xylonate production rate by tenfold (Toivari et al. 2012). Co-expression of the lactonase gene xylC did not significantly increase d-xylonic acid accumulation but produced higher linear/lactone ratio of d-xylonic acid (Nygård et al. 2014). Hydrolysis of the lactonic d-xylonic acid was found critical in achieving higher production rates at the expense of cellular viability (Nygård et al. 2014).
Recent studies have also demonstrated successful engineering of the XOP in S. cerevisiae (Salusjärvi et al. 2017; Wasserstrom et al. 2018). Through the Dahms pathway, ethylene glycol and glycolic acid were successfully produced in the prototype S. cerevisiae strain expressing xylB and xylD from C. crescentus and yagE or yjhH and aldA from E. coli (Salusjärvi et al. 2017). Furthermore, the xylonate dehydratase activity was identified as the bottleneck of the pathway, which is consistent with E. coli host systems discussed previously. This was due to the low supply of Fe-S clusters in the cytosol of the host. This issue was circumvented by expressing XylD in the mitochondria, along with the inactivation of the FRA2 gene involved in the regulation of the iron regulon (Kumánovics et al. 2008; Salusjärvi et al. 2017). It is interesting to note that YjhH overexpression facilitated ethylene glycol production while YagE for glycolic acid formation (Salusjärvi et al. 2017). Meanwhile, d-xylose assimilation through the Weimberg pathway in S. cerevisiae was proven to be challenging (Wasserstrom et al. 2018). The complete set of enzymes from the xyl operon of C. crescentus was successfully expressed in the host yeast strain. However, XylD and XylX dehydratases did not show any activity in vivo, which is mainly due to the deficiency in Fe-S cluster that is required for them to form a mature and active apoprotein (Wasserstrom et al. 2018).
Corynebacterium
The integration of XOP in a heterologous host was also accomplished in Corynebacterium glutamicum by recruiting the complete xyl operon from C. crescentus (Radek et al. 2014). The highlighted advantage of choosing XOP for this microorganism was its capability to directly convert d-xylose to ⍺-ketoglutaric acid (while avoiding the TCA cycle). ⍺-Ketoglutaric acid is a known intermediate during the biosynthesis of amino acids, which are popularly produced by C. glutamicum at industrial scales. The maximum growth rate of the final engineered strain was at 0.07 h−1 and it secreted d-xylonic acid as a major by-product (Radek et al. 2014). Low xylitol accumulation was also observed during cultivation of the engineered strain, which is favorable since this compound is known to inhibit bacterial growth (Radek et al. 2016). Noting that xylitol accumulation is more pronounced in C. glutamicum when d-xylose is metabolized through the XIP, the XOP was proposed to be a more suitable xylose metabolic pathway for C. glutamicum (Radek et al. 2016). Adaptive evolution technique was also implemented in C. glutamicum to further improve d-xylose utilization through the XOP (Radek et al. 2017). The resulting strain, WMB2evo, was capable of growing on d-xylose at a maximum rate of 0.26 h−1 which is almost four-fold higher than the parental strain. The evolved strain was shown to carry multiple genome sequence mutations identified through DNA sequencing (Radek et al. 2017). In-frame deletion of iolR gene encoding for a repressor molecule of myo-inositol utilization genes was singled out as the major contributor of the improved growth phenotype in WMB2evo. Loss of IolR expression resulted in the de-repression of myo-inositol utilization genes including the permease (transporter), IolT1, which is believed to enhance d-xylose uptake in the WMB2evo (Brüsseler et al. 2018). Alternatively, a double nucleotide substitution at the IolR-binding site of the iolT1 promoter sequence confers the same improved growth phenotype.
Similar to E. coli, the main challenge for the XOP in C. glutamicum was the transient accumulation of d-xylonic acid. However, a recent study exploited such handicap and viewed it as a potential for C. glutamicum strains to be used as microbial catalysts for the production of d-xylonic acid from hemicellulosic biomass. This was demonstrated in a recent study wherein C. glutamicum was engineered to express XylB (xylose dehydrogenase) from C. crescentus, XylE (xylose transporter) from E. coli, XlnA (endoxylanase) from Streptomyces coelicolor, and XynB (xylosidase) from Bacillus pumilus (Yim et al. 2017). The final strain was capable of producing up to 6.23 g/L of d-xylonic acid from 20 g/L xylan.
Future perspectives
It is apparent that the XOP is potentially an excellent alternative to the traditional xylose metabolic pathways in microorganisms. The XOP has been successfully recruited in different industrial strains, some of which are capable of producing the target compounds at titers near industrial viability. For example, d-xylonic acid production in recombinant bacteria and yeasts can reach from 50 to 150 g/L in batch or fed-batch modes (Liu et al. 2012; Toivari et al. 2013; Cao et al. 2013). On the other hand, production of smaller compounds such as BTO, BDO, DHBA, EG, and GA would still require further optimization and engineering.
To streamline the conversion of d-xylose to any target compound via the XOP, two prominent drawbacks must be addressed. First, the accumulation of pathway intermediates such as d-xylonic acid and KDX should be circumvented, if not minimized. Second, bottlenecks of the pathway such as the dehydratase enzymes should be optimized in terms of expression and catalytic efficiency.
Tackling the issues on pathway intermediate secretion and accumulation may be complex. It has been suggested that phosphorylation of metabolites increases their charges, thus restricting cell membrane diffusion as well as increasing enzyme affinity (Bar-Even et al. 2012). Since the XOP intermediates are non-phosphorylated, these compounds have increased chances of permeating outside the host cell. Furthermore, the specific transporters of these intermediates remain unknown, which makes it difficult to select the proper transporter proteins for rational engineering.
Currently, the optimization of the XOP bottlenecks remains challenging. Pathway engineering through enzyme screening or protein engineering is a good strategy. However, the available pieces of information for alternative enzymes and their corresponding biochemical data are limited. Further studies on the XOP pathway in their native hosts are required to understand the whole pathway and how it interacts with the global cellular network.
As described in the previous sections, most of the studies that recruited the XOP for d-xylose assimilation in engineered strains employed the simple overexpression–deletion strategies. The emerging field of synthetic biology has developed genetic tools and techniques that can improve the XOP performance in the recombinant hosts. These tools can allow dynamic control of target enzyme expression and can be used to alleviate intermediate accumulation. Some of these tools can also be used for high throughput screening of candidate enzymes for the optimization of the pathway bottleneck.
Further development of d-xylose metabolizing recombinant strains operating through the XOP must include co-utilization of other carbohydrates to circumvent the “glucose effect” (carbon catabolite repression). This issue must be considered particularly at the industrial scale since microbial cell factories typically use feedstocks containing several substrates. Industrial strains that are capable of rapid simultaneous consumption of all sugars would be ideal and will eventually lead to higher productivity and cost efficiency.
References
Andberg M, Aro-Kärkkäinen N, Carlson P, Oja M, Bozonnet S, Toivari M, Hakulinen N, O'Donohue M, Penttilä M, Koivula A (2016) Characterization and mutagenesis of two novel iron-sulphur cluster pentonate dehydratases. Appl Microbiol Biotechnol 100:7549–7563. https://doi.org/10.1007/s00253-016-7530-8
Archer RM, Royer SF, Mahy W, Winn CL, Danson MJ, Bull SD (2013) Syntheses of 2-keto-3-deoxy-D-xylonate and 2-keto-3-deoxy-L-arabinonate as stereochemical probes for demonstrating the metabolic promiscuity of Sulfolobus solfataricus towards D-xylose and L-arabinose. Chemistry 19:2895–2902. https://doi.org/10.1002/chem.201203489
Atsumi S, Wu T-Y, Eckl E-M, Hawkins SD, Buelter T, Liao JC (2010) Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol 85:651–657. https://doi.org/10.1007/s00253-009-2085-6
Baliga NS, Bonneau R, Facciotti MT, Pan M, Glusman G, Deutsch EW, Shannon P, Chiu Y, Weng RS, Gan RR, Hung P, Date SV, Marcotte E, Hood L, Ng WV (2004) Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res 14:2221–2234. https://doi.org/10.1101/gr.2700304
Bar-Even A, Flamholz A, Noor E, Milo R (2012) Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat Chem Biol 8:509–517. https://doi.org/10.1038/nchembio.971
Berghäll S, Hilditch S, Penttilä M, Richard P (2007) Identification in the mould Hypocrea jecorina of a gene encoding an NADP+: D-xylose dehydrogenase. FEMS Microbiol Lett 277:249–253. https://doi.org/10.1111/j.1574-6968.2007.00969.x
Bettiga M, Hahn-Hägerdal B, Gorwa-Grauslund MF (2008) Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol Biofuels 1:16. https://doi.org/10.1186/1754-6834-1-16
Blaschek HP, Ezeji T, Price ND (2010) Present and future possibilities for the deconstruction and utilization of lignocellulosic biomass. In: Khanna M, Scheffran J, Zilberman D (eds) Handbook of bioenergy economics and policy. Springer, New York, NY, pp 39–51
Brouns SJJ, Walther J, Snijders APL, van de Werken HJG, Willemen HLDM, Worm P, de Vos MGJ, Andersson A, Lundgren M, Mazon HFM, van den Heuvel RHH, Nilsson P, Salmon L, de Vos WM, Wright PC, Bernander R, van der Oost J (2006) Identification of the missing links in prokaryotic pentose oxidation pathways: evidence for enzyme recruitment. J Biol Chem 281:27378–27388. https://doi.org/10.1074/jbc.M605549200
Brüsseler C, Radek A, Tenhaef N, Krumbach K, Noack S, Marienhagen J (2018) The myo-inositol/proton symporter IolT1 contributes to D-xylose uptake in Corynebacterium glutamicum. Bioresour Technol 249:953–961. https://doi.org/10.1016/j.biortech.2017.10.098
Buchert J, Viikari L (1988) Oxidative D-xylose metabolism of Gluconobacter oxydans. Appl Microbiol Biotechnol 29:375–379. https://doi.org/10.1007/BF00265822
Cabulong RB, Lee W-K, Bañares AB, Ramos KRM, Nisola GM, Valdehuesa KNG, Chung W-J (2018) Engineering Escherichia coli for glycolic acid production from D-xylose through the Dahms pathway and glyoxylate bypass. Appl Microbiol Biotechnol 102:2179–2189. https://doi.org/10.1007/s00253-018-8744-8
Cabulong RB, Valdehuesa KNG, Ramos KRM, Nisola GM, Lee W-K, Lee CR, Chung W-J (2017) Enhanced yield of ethylene glycol production from D-xylose by pathway optimization in Escherichia coli. Enzym Microb Technol 97:11–20. https://doi.org/10.1016/j.enzmictec.2016.10.020
Cao Y, Niu W, Guo J, Xian M, Liu H (2015) Biotechnological production of 1,2,4-butanetriol: an efficient process to synthesize energetic material precursor from renewable biomass. Sci Rep 5:18149. https://doi.org/10.1038/srep18149
Cao Y, Xian M, Zou H, Zhang H (2013) Metabolic engineering of Escherichia coli for the production of xylonate. PLoS One 8:e67305. https://doi.org/10.1371/journal.pone.0067305
Carter AT, Pearson BM, Dickinson JR, Lancashire WE (1993) Sequence of the Escherichia coli K-12 edd and eda genes of the Entner-Doudoroff pathway. Gene 130:155–156. https://doi.org/10.1016/0378-1119(93)90362-7
Cheng JJ, Timilsina GR (2011) Status and barriers of advanced biofuel technologies: a review. Renew Energy 36:3541–3549. https://doi.org/10.1016/j.renene.2011.04.031
Dahms AS (1974) 3-Deoxy-D-pentulosonic acid aldolase and its role in a new pathway of D-xylose degradation. Biochem Biophys Res Comm 60:1433–1439. https://doi.org/10.1016/0006-291X(74)90358-1
Danson MJ, Lamble HJ, Hough DW (2007) Central metabolism. In: Cavicchioli R (ed) Archaea. American Society of Microbiology, Washington, DC, pp 260–287
Dumon C, Song L, Bozonnet S, Fauré R, O’Donohue MJ (2012) Progress and future prospects for pentose-specific biocatalysts in biorefining. Process Biochem 47:346–357. https://doi.org/10.1016/j.procbio.2011.06.017
Flamholz A, Noor E, Bar-Even A, Milo R (2012) eQuilibrator—the biochemical thermodynamics calculator. Nucl Acids Res 40:D770–D775. https://doi.org/10.1093/nar/gkr874
Gao H, Gao Y, Dong R (2017) Enhanced biosynthesis of 3,4-dihydroxybutyric acid by engineered Escherichia coli in a dual-substrate system. Bioresour Technol 245:794–800. https://doi.org/10.1016/j.biortech.2017.09.017
Hottes AK, Meewan M, Yang D, Arana N, Romero P, McAdams HH, Stephens C (2004) Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J Bacteriol 186:1448–1461. https://doi.org/10.1128/JB.186.5.1448-1461.2004
Jiang Y, Liu W, Cheng T, Cao Y, Zhang R, Xian M (2015) Characterization of D-xylonate dehydratase YjhG from Escherichia coli. Bioeng 6:227–232. https://doi.org/10.1080/21655979.2015.1040208
Johnsen U, Dambeck M, Zaiß H, Fuhrer T, Soppa J, Sauer U, Schönheit P (2009) D-xylose degradation pathway in the halophilic archaeon Haloferax volcanii. J Biol Chem 284:27290–27303. https://doi.org/10.1074/jbc.M109.003814
Johnsen U, Schönheit P (2004) Novel xylose dehydrogenase in the halophilic archaeon Haloarcula marismortui. J Bacteriol 186:6198–6207. https://doi.org/10.1128/JB.186.18.6198-6207.2004
Johnsen U, Sutter J-M, Schulz A-C, Tästensen J-B, Schönheit P (2015) XacR—a novel transcriptional regulator of D-xylose and L-arabinose catabolism in the haloarchaeon Haloferax volcanii. Environ Microbiol 17:1663–1676. https://doi.org/10.1111/1462-2920.12603
Johnsen U, Sutter J-M, Zaiß H, Schönheit P (2013) L-Arabinose degradation pathway in the haloarchaeon Haloferax volcanii involves a novel type of L-arabinose dehydrogenase. Extremophiles 17:897–909. https://doi.org/10.1007/s00792-013-0572-2
Karhumaa K, Garcia Sanchez R, Hahn-Hägerdal B, Gorwa-Grauslund MF (2007) Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Factories 6(5):5. https://doi.org/10.1186/1475-2859-6-5
Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF (2005) Investigation of limiting metabolic steps in the utilization of xylose by recombinant Saccharomyces cerevisiae using metabolic engineering. Yeast 22:359–368. https://doi.org/10.1002/yea.1216
Kobayashi H, Fukuoka A (2013) Synthesis and utilisation of sugar compounds derived from lignocellulosic biomass. Green Chem 15:1740–1763. https://doi.org/10.1039/c3gc00060e
Köhler KAK, Blank LM, Frick O, Schmid A (2015) D-Xylose assimilation via the Weimberg pathway by solvent-tolerant Pseudomonas taiwanensis VLB120. Environ Microbiol 17:156–170. https://doi.org/10.1111/1462-2920.12537
Kumánovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J (2008) Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem 283:10276–10286. https://doi.org/10.1074/jbc.M801160200
Lamble HJ, Heyer NI, Bull SD, Hough DW, Danson MJ (2003) Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J Biol Chem 278:34066–34072. https://doi.org/10.1074/jbc.M305818200
Liu H, Lu T (2015) Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli. Metab Eng 29:135–141. https://doi.org/10.1016/j.ymben.2015.03.009
Liu H, Ramos KRM, Valdehuesa KNG, Nisola GM, Lee W-K, Chung W-J (2013) Biosynthesis of ethylene glycol in Escherichia coli. Appl Microbiol Biotechnol 97:3409–3417. https://doi.org/10.1007/s00253-012-4618-7
Liu H, Valdehuesa KNG, Nisola GM, Ramos KRM, Chung W-J (2012) High yield production of D-xylonic acid from D-xylose using engineered Escherichia coli. Bioresour Technol 115:244–248. https://doi.org/10.1016/j.biortech.2011.08.065
Liu M, Ding Y, Xian M, Zhao G (2018) Metabolic engineering of a xylose pathway for biotechnological production of glycolate in Escherichia coli. Microb Cell Factories 17:51. https://doi.org/10.1186/s12934-018-0900-4
Lockwood LB, Nelson GE (1946) The oxidation of pentoses by Pseudomonas. J Bacteriol 52:581–586
Lu X, He S, Zong H, Song J, Chen W, Zhuge B (2016) Improved 1,2,4-butanetriol production from an engineered Escherichia coli by co-expression of different chaperone proteins. World J Microbiol Biotechnol 32:149. https://doi.org/10.1007/s11274-016-2085-5
Manicka S, Peleg Y, Unger T, Albeck S, Dym O, Greenblatt HM, Bourenkov G, Lamzin V, Krishnaswamy S, Sussman JL (2008) Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12. Proteins 71:2102–2108. https://doi.org/10.1002/prot.22023
Matsushika A, Inoue H, Kodaki T, Sawayama S (2009) Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84:37–53. https://doi.org/10.1007/s00253-009-2101-x
Meijnen J-P, de Winde JH, Ruijssenaars HJ (2009) Establishment of oxidative D-xylose metabolism in Pseudomonas putida S12. Appl Environ Microbiol 75:2784–2791. https://doi.org/10.1128/AEM.02713-08
Meijnen J-P, de Winde JH, Ruijssenaars HJ (2012) Metabolic and regulatory rearrangements underlying efficient D-xylose utilization in engineered Pseudomonas putida S12. J Biol Chem 287:14606–14614. https://doi.org/10.1074/jbc.M111.337501
Meisenzahl AC, Shapiro L, Jenal U (1997) Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J Bacteriol 179:592–600
Mihasan M, Stefan M, Hritcu L, Artenie V, Brandsch R (2013) Evidence of a plasmid-encoded oxidative xylose-catabolic pathway in Arthrobacter nicotinovorans pAO1. Res Microbiol 164:22–30. https://doi.org/10.1016/j.resmic.2012.10.003
Milanesio P, Arce-Rodríguez A, Muñoz A, Calles B, de Lorenzo V (2011) Regulatory exaptation of the catabolite repression protein (Crp)-cAMP system in Pseudomonas putida. Environ Microbiol 13:324–339. https://doi.org/10.1111/j.1462-2920.2010.02331.x
Naik SN, Goud VV, Rout PK, Dalai AK (2010) Production of first and second generation biofuels: a comprehensive review. Renew Sust Energy Rev 14:578–597. https://doi.org/10.1016/j.rser.2009.10.003
Nakamura CE, Whited GM (2003) Metabolic engineering for the microbial production of 1,3-propanediol. Curr Opin Biotechnol 14:454–459
Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM, Eisen J (2001) Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98:4136–4141. https://doi.org/10.1073/pnas.061029298
Niu W, Molefe MN, Frost JW (2003) Microbial synthesis of the energetic material precursor 1,2,4-butanetriol. J Am Chem Soc. https://doi.org/10.1021/ja036391
Nunn CEM, Johnsen U, Schonheit P, Fuhrer T, Sauer U, Hough DW, Danson MJ (2010) Metabolism of pentose sugars in the hyperthermophilic archaea Sulfolobus solfataricus and Sulfolobus acidocaldarius. J Biol Chem 285:33701–33709. https://doi.org/10.1074/jbc.M110.146332
Nygård Y, Maaheimo H, Mojzita D, Toivari M, Wiebe M, Resnekov O, Gustavo Pesce C, Ruohonen L, Penttilä M (2014) Single cell and in vivo analyses elucidate the effect of xylC lactonase during production of D-xylonate in Saccharomyces cerevisiae. Metab Eng 25:238–247. https://doi.org/10.1016/j.ymben.2014.07.005
Rabinovitch-Deere CA, Oliver JWK, Rodriguez GM, Atsumi S (2013) Synthetic biology and metabolic engineering approaches to produce biofuels. Chem Rev 113:4611–4632. https://doi.org/10.1021/cr300361t
Radek A, Krumbach K, Gätgens J, Wendisch VF, Wiechert W, Bott M, Noack S, Marienhagen J (2014) Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D-xylose containing substrates. J Biotechnol 192 Pt A 192:156–160. https://doi.org/10.1016/j.jbiotec.2014.09.026
Radek A, Müller M-F, Gätgens J, Eggeling L, Krumbach K, Marienhagen J, Noack S (2016) Formation of xylitol and xylitol-5-phosphate and its impact in growth of D-xylose-utilizing Corynebacterium glutamicum strains. J Biotechnol 231:160–166. https://doi.org/10.1016/j.jbiotec.2016.06.009
Radek A, Tenhaef N, Müller MF, Brüsseler C, Wiechert W, Marienhagen J, Polen T, Noack S (2017) Miniaturized and automated adaptive laboratory evolution: evolving Corynebacterium glutamicum towards an improved D-xylose utilization. Bioresour Technol 245:1377–1385. https://doi.org/10.1016/j.biortech.2017.05.055
Rahman MM, Andberg M, Koivula A, Rouvinen J, Hakulinen N (2016) Crystallization and X-ray diffraction analysis of an L-arabinonate dehydratase from Rhizobium leguminosarum bv. trifolii and a D-xylonate dehydratase from Caulobacter crescentus. Acta Cryst 72:604–608. https://doi.org/10.1107/S2053230X16010311
Rahman MM, Andberg M, Koivula A, Rouvinen J, Hakulinen N (2018) The crystal structure of D-xylonate dehydratase reveals functional features of enzymes from the Ilv/ED dehydratase family. Sci Rep 8:865. https://doi.org/10.1038/s41598-018-19192-6
Rossoni L, Carr R, Baxter S, Cortis R, Thorpe T, Eastham G, Stephens G (2018) Engineering Escherichia coli to grow constitutively on D-xylose using the carbon-efficient Weimberg pathway. Microbiol 164:287–298. https://doi.org/10.1099/mic.0.000611
Salusjärvi L, Toivari M, Vehkomäki M-L, Koivistoinen O, Mojzita D, Niemelä K, Penttilä M, Ruohonen L (2017) Production of ethylene glycol or glycolic acid from D-xylose in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 14:127–113. https://doi.org/10.1007/s00253-017-8547-3
Shimada T, Momiyama E, Yamanaka Y, Watanabe H, Yamamoto K, Ishihama A (2017) Regulatory role of XynR (YagI) in catabolism of xylonate in Escherichia coli K-12. FEMS Microbiol Lett 364:2926. https://doi.org/10.1093/femsle/fnx220
Shimada T, Ogasawara H, Ishihama A (2018) Single-target regulators form a minor group of transcription factors in Escherichia coli K-12. Nucl Acids Res 34:gky138. https://doi.org/10.1093/nar/gky138
Singh A, Nigam PS, Murphy JD (2011) Renewable fuels from algae: an answer to debatable land based fuels. Bioresour Technol 102:10–16. https://doi.org/10.1016/j.biortech.2010.06.032
Stephens C, Christen B, Fuchs T, Sundaram V, Watanabe K, Jenal U (2007a) Genetic analysis of a novel pathway for D-xylose metabolism in Caulobacter crescentus. J Bacteriol 189:2181–2185. https://doi.org/10.1128/JB.01438-06
Stephens C, Christen B, Watanabe K, Fuchs T, Jenal U (2007b) Regulation of D-xylose metabolism in Caulobacter crescentus by a LacI-type repressor. J Bacteriol 189:8828–8834. https://doi.org/10.1128/JB.01342-07
Sun L, Yang F, Sun H, Zhu T, Li X, Li Y, Xu Z, Zhang Y (2016) Synthetic pathway optimization for improved 1,2,4-butanetriol production. J Ind Microbiol Biotechnol 43:67–78. https://doi.org/10.1007/s10295-015-1693-7
Suzuki T, Onishi H (1973) Oxidation and reduction of D-xylose by cell-free extract of Pichia quercuum. Appl Microbiol 25:850–852
Tai Y-S, Xiong M, Jambunathan P, Wang J, Wang J, Stapleton C, Zhang K (2016) Engineering nonphosphorylative metabolism to generate lignocellulose-derived products. Nat Chem Biol 12:1–10. https://doi.org/10.1038/nchembio.2020
Toivari M, Nygård Y, Kumpula E-P, Vehkomäki M-L, Benčina M, Valkonen M, Maaheimo H, Andberg M, Koivula A, Ruohonen L, Penttilä M, Wiebe MG (2012) Metabolic engineering of Saccharomyces cerevisiae for bioconversion of D-xylose to D-xylonate. Metab Eng 14:427–436. https://doi.org/10.1016/j.ymben.2012.03.002
Toivari M, Vehkomäki M-L, Nygård Y, Penttilä M, Ruohonen L, Wiebe MG (2013) Low pH D-xylonate production with Pichia kudriavzevii. Bioresour Technol 133:555–562. https://doi.org/10.1016/j.biortech.2013.01.157
Toivari MH, Ruohonen L, Richard P, Penttilä M, Wiebe MG (2010) Saccharomyces cerevisiae engineered to produce D-xylonate. Appl Microbiol Biotechnol 88:751–760. https://doi.org/10.1007/s00253-010-2787-9
Valdehuesa KNG, Lee W-K, Ramos KRM, Cabulong RB, Choi J, Liu H, Nisola GM, Chung W-J (2015) Identification of aldehyde reductase catalyzing the terminal step for conversion of xylose to butanetriol in engineered Escherichia coli. Bioprocess Biosyst Eng 38:1761–1772. https://doi.org/10.1007/s00449-015-1417-4
Valdehuesa KNG, Liu H, Ramos KRM, Park SJ, Nisola GM, Lee W-K, Chung W-J (2014) Direct bioconversion of D-xylose to 1,2,4-butanetriol in an engineered Escherichia coli. Process Biochem 49:25–32. https://doi.org/10.1016/j.procbio.2013.10.002
Wang J, Jain R, Shen X, Sun X, Cheng M, Liao JC, Yuan Q, Yan Y (2017a) Rational engineering of diol dehydratase enables 1,4-butanediol biosynthesis from xylose. Metab Eng 40:148–156. https://doi.org/10.1016/j.ymben.2017.02.003
Wang J, Shen X, Jain R, Wang J, Yuan Q, Yan Y (2017b) Establishing a novel biosynthetic pathway for the production of 3,4-dihydroxybutyric acid from xylose in Escherichia coli. Metab Eng 41:39–45. https://doi.org/10.1016/j.ymben.2017.03.003
Wang X, Xu N, Hu S, Yang J, Gao Q, Xu S, Chen K, Ouyang P (2018) D-1,2,4-Butanetriol production from renewable biomass with optimization of synthetic pathway in engineered Escherichia coli. Bioresour Technol 250:406–412. https://doi.org/10.1016/j.biortech.2017.11.062
Wasserstrom L, Portugal-Nunes D, Almqvist H, Sandström AG, Lidén G, Gorwa-Grauslund MF (2018) Exploring D-xylose oxidation in Saccharomyces cerevisiae through the Weimberg pathway. AMB Express 8:33. https://doi.org/10.1186/s13568-018-0564-9
Watanabe S, Kodaki T, Kodak T, Makino K (2006a) Cloning, expression, and characterization of bacterial L-arabinose 1-dehydrogenase involved in an alternative pathway of L-arabinose metabolism. J Biol Chem 281:2612–2623. https://doi.org/10.1074/jbc.M506477200
Watanabe S, Kodaki T, Makino K (2006b) A novel ⍺-ketoglutaric semialdehyde dehydrogenase: evolutionary insight into an alternative pathway of bacterial L-arabinose metabolism. J Biol Chem 281:28876–28888. https://doi.org/10.1074/jbc.M602585200
Watanabe S, Shimada N, Tajima K, Kodaki T, Makino K (2006c) Identification and characterization of L-arabonate dehydratase, L-2-keto-3-deoxyarabonate dehydratase, and L-arabinolactonase involved in an alternative pathway of L-arabinose metabolism: novel evolutionary insight into sugar metabolism. J Biol Chem 281:33521–33536. https://doi.org/10.1074/jbc.M606727200
Weimberg R (1961) Pentose oxidation by Pseudomonas fragi. J Biol Chem 236:629–635
Yim H, Haselbeck R, Niu W, Pujol-Baxley C, Burgard A, Boldt J, Khandurina J, Trawick JD, Osterhout RE, Stephen R, Estadilla J, Teisan S, Schreyer HB, Andrae S, Yang TH, Lee SY, Burk MJ, Van Dien S (2011) Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol 7:445–452. https://doi.org/10.1038/nchembio.580
Yim SS, Choi JW, Lee SH, Jeon EJ, Chung W-J, Jeong KJ (2017) Engineering of Corynebacterium glutamicum for consolidated conversion of hemicellulosic biomass into xylonic acid. Biotechnol J 96:1700040. https://doi.org/10.1002/biot.201700040
Zhang M, Wei L, Zhou Y, Du L, Imanaka T, Hua Q (2013) Genetic analysis of D-xylose metabolism pathways in Gluconobacter oxydans 621H. J Ind Microbiol Biotechnol 40:379–388. https://doi.org/10.1007/s10295-013-1231-4
Zhang N, Wang J, Zhang Y, Gao H (2016) Metabolic pathway optimization for biosynthesis of 1,2,4-butanetriol from xylose by engineered Escherichia coli. Enzym Microb Technol 93-94:51–58. https://doi.org/10.1016/j.enzmictec.2016.07.007
Zhao A, Hu X, Wang X (2017) Metabolic engineering of Escherichia coli to produce gamma-aminobutyric acid using xylose. Appl Microbiol Biotechnol 101:3587–3603. https://doi.org/10.1007/s00253-017-8162-3
Funding
This work was supported by Korea Research Fellowship Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2015H1D3A1062172 and 2016R1C1B1013252) and by the Ministry of Education (No. 2018R1D1A1B07043993 and No. 2009-0093816).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Valdehuesa, K.N.G., Ramos, K.R.M., Nisola, G.M. et al. Everyone loves an underdog: metabolic engineering of the xylose oxidative pathway in recombinant microorganisms. Appl Microbiol Biotechnol 102, 7703–7716 (2018). https://doi.org/10.1007/s00253-018-9186-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9186-z