Abstract
1, 2, 4-Butanetriol (BT) is a high-value non-natural chemical and has important applications in polymers, medical production and military industry. In the constructed BT biosynthesis pathway from xylose in Escherichia coli, the xylose dehydrogenase (Xdh) and the benzoylformate decarboxylase (MdlC) are heterologous enzymes and the activity of MdlC is the key limiting factor for BT production. In this study, six chaperone protein systems were introduced into the engineered E. coli harboring the recombinant BT pathway. The chaperone GroES–GroEL was beneficial to Xdh activity but had a negative effect on MdlC activity and BT titer. The plasmid pTf16 containing the tig gene (trigger factor) was beneficial to Xdh and MdlC activities and improved the BT titer from 0.42 to 0.56 g/l from 20 g/l xylose. However, co-expression of trigger factor and GroES–GroEL simultaneously reduced the activity of MdlC and had no effect on the BT production. The plasmid pKJE7 harboring dnaK–dnaJ–grpE showed significant negative effects on these enzyme activities and cell growth, leading to completely restrained the BT production. Similarly, co-expression of DnaKJ–GrpPE and GroES–GroEL simultaneously reduced Xdh and MdlC activities and decreased the BT titer by 45.2 %. The BT production of the engineered E. coli harboring pTf16 was further improved to the highest level at 1.01 g/l under pH control (pH 7). This work showed the potential application of chaperone proteins in microorganism engineering to get high production of target compounds as an effective and valuable tool.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
1, 2, 4-butanetriol (BT) is a high-value non-natural chemical with the properties resemble to glycerol. It has an important application in the military industry as the precursor of 1, 2, 4-butanetriol trinitrate (BTTN), a novel energetic plasticizer in propellant and explosive with less sensitivity to shock and better thermostability than nitroglycerin (Gouranlou and Kohsary 2010). Although several chemical synthesis routes have been developed (Paul and Hilly 1939), the harsh conditions (Adkins and Billica 1948), abundant byproducts, and potential environmental pollution restrict the commercial production of BT by these chemical strategies.
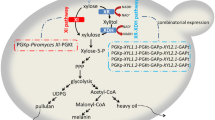
Due to the problems of these chemical strategies, microbial synthesis of BT is selected as an alternative route. There are two artificial designed pathways available for the biosynthesis of BT, the CoA-dependent pathway (Fig. 1a) (Li et al. 2014) and the CoA-independent pathway (Fig. 1b) (Niu et al. 2003). In the CoA-dependent pathway, glucose is converted to malate by glycolysis and then transformed to BT by six enzymatic reactions, two of which are CoA-dependent reactions (Li et al. 2014). Owing to the complexity and low efficiency of the CoA-dependent reactions, the BT production from glucose or malate is only 120 or 180 ng/l, respectively. In the CoA-independent pathway, xylose is converted to xylonic acid by xylose dehydrogenase (Xdh) and then dehydrated to 2-keto-3-deoxy-xylonate (KDX) by xylonate dehydratase (YjhG). Subsequently, KDX is converted to 3, 4-dihydroxybutanal (DHB) by benzoylformate decarboxylase (MdlC) isolated from Pseudomonas putida and then transformed to BT by alcohol dehydrogenases (Adh). The four enzymes responsible for the bioproduction of BT have been integrated into Escherichia coli and produced BT with a titer of 0.88 g/l which is much higher than that obtained from the CoA-dependent pathway (Valdehuesa et al. 2014). The decarboxylation of KDX by MdlC is the crucial step in the synthesis of BT. However, the catalytic activity of MdlC on the non-natural substrate KDX is unsatisfactory (Lau 2007). In order to increase the titer of BT, improving the catalytic activity of the MdlC is imperative.
Generally, there are two strategies to increase the activity of a metabolic enzyme: increasing its catalytic efficiency to a specific substrate by protein engineering or improving its expression level. A previous report failed to achieve the evolution of MdlC by gene shuffling and directed mutagenesis (Lau 2007). Similarly, codon optimization and increasing promoter strength to increase the expression level of MdlC also resulted in the decreased BT titer (Lau 2007). Improving enzyme folding efficiency is another valuable strategy used to increase the activity of heterologously expressed enzymes. It is known that chaperones can assist the correct folding of foreign proteins (Schlieker et al. 2002), for instance, introduction of trigger factor resulted in positive effects on the soluble expressions of mouse endostatin and human lysozyme (Nishihara et al. 2000).
There are three main elements of the chaperone systems in E. coli: trigger factor, GroEL–GroES, and DnaK–DnaJ–GrpE (Baneyx and Mujacic 2004), which can enable the correct folding of polypeptides and prevent the formation of inclusion bodies (Schlieker et al. 2002). Trigger factor is a three domain protein that can associate with the nascent polypeptide and exhibits prolyl-isomerase cis/trans activity (Valent et al. 1997). The GroESL is a member of Hsp60 chaperone family and can mediate protein transition between soluble and insoluble status (Kerner et al. 2005). The DnaK (Hsp70 chaperone family) is targeted to high-affinity binding sites on nascent proteins by chaperone DnaJ, and a new synthesized protein in a native conformation is released from this complex by GrpE (Ulrich and Manajit 2002). In this study, five plasmids, pTf16, pGro7, pG-Tf2, pKJE7 and pG-KJE8 (Table 1), harboring different chaperone proteins were introduced into the engineered E. coli containing the CoA-independent BT pathway. The effects of different chaperones on the catalytic activities of the metabolic enzymes and the production of BT and the potential mechanisms were analyzed.
Materials and methods
Materials
DNA polymerase, restriction endonuclease, T4 ligase, protein marker were purchased from Takara (Dalian, China). Genomic DNA Purification and Gel DNA Purification Kits were purchased from Jierui (Shanghai, China). Isopropyl β-D-1-thiogalactopyranoside (IPTG), kanamycin sulfate, chloramphenicol, and arabinose were purchased from Sangon (Shanghai, China). All primers used were synthesized by Sangon (Shanghai, China). Tryptone and yeast extract were purchased from Oxoid (Basingstoke, UK).
Gene cloning and plasmids construction
Pseudomonas putida (CCTCC AB92019) was used as the source of mdlC (Gene ID: 882177). Caulobacter crescentus (ATCC BAA-2331) was used as the source of xdh (Gene ID: 7329904). E. coli JM109 (Novagen, USA) and pMD19-T (simple) vector (Takara, China) were used for the cloning of PCR fragments. E. coli W3110 (ATCC 27325) and pEtac (Sheng et al. 2003) were used for the protein expression. The plasmids harboring chaperones are listed in Table 1.
The genomic DNA of C. crescentus and P. putida were extracted and used as the PCR template. All primers used in this work are listed in Table 2. Primers P1 and P2 were designed based upon the C. crescentus xdh. Primers P3 and P4 were designed based upon the P. putida mdlC. The amplified fragments were inserted into the plasmid pMD19-T (simple). The resulted derivatives were named as pMD-xdh and pMD-mdlC, respectively. Plasmid pMD-xdh was double-digested with EcoRI and HindIII and the resultant fragment was inserted into the EcoRI–HindIII sites of pEtac to generate pEtac-xdh. Plasmid pMD-mdlC was digested with EcoRI and the resultant mdlC fragment was cloned into the EcoRI site of the plasmid pEtac to generate pEtac-mdlC. Primers P5 and P6 were used to amplify tac-xdh fragment from plasmid pEtac-xdh and the resultant product was digested with XhoI and then inserted into the XhoI site of pEtac-mdlC to generate pEtac-mdlC–tac-xdh. Plasmid pEtac-mdlC–tac-xdh was transformed into E. coli W3110 to generate a BT-production strain termed E. coli (BT). The plasmids containing chaperone proteins were transformed into E. coli (BT), generating the recombinants E. coli (BT)/(pG-KJE8), E. coli (BT)/(pGro7), E. coli (BT)/(pTf16), E. coli (BT)/(pKJE7), and E. coli (BT)/(pG-Tf2), respectively.
Media and growth conditions
A single colony of BT-production strain was inoculated into 10 ml Luria–Bertani medium at 37 °C. About 500 μl overnight culture of the recombinant strain was inoculated into 50 ml LB medium containing 20 g/l xylose and 1.0 mg/l thiamine pyrophosphate in a 250-ml shaker and incubated on a rotary shaker at 37 °C. All the seeds and fermentation media were supplemented with 50 μg/ml kanamycin sulfate or 20 μg/ml chloramphenicol. IPTG, l-arabinose or tetracycline were added at the final concentration of 0.5 mM, 2 mg/ml or 10 ng/ml, respectively, as necessary for the induction of BT pathway or the expressions of chaperones until cell density (OD600) reached 0.6 (about 2 h after inoculation). In the pH-controlled fermentation, the pH of broth was monitored by a pH indicator paper and maintained at 7.0 ± 0.5 by adding 4 M NaOH every 2 h.
Protein analysis
Protein fractions were analyzed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) on 12 % gel slabs and stained with Coomassie blue. The protein concentration was determined by binding the protein to Coomassie Brilliant Blue G-250 and monitoring the absorption at 595 nm, as described by Bradford (Bradford 1976).
Enzyme activity assay
The recombinant cell was collected by centrifugation at 10,000×g for 20 min and washed twice by 50 mM potassium phosphate buffer (pH 7.0). The cell lysis was performed by sonication on ice and the supernatant was obtained by centrifugation (10,000×g; 20 min; 4 °C) for further analysis.
Xdh activity was measured by adding protein extracts to the substrate (containing 100 mM Tris (pH 8.1), 2 mM MgCl2, 100 mM xylose, and 1 mM NAD+). The formation of NADH was measured at 340 nm at 30 °C. One unit of activity is defined as the enzyme required to convert 1 μmol NAD+ to NADH per minute (Berghall et al. 2008; Cao et al. 2013). The measure of MdlC activity was determined by a coupled spectrophotometric assay performed at 30 °C in 50 mM potassium phosphate buffer (pH 6.0) containing 0.1 mM thiamine pyrophosphate, 10 mM MgCl2, 10 mM benzoyl formate, 0.35 mM NADH and 5 U/ml horse liver alcohol dehydrogenase. The assay was performed in a total volume of 200 μl in a 96-well plate. One unit is defined as the enzyme required to catalyze the decarboxylation of 1 μmol of benzoyl formate per minute under standard conditions (Siegert et al. 2005).
Analytical method
Biomass was monitored at 600 nm. The total amount of compounds was measured by High Performance Liquid Chromatography (HPLC) with a refractive index detector and a Bio-Rad Aminex HPX-87 column (300 mm × 7.8 mm) using 5 mM H2SO4 as mobile phase with a flow rate of 0.6 ml/min at 60 °C.
GC–MS analysis performed on BRUKER SCIONSQ-456 gas chromatograph coupled to mass spectrometer. The GC separation was achieved on DB-WAX (30 m × 0.25 mm × 0.25 μm) with helium working as carrier gas at 0.9 ml/min. The temperature program was set as follows: 70 °C for 0.5 min, increasing to 230 at 15 °C/min, then held at 230 °C for 6 min. Ionization was performed by electron impact (EI+; 70 eV; emission current, 80 μA) and masses were scanned from 33 to 300 amu.
Statistical analysis
To investigate statistical differences, non-parametric Mann–Whitney U test (for pH-controlled and uncontrolled fermentation) and ANOVA with multiple comparisons (for other results) were conducted on Statistical Package for Social Scientists (SPSS) version 20.0 (IBM, Armonk, New York, USA). Samples with P values of <0.05 were considered statistically different.
Results
Construction of BT synthesis pathway in E. coli
To construct the BT pathway, the heterologous xdh and mdlC were cloned from C. crescentus and P. putida, respectively. The resultant DNA fragments were fused with tac promoter and then inserted into the plasmid pEtac to form the recombinant plasmid pEtac-mdlC–tac-xdh (Fig. 2). The soluble expressions of Xdh and MdlC were confirmed by SDS-PAGE analysis and enzyme activity assay. SDS-PAGE analysis (Fig. 3) showed significant amount of 57 and 28 kDa protein bands which are similar to the predicted molecular weight of MdlC and Xdh in the intracellular soluble fractions of the cells carrying pEtac-mdlC–tac-xdh. The activities of Xdh and MdlC were found to be 0.59 and 0.82 U/mg protein, respectively. These results suggest that the introduced xdh and mdlC genes are successfully expressed in E. coli.
SDS-PAGE analysis of the intracellular soluble protein of recombinant E. coli. (M) Protein marker; 1 E. coli W3110/pEtac; 2 E. coli (BT); 3 E. coli (BT)/(pG-KJE8) (co-expressed with DnaK–DnaJ–GrpE and GroES–GroEL); 4 E. coli (BT)/(pGro7) (co-expressed with GroES–GroEL); 5 E. coli (BT)/(pTf16) (co-expressed with trigger factor); 6 E. coli (BT)/(pKJE7) (co-expressed with DnaK–DnaJ–GrpE); 7 E. coli (BT)/(pG-Tf2) (co-expressed with GroES–GroEL and trigger factor); 8 E. coli (BT) induced with arabinose and IPTG; 9 E. coli (BT) induced with tetracycline and IPTG. Note The expressed GroEL is indicated by triangle. The trigger factor (56 kDa) shares similar molecular weight with MdlC (56 kDa), which makes it difficultly observed. All other recombinant proteins are indicated by arrows. GroES (10 kDa) is not detected
To further analyze the BT synthesis capability of E. coli (BT), the recombinant strain was incubated in LB medium with the addition of xylose as precursor. As shown in Fig. 4, about 418 mg/l BT was detected in the culture supernatant by HPLC analysis. The GC–MS analysis further indicated the presence of BT in fermentation broth. In contrast, no BT was detected in the culture broth of E. coli/pEtac. These results suggest that the constructed artificial pathway has the ability to synthesize BT from xylose.
Effects of co-expression of chaperones on protein expressions and enzyme activities
To improve the production of BT, plasmids encoding five different combinations of chaperones were transformed into E. coli (BT) (Table 1). As shown in Fig. 3, the soluble fractions of Xdh and MdlC were dramatically reduced by expression of DnaK–DnaJ–GrpE proteins. The activities of Xdh and MdlC were reduced by 96.6 % and 54.8 % (Table 3), respectively, suggesting significant negative effect of DnaK–DnaJ–GrpE proteins on the soluble expressions of Xdh and MdlC. Similarly, combined expressions of DnaK–DnaJ–GrpE and GroES–GroEL proteins also had the negative effect on the activities of both enzymes. In contrast, introduction of trigger factor alone increased the activities of Xdh and MdlC by about 74.6 % and 11.0 % (Table 3), respectively. Expression of GroES–GroEL proteins were beneficial to the production of soluble Xdh but not to that of MdlC. Combined expression of GroES–GroEL and trigger factor proteins in recombinant strain showed slightly improvement on the Xdh activity by 11.9 % but reduced the activity of MdlC by 32.9 % (Table 3). These results demonstrate that the effects of these chaperones on the soluble expressions and the activities of these two enzymes are varied.
Effects of co-expression of chaperones on cell growth and BT production
The effects of different chaperones on cell growth and BT production were also investigated (Fig. 5). There was no obvious difference between these six recombinant strains in the first two hours. However, after the addition of IPTG, the growth rates of recombinant strains containing DnaK–DnaJ–GrpE clearly decreased leading to the final biomass reductions of 50.4 % and 75.6 %, respectively. This result suggests that the expression of DnaK–DnaJ–GrpE is not conducive to cell growth. In contrast, the biomass of the recombinants harboring GroES–GroEL or trigger factor were improved by 23.6 % and 68.4 %, respectively. The plasmid pG-Tf2 containing groES–groEL and tig genes also showed slightly positive effect on cell growth.
As shown in Fig. 5, no BT was detected before 10 h in all the broth of strains tested. BT synthesis was completely inhibited by the expression of DnaK–DnaJ–GrpE and a large amount of residual xylose remained (data not shown). Co-expression of DnaK–DnaJ–GrpE and GroES–GroEL also reduced the BT titer by 45.2 %. In comparison, BT titer of the recombinant strain harboring trigger factor was increased from 0.41 to 0.56 g/l. Compared with the parent strain, the final BT titers produced by E. coli (BT)/(pGro7) and E. coli (BT)/(pG-Tf2) were similar.
Improvement of BT production by pH control strategy
The accumulation of xylonic acid and significant acidification of broth were observed (data no shown). Generally, acidification of culture broth can be an important limitation for cell growth and product synthesis in microbial fermentation (Simango 1995). To reduce the acidic stress, a pH control strategy was applied to maintain the culture pH at 7.0 ± 0.5 (Table 4). The biomass and BT titers of recombinant E. coli (BT) and E. coli (BT)/(pG-Tf2) were improved by about 75–90 % using the pH control strategy. Similarly, the biomass and BT titer of recombinant E. coli (BT)/(pTf16) were increased by 27.9 % and 80.4 %, respectively. The biomass and BT titer of recombinant E. coli (BT)/(pGro7) were enhanced by 57.0 % and 15.8 %, respectively. However, there was no obvious difference detected in the cell growth or BT production of recombinants harboring pKJE7 and pG-KJE8 after pH-controlled optimization.
Discussion
Generally, the improvement of enzyme activity is the first priority in metabolic engineering to increase production (Shu et al. 2010). In this study, different protein chaperone systems were co-expressed in a recombinant E. coli BT producer strain to try and increase both enzyme activity and BT titer. The plasmid pTf16 harboring trigger factor alone was the only one that exhibited a positive effect on the activities of the heterologous enzymes and the titer of BT produced. It has been reported that trigger factor associates with ribosomes and nascent polypeptide chains to assist folding of newly synthesized proteins (Hesterkamp et al. 1996), this may be the main reason of the improved activity of MdlC and the BT production.
The groES–groEL gene was beneficial to Xdh activity but not conducive to MdlC activity. GroESL is known to assist the folding of only a minority of protein within the cell which have a molecular mass in the range of 10–60 kDa (Houry et al. 1999). Interestingly, MdlC is a tetramer which is too large to enter the cavity formed within GroEL. In the engineered BT pathway, the activity of MdlC is the limiting factor. Although the Xdh activity was improved by the introduction of GroES–GroEL, the reduced MdlC activity may also lead to the accumulation of KDX, which is not beneficial to the biosynthesis of BT from xylose. DnaK–DnaJ–GrpE is significantly detrimental to the enzyme activities and cell growth. Similar result is also reported in the co-expression of horseradish peroxidase with DnaK–DnaJ–GrpE (Kondo et al. 2000). It has been demonstrated that the positive effect of the DnaKJ system is mostly restricted to target proteins with a molecular weight greater than 60 kDa (Pfeffer et al. 2007). Compared with MdlC, the lower molecular weight of Xdh may be the part of the reasons of the almost completely inhibited enzyme activity. Besides assisting protein folding, the DnaK may also act as a proteolytic enhancer and improve the proteolytic degradation of target enzymes (Mónica et al. 2007), as reported in the study of aggregation-prone GFP (Rinas et al. 2007). In the process of cell growth, partially folded protein complexed with DnaK–DnaJ–GrpE may accumulate, while the other proteins may be degraded by protease (Kondo et al. 2000). These factors may be responsible for the reduced enzyme activities and cell growth and lead to the completely inhibited BT production. Although groES–groEL showed slightly improvement on Xdh activity, the plasmid pG-KJE8 harboring dnaK–dnaJ–grpE and groES–groEL also significantly reduced the enzyme activities and BT production, which further supports the strongly negative effect of dnaK–dnaJ–grpE on BT synthesis. Similarly, co-expression of trigger factor and GroES–GroEL proteins simultaneously by introduction of pG-Tf2 did not exhibit positive synergistic effect on BT production.
In microbial fermentation, acidification of broth will cause the inhibition of the cell growth and the products synthesis (Nakano et al. 1997). The pH control strategy reduced adverse effect of low pH stress on recombinants harboring pGro7, pTf16 and pG-Tf2 and improved the BT titer up to 1 g/l. However, it had no effect on the cell growth and BT production of recombinants containing the plasmid pKJE7 or pG-KJE8. These results demonstrate that the stress induced by expression of dnaK–dnaJ–grpE is much stronger than that from the acidification of surroundings. Actually, these introduced chaperones may not only have effect on Xdh and MdlC but also act as the global regulatory proteins (And and Bouché 1992). Thus, detailed mechanisms involved in the varied performances of the chaperone proteins applied here need further study.
In conclusion, six chaperone proteins were applied to try and improve enzyme activity and BT production in a recombinant E. coli BT strain. Even though only trigger factor showed a positive effect on BT production, the application of chaperones in microorganism engineering to improve the production of target compounds is still an effective and valuable strategy. Further attention can be put on the screening of other effective regulatory chaperones and the disruption of byproducts pathway to improve the biosynthesis of BT from xylose.
References
Adkins H, Billica HR (1948) The hydrogenation of Esters to Alcohols at 25–150. J Am Chem Soc 70:3121–3125
And FT, Bouché JP (1992) Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol 6:615–620
Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 22:1399–1408
Berghall S, Hilditch SM, Richard P (2008) Identification in the mould Hypocrea jecorina of a gene encoding an NADP+: d-xylose dehydrogenase. FEMS Microbiol Lett 277:249–253
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cao Y, Xian M, Zou H, Zhang H (2013) Metabolic engineering of Escherichia coli for the production of xylonate. PLoS One 8:e67305
Gouranlou F, Kohsary I (2010) Synthesis and characterization of 1, 2, 4-butanetrioltrinitrate. Asian J Chem 22:4221
Hesterkamp T, Hauser S, Lütcke H, Bukau B (1996) Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci 93:4437–4441
Houry WA, Frishman D, Eckerskorn C, Lottspeich F et al (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402:147–154
Kerner MJ, Naylor DJ, Ishihama Y, Maier T et al (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122:209–220
Kondo A, Kohda J, Endo Y, Shiromizu T et al (2000) Improvement of productivity of active horseradish peroxidase in Escherichia coli by coexpression of Dsb proteins. J Biosci Bioeng 90:600–606
Lau MK (2007) Syntheses and downstream purification of 1,2,4-butanetriol. Dissertation, Michigan State University
Li XH, Cai Z, Li Y, Zhang YP (2014) Design and construction of a non-natural malate to 1,2,4-butanetriol pathway creates possibility to produce 1,2,4-butanetriol from glucose. Sci Rep 4:9
Mónica MA, Andrea V, Antonio V (2007) Role of the chaperone DnaK in protein solubility and conformational quality in inclusion body-forming Escherichia coli cells. FEMS Microbiol Lett 273:187–195
Nakano K, Rischke M, Sato S, Märkl H (1997) Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Appl Microbiol Biotechnol 48:597–601
Nishihara K, Kanemori M, Yanagi H, Yura T (2000) Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl Environ Microbiol 66:884–889
Niu W, Molefe MN, Frost JW (2003) Microbial synthesis of the energetic material precursor 1,2,4-butanetriol. J Am Chem Soc 125:12998–12999
Paul R, Hilly G (1939) Preparation of an active iron and its application to the semihydrogenation of acetylene derivatives. Bull Soc Chim Fr 6:218–223
Pfeffer J, Rusnak M, Hansen CE, Rhlid RB et al (2007) Functional expression of lipase A from Candida antarctica in Escherichia coli: a prerequisite for high-throughput screening and directed evolution. J Mol Catal B Enzym 45:62–67
Rinas U, Hoffmann F, Betiku E, Estapé D et al (2007) Inclusion body anatomy and functioning of chaperone-mediated in vivo inclusion body disassembly during high-level recombinant protein production in Escherichia coli. J Biotechnol 127:244–257
Schlieker C, Bukau B, Mogk A (2002) Prevention and reversion of protein aggregation by molecular chaperones in the Escherichia coli cytosol: implications for their applicability in biotechnology. J Biotechnol 96:13–21
Sheng W, Wang ZX, Shao WL, Liu JQ et al (2003) Secretory expression of high thermophilic α-amylase gene of Pyrococcus furiosus, a kind of Earchaic bacterium. China Brew 22:12–14
Shu ZY, Jiang H, Lin RF, Jiang YM et al (2010) Technical methods to improve yield, activity and stability in the development of microbial lipases. J Mol Catal B Enzym 62:1–8
Siegert P, McLeish MJ, Baumann M, Iding H et al (2005) Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein Eng Des Sel 18:345–357
Simango C (1995) Effective acidification of traditional fermented foods. J Trop Med Hyg 98:465–468
Ulrich FH, Manajit HH (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858
Valdehuesa KNG, Liu H, Ramos KRM, Park SJ et al (2014) Direct bioconversion of D-xylose to 1,2,4-butanetriol in an engineered Escherichia coli. Process Biochem 49:25–32
Valent QA, De Gier JWL, Heijne GV, Kendall DA et al (1997) Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol 25:53–64
Acknowledgments
This work was supported by the Natural Science Foundation of Jiangsu Province (No. BK20140138), the Six Talent Peaks Project in Jiangsu Province (No. 2014-XCL-017), Natural Science Foundation of China (No. 31270080), the National High Technology Research and Development Program of China (863 Program, No. 2012AA021201), the Fundamental Research Funds for the Central Universities (No. JUSRP11431) and the 111 project (No. 111-2-06).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xinyao Lu and Shuying He have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Lu, X., He, S., Zong, H. et al. Improved 1, 2, 4-butanetriol production from an engineered Escherichia coli by co-expression of different chaperone proteins. World J Microbiol Biotechnol 32, 149 (2016). https://doi.org/10.1007/s11274-016-2085-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2085-5