Abstract
Pyruvate decarboxylase (Pdc) is a cytosolic enzyme located at the branch point between fermentative and respiratory sugar catabolism. Here, we identified and functionally characterized KmPDC1 and KmPDC5 encoding two homologs of Pdc in the thermotolerant yeast Kluyveromyces marxianus KCTC 17555. Despite the conservation of important Pdc domains, a few amino acid sequences essential for enzymatic activity are not conserved in KmPdc5p. Deletion of KmPDC1 alone eliminated most of Pdc activity, but the growth of the Kmpdc1Δ strain on glucose was comparable to that of the wild type (WT) strain under aerobic conditions. In contrast to the WT, Kmpdc1Δ could not grow on glucose under oxygen-limited conditions. The KmPDC5 deletion did not generate any apparent change in Pdc activity or growth patterns under several tested conditions. Whereas the expression of KmPDC1 was enhanced by glucose, the basic expression levels of KmPDC5 were very low, without a detectable difference between glucose and nonfermentable carbon sources. Moreover, KmPDC5 overexpression was unable to complement the growth defect of Kmpdc1Δ in the presence of antimycin A, and the purified recombinant KmPdc5p was inactive in Pdc activity assay, supporting the notion that KmPdc5p may lack Pdc enzymatic activity. Notably, compared to the WT, Kmpdc1Δ single and Kmpdc1Δpdc5Δ double mutants produced significantly less glycerol, acetate, and ethanol while accumulating pyruvate. Altogether, our data indicate that a single deletion of KmPDC1 is sufficient in Crabtree-negative K. marxianus strains to generate a starting host strain for engineering of production of high-value biomaterials derived from pyruvate without byproduct formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pyruvate decarboxylase (Pdc) is a cytosolic enzyme that decarboxylates mainly pyruvate, producing acetaldehyde and carbon dioxide (Pronk et al. 1996). Pdc forms a homotetramer via interaction between two homodimers, and thus carries four active sites requiring thiamine pyrophosphate (TPP) and a magnesium ion as cofactors (Dyda et al. 1993). Pdc, located at the branch point of the respiratory and fermentative pathways, is widely conserved in plants and fungi, rare in bacteria, and is absent in most animals (Konig 1998). In yeasts of the genus Saccharomyces, the efficiency of this enzyme is a key factor in directing the flow of pyruvate toward alcoholic fermentation. In the Crabtree-positive yeast Saccharomyces cerevisiae, six PDC genes related to Pdc activity (PDC1, PDC2, PDC3, PDC4, PDC5, and PDC6) have been reported. While PDC1, PDC5, and PDC6 code for Pdc enzymes, PDC2 encodes a transcription factor regulating PDC5 expression (Nosaka et al. 2012). Pdc enzymatic activity is still substantially retained in the PDC1-null S. cerevisiae; nonetheless, Pdc1p, the enzyme encoded by PDC1, is responsible for most Pdc activity in this yeast (Schaaff et al. 1989). It was later shown that PDC5 expression is induced by Pdc2p when PDC1 is disrupted, thus recovering Pdc enzyme activity to ~ 60–70% in S. cerevisiae (Nosaka et al. 2012; Seeboth et al. 1990). Expression of ScPDC6 is not observed in yeast cells under normal physiological conditions, but is detected only under sulfur-limiting conditions and during high-sugar stress conditions (Erasmus et al. 2003). Saccharomyces kluyveri, a distant relative of S. cerevisiae, was also reported to have three PDC genes coding for functional Pdc enzymes (Moller et al. 2004).
PDC homologs have also been reported in Crabtree-negative yeasts such as Kluyveromyces lactis (Bianchi et al. 1996), Ogataea polymorpha (Ishchuk et al. 2008), Scheffersomyces stipitis (Lu et al. 1998), Wickerhamomyces anomalus (Fredlund et al. 2006), Pichia pastoris (Agarwal et al. 2013), and Candida utilis (Franzblau and Sinclair 1983). In the case of these respiratory yeasts, only one PDC gene has been mostly reported except in S. stipitis. Although Pdc-deficient S. cerevisiae cannot grow on glucose as a sole carbon source due to C2-auxotrophy and a redox imbalance in the cytosol (van Maris et al. 2004), disruption of the PDC1 gene blocks ethanol fermentation but does not decrease growth in Crabtree-negative yeasts like K. lactis and C. utilis (Ikushima et al. 2009; Porro et al. 1999).
Kluyveromyces marxianus is homothallic hemiascomycetous yeast and is taxonomically related to S. cerevisiae and is a sister species of K. lactis (Lane and Morrissey 2010). K. marxianus is a GRAS (Generally Recognized as Safe) organism based on its safe association with food production for long periods. K. marxianus has drawn attention as potential industrial host strains in the biotechnology industry, particularly owing to its high thermotolerance, broad substrate utilization, and a high growth rate (Fonseca et al. 2008; Lane and Morrissey 2010). K. marxianus is classified as the so-called aerobic-respiring or Crabtree-negative yeast, in which a high glucose concentration does not affect the respiratory pathway (Lane and Morrissey 2010), although some strains of K. marxianus have Crabtree-positive yeast-like growth phenotypes, such as a relatively high fermentation ability (Lane et al. 2011; Merico et al. 2007). In the present study, we identified two K. marxianus PDC homologs, KmPDC1 and KmPDC5, in the recently sequenced genome of K. marxianus KCTC 17555 (Jeong et al. 2012). We were interested in investigating whether deletion of both two KmPDC homologs might be required to generate a pyruvate-accumulating precursor strain in K. marxianus, whereas a single knockout of PDC1 is shown to be sufficient in most other Crabtree-negative yeasts. Thus, we carried out functional analysis of the two KmPDC genes, focusing on their physiological functions in fermentative and respiratory carbon catabolism, and evaluated the potential of Pdc-deficient K. marxianus as a starting host strain for subsequent metabolic engineering aimed at production of high-value biomaterials derived from pyruvate.
Materials and methods
Strains and media
Strains, including Escherichia coli and yeast, and plasmids used in this study are listed in Table 1. Yeast cells were routinely grown at 37 °C in YPD (1% yeast extract, 2% bacto-peptone, and 2% glucose) or in selective synthetic complete media without uracil (SC-URA, 0.67% yeast nitrogen base without amino acids, 2% glucose, and the drop-out amino acid mixture without uracil). To pop out the URA3 selection marker, the yeast cells were cultivated in the 5-fluoroorotic acid (5-FOA) medium, consisting of 0.67% yeast nitrogen base without amino acids, an amino acid mixture containing 90 mg uracil/l, 2% dextrose, and 0.5 μg 5-FOA/l. Escherichia coli DH5α (Invitrogen) was used for the general recombinant DNA techniques, and BL21(DE3) (New England Biolabs) was used for recombinant protein expression. E. coli transformants were cultured in the Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 1% NaCl) supplemented with 100 μg/ml ampicillin.
Growth analysis under aerobic and oxygen-limited conditions
Yeast cells were cultivated in YPD overnight, their concentration was adjusted to initial optical density at 600 nm (OD600) of 0.1 in 50 ml of YPD in a 500-ml flask, and the cells were incubated at 37 °C and 220 rpm for aerobic cultivation. In oxygen-limited cultures, the concentration of yeast cells was adjusted to initial OD600 of 0.5 in 50 ml of YPD, and then 2-ml culture aliquots were placed into 2-ml microcentrifuge tubes, which were tightly sealed by wrapping five times with parafilm (Bemis Company, Inc.) and incubated at 37 °C and 50 rpm. OD600 of each sample was measured for growth analysis twice in two independent experiments.
In silico analysis
Multiple sequence alignments were performed using the ClustalW software (http://align.genome.jp) and were shaded using Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). Phylogenetic tree was constructed by the ClustalW function in the DNASTAR MegAlign software (Thompson et al. 1994). Amino acid sequences of S. cerevisiae Pdc proteins were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Amino acid sequences of K. marxianus and K. lactis Pdc proteins were derived from ERGO (http://igenbio.com/ergo_bioinformatics_and_analysis) provided by SAIT. Protein motif analysis was conducted using CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
Construction of the K. marxianus PDC1 and PDC5 double-deletion strain
To construct the KmPDC1 and KmPDC5 double-deletion strain (Kmpdc1Δpdc5Δ), the Km05ku80Δpdc5Δ::ScURA3 strain (Choo et al. 2014) served as a parent strain. After recovering the Ura− auxotrophic phenotype by popping out the ScURA3 gene from the KmPDC5::hisG-ScURA3-hisG locus in the 5-FOA medium, the resultant Km05ku80Δpdc5Δ was transformed by the modified lithium acetate-dimethyl sulfoxide (DMSO) method (Hill et al. 1991) with the KmPDC1::hisG-ScURA3-hisG disruption cassette, which was generated by digestion of pKI-KmPDC1DU2 with XhoI/SacI (Choo et al. 2014). Ura+ transformants were selected on SC-URA plates, and correct deletion of the KmPDC1 gene via double homologous recombination was confirmed by PCR using the primer sets listed in Table S1. The Ura− auxotrophic phenotype was recovered from the of Km05ku80Δpdc1::ScURA3pdc5Δ strain by popping out the ScURA3 gene to generate Km05ku80Δpdc1Δpdc5Δ strain. The pop-out of ScURA3 was also confirmed by PCR using the primer set listed in Table S1.
Construction of the KmPDC1- and KmPDC5-complemented strains
To express KmPDC1 and KmPDC5 under the control of the KmPDC1 promoter in K. miarxianus, the DNA fragments containing the full-length KmPDC1 and KmPDC5 ORFs with their 500-bp native terminators were amplified by PCR with the following primer sets: KmPDC1 5D 1F Xho/KmPDC1 3D2B Not for KmPDC1, and KmPDC5 1F Xho/KmPDC5 3D 2B Not for KmPDC5 (Table S1), respectively, from the genomic DNA of K. marxianus KCTC 17555. The 2-kb KmPDC1 promoter was amplified by PCR with primers KmPDC1 5D 1F Xba/KmPDC1 5D 2B Xho (Table S1). The obtained amplicons of KmPDC1 and KmPDC5 were digested with XhoI/NotI and ligated with the XbaI/NotI-digested pKI vector (Table 1) and the XbaI/XhoI digested KmPDC1 promoter, resulting in plasmids pKI-PDC1p-KmPDC1 and pKI-PDC1p-KmPDC5, respectively. To integrate the resultant KmPDC expression vectors at the KmPDC1 promoter locus by a single homologous recombination, the vectors pKI-PDC1p-KmPDC1 and pKI-PDC1p-KmPDC5 were linearized by digestion at the SphI site within KmPDC1 promoter and introduced into the Kmpdc1Δpdc5Δ mutant strain. The correct integration of KmPDC vectors at the KmPDC1 promoter locus was confirmed by PCR analysis using the primer sets, Iden KmPDC1 5D 0F/KmPDC1 3D 2B Not and Iden KmPDC1 5D 0F/KmPDC5 3D 2B Not, respectively.
Construction of E. coli expression vectors for MBP-fused KmPdc proteins
The E. coli vectors pET-MBP-KmPDC1 and pET-MBP-KmPDC5, expressing KmPdc1 and KmPdc5 proteins as a recombinant protein fused with maltose binding protein (MBP), were constructed as follows. The DNA fragments of the full-length KmPDC1 and KmPDC5 ORFs containing the 5’ TEV (Tobacco Etch Virus) protease cleavage sequence were amplified by PCR using the two primer sets, KmPDC1 tev 1F Bam/KmPDC1 2B xho and KmPDC5 tev 1F Bam/KmPDC5 2B Xho (Table S1), respectively, from the genomic DNA of K. marxianus KCTC 17555. The DNA fragment encoding MBP was amplified by PCR using the primers MBP Nde 1F/MBP BamH 2B (Table S1) from the pMAL-c5X (New England Biolabs). The NdeI/BamHI digested MBP DNA fragment and the BamHI/XhoI digested DNA fragments of TEV-KmPDC1 and TEV-KmPDC5 were ligated with the XhoI/NdeI-digested pET28b (+) (Novagen), resulting in pET-MBP-KmPDC1 and pET-MBP-KmPDC5, respectively.
Purification of recombinant MBP-KmPdc fusion proteins
The E. coli BL21(DE3) (New England Biolabs) cells harboring pET-MBP-KmPDC1 and pET-MBP-KmPDC5 were grown in the auto-induction medium (1% peptone, 0.5% yeast extract, 171 mM NaCl, 25 mM Na2HPO4, 25 mM KH2PO4, 50 mM NH4Cl, 5 mM Na2SO4,2 mM MgSO4, 20 mM FeCl3, 0.5% glycerol, 0.05% D-glucose, 0.2% lactose, pH 7.0). The cell pellets were obtained by centrifugation and lysed by sonication in the phosphate-buffered saline (PBS, pH 6.0). The MBP-tagged KmPdc1 and KmPdc5 proteins were purified using MBPTrap™ HP Prepacked Columns (GE Healthcare Life Sciences).
Pdc enzymatic activity analysis
Pdc activity of the yeast cell extracts and the purified recombinant KmPdc proteins was assayed by the method coupled with the reaction mediated by alcohol dehydrogenase, as previously described (Gounaris et al. 1971). Yeast cells were lysed in a buffer consisting of 180 mM sodium citrate and 10 mM magnesium chloride, pH 6.8, by vortexing with glass beads. Total protein concentrations were analyzed by the Bradford Assay Kit (Bio-Rad). Briefly, the cell extracts or the purified Pdc proteins were mixed with the reaction buffer (pH 6.8) consisting of 180 mM citric acid, 10 mM magnesium chloride, 33 mM pyruvate, 1 mM thiamine pyrophosphate, 110 nM β-NADH, and 10 U of alcohol dehydrogenase (Sigma). The reactions were initiated by adding pyruvate, and a decrease in absorbance at 340 nm was measured. One unit of activity was defined as the amount of enzyme oxidizing 1 μmol of NADH per minute under given reaction conditions. The specific Pdc activity was calculated by dividing the units of activity by the concentration of proteins used in the reactions.
RNA preparation and quantitative real-time PCR analysis (qRT-PCR)
Yeast cells were inoculated at initial OD600 of 0.1 and grown to the early logarithmic phase (OD600 = 0.3) in the YP medium containing different carbon sources. The cells were harvested and immediately frozen in liquid nitrogen. Total RNA was isolated by the hot phenol extraction method (Chen et al. 2003) and treated with DNase I according to the protocol of the manufacturer (TAKARA). cDNA was generated from 1 μg of total RNA using the SuperScript™ III First Strand Synthesis system (Invitrogen) and oligo(dT) primers. qRT-PCR was conducted on a CFX96 Real-Time PCR detection system (Bio-Rad) using 1 ng of the synthesized cDNA and KmPDC-specific primers (Table S1) with the Maxima SYBR Green qPCR Master Mix (Fermentas). Specificity of the amplification was confirmed by melting curve analysis with a single peak. Each sample was analyzed in triplicate, and data were normalized to the endogenous control, KmACT1, using the primers KmACT1 RT 1F/KmACT1 RT 2B.
High-pressure liquid chromatography (HPLC) analysis of metabolites
Yeast cells, grown overnight, were inoculated at initial OD600 of 1.0 into 50 ml of YPD in a 500-ml flask and cultivated at 37 °C and 220 rpm. The culture supernatants were collected by centrifugation and used for HPLC analysis. Concentrations of glucose, ethanol, acetate, and glycerol were measured by HPLC (Agilent Technologies, Inc.). Separation was implemented on an Aminex HPX-87H 300-by-7.8-mm column (Bio-Rad) using 0.9% acetonitrile as a solvent at 60 °C and a 0.6 ml⋅min−1 flow rate. Peaks were detected with a Refractive Index detector (Agilent Technologies, Inc.) and a Photodiode Array detector.
Results
Domain and sequence analysis of KmPDC1 and KmPDC5
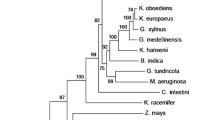
Based on their significant homology with S. cerevisiae Pdc enzymes, two K. marxianus ORFs, named KmPDC1 and KmPDC5 (Table S1), were identified in the K. marxianus KCTC 17555 genome (Jeong et al. 2012). Although K. lactis has been reported to possess only one PDC gene (Bianchi et al. 1996), we identified another K. lactis PDC homolog, named KlPDC5, by means of a BLAST search using the amino acid sequence of S. cerevisiae Pdc1p as a query. Crabtree-negative yeasts have been mostly reported to possess only one PDC gene, PDC1, except S. stipitis (Lu et al. 1998), in contrast to the multiple PDC genes encoding functional isoforms of Pdc1 in Crabtree-positive yeasts. Thus, it was interesting to investigate whether the putative KmPDC5 gene, found in a Crabtree-negative yeast K. marxianus, encodes a functional homolog of Pdc1p, as reported in S. cerevisiae. KmPDC1 codes for a protein of 564 amino acid residues, whereas KmPDC5 encodes a protein of 578 amino acid residues (Fig. 1a). KmPdc1p showed 86 and 91.7% identity to S. cerevisiae Pdc1 and K. lactis Pdc1 proteins, respectively, while KmPdc5p was found to share 33.2 and 76.2% identity with S. cerevisiae Pdc5 and K. lactis Pdc5 proteins, respectively (Fig. S2). The domain analysis indicated the presence of the conserved thiamine pyrophosphate (TPP)-dependent pyrimidine-binding domain (N: TPP enzyme N), TPP enzyme central domain (TPP enzyme M), and TPP enzyme PDC domain (C: TPP enzyme C) in both KmPdc1 and KmPdc5 proteins (Fig. 1a, Fig. S3). Noticeably, the alignment of amino acid sequences in the active sites of S. cerevisiae, K. lactis, and K. marxianus Pdc enzymes revealed that several amino acid residues in the active site, which were reported to have critical roles in the TPP-binding and pyruvate-binding activity of ScPdc1, were changed in KmPdc5 and KlPdc5, whereas they are well conserved in other yeast Pdc proteins (Fig. 1b). The phylogenetic analysis further indicated that K. lactis and K. marxianus Pdc5 proteins have diverged substantially not only from the S. cerevisiae Pdc proteins but also from their own Pdc1 proteins (Fig. 1c), implying the possibility that KmPdc5p may be distinctive from KmPdc1p for Pdc enzymatic activity and expression pattern.
Structural characterization of K. marxianus Pdc1 and Pdc5 proteins. a Comparison of domain organization among yeast Pdc homologs: S. cerevisiae Pdc1p and K. marxianus Pdc1p and Pdc5p. Conserved thiamin pyrophosphate (TPP)-dependent pyrimidine-binding domain (N: TPP enzyme N), TPP enzyme central domain (TPP enzyme central), TPP enzyme PDC domain (C: TPP enzyme C). b Alignment of partial amino acid sequences of yeast Pdc homologs from S. cerevisiae (ScPdc1, ScPdc5, and ScPdc6), K. lactis (KlPdc1 and KlPdc5), and K. marxianus (KmPdc1 and KmPdc5). Amino acid residues known to be important for Pdc activity are indicated (▼). c The phylogenetic tree of yeast Pdc structural proteins. Protein ID numbers from GenBank or NCBI Reference Sequence are shown in parenthesis. *The sequences of KmPDC1 and KmPDC5 were from the genome sequence of K. marxianus KCTC 17555 (Fig. S1)
Growth features of K. marxianus PDC deletion mutant strains
To investigate the physiological functions of KmPDC1 and KmPDC5 in K. marxianus, we analyzed the growth phenotypes of the single-deletion strains of KmPDC1 and KmPDC5 (Kmpdc1Δ and Kmpdc5Δ) of K. marxianus KCTC 17555 (Choo et al. 2014), along with the KmPDC1 and KmPDC5 double-deletion strain (Kmpdc1Δpdc5Δ), which was constructed in the present study (Fig. 2a). The correct deletion of the KmPDC1 and KmPDC5 genes was confirmed by PCR analysis (Fig. 2b). Spotting analysis under aerobic conditions indicated that the growth of all the tested K. marxianus pdc mutant strains, Kmpdc1Δ, Kmpdc5Δ, and Kmpdc1Δpdc5Δ, was comparable to that of the wild-type strain (WT) except for subtle growth retardation in Kmpdc1Δ and Kmpdc1Δpdc5Δ compared to the WT. All the K. marxianus pdc-null strains grew well not only on fermentable carbon sources, such as glucose (YPD) and galactose (YPGal), but also on nonfermentable carbon sources, such as glycerol (YPGly) and ethanol (YPE). In contrast, the growth of the Kmpdc1Δ single-deletion and Kmpdc1Δpdc5Δ double-deletion strains was severely inhibited on the YPD plate containing antimycin A (Fig. 3a, YPD + Anti A), which is a chemical compound binding to the Qi site of Complex III in the mitochondrial inner membrane, thus blocking proton transport of the electron transport system (Xu et al. 2011). The absence of growth on glucose in the presence of respiratory inhibitors was also observed in the pdc1Δ mutant strain of the sister species K. lactis, a model Crabtree-negative yeast (Bianchi et al. 1996). In contrast, no growth defect was detected in the Kmpdc5Δ single-deletion strain not only on the tested carbon sources but also even in the presence of antimycin A.
Construction of the K. marxianus pdc double-mutant strain based on homologous recombination. a The construction scheme of the K. marxianus pdc1Δpdc5Δ double deletion strain using the ura3 pop-out cassette to recover a uracil auxotroph. b PCR analysis of the K. marxianus pdc1Δpdc5Δ strain. Lane W, the wild-type strain (ku80Δ); Lanes 1–4, candidates of the pdc1Δpdc5Δ strain; PCR1 (primers Iden KmPDC1 1F/Iden KmPDC1 2B), PCR2 (primers Iden KmPDC5 1F/Iden KmPDC5 2B), PCR3 (primers ScURA3 1F 47/ScURA3 2B 48) and PCR4 (primers Iden His-G 1F 48/Iden His-G 2B 48) were carried out to confirm the absence of KmPDC1, KmPDC5, and ScURA3, and the presence of hisG, respectively, with the primers listed in Table S1
Growth analysis of K. marxianus pdc-null strains. a Spotting analysis of K. marxianus pdc mutants on YP plates containing different carbon sources, such as 2% glucose (YPD), 2% galactose (YPGal), 3% glycerol (YPGly), and 3% ethanol (YPE), and YPD plate supplemented with 5 μM antimycin A (YPD + Anti A). Yeast cells were precultured overnight in YPD, and 3 μl of the serially diluted cells corresponding to an OD600 of 1, 0.1, 0.01, and 0.001 were spotted. b Growth curves of K. marxianus strains (○: WT, □: pdc1Δ, △: pdc5∆, and ◇: pdc1Δpdc5∆) cultivated in YPD and YPE broth under aerobic (top) and oxygen-limited (bottom) conditions, respectively
For more detailed growth analysis under aerobic and oxygen-limited conditions, time course measurement of cell growth during shake flask cultivation was performed on the K. marxianus pdc mutant strains. Under aerobic conditions, all the tested strains grew well, with similar growth patterns in YPD and YPE cultures (Fig. 3b, left panel). In contrast, it is notable that under oxygen-limited conditions, the Kmpdc1Δ and Kmpdc1Δpdc5Δ mutant strains hardly grew, as opposed to the growth of the WT and Kmpdc5Δ single-deletion strains during YPD cultivation. On the contrary, even the WT strain of K. marxianus was unable to grow when a nonfermentable carbon source, such as ethanol, was provided as a sole carbon source under oxygen-limited conditions (Fig. 3b, right panel). Along with the data from growth analysis on YPD plate in the presence of antimycin A (Fig. 3a), the results strongly indicated that K. marxianus KCTC 17555 has a typical phenotype of respiratory yeasts that cannot grow on nonfermentable carbon sources without respiration activity. Thus, like other Crabtree-negative yeasts, the Pdc-deficient mutants of K. marxianus KCTC 17555 can grow without severe growth retardation on glucose under aerobic conditions but cannot grow under respiration-limited conditions.
Transcriptional analysis of KmPDC1 and KmPDC5 in the presence of different carbon sources
To determine how the various carbon sources affect the expression of KmPDC at the transcriptional level, qRT-PCR analysis was performed on total RNA samples obtained from the K. marxianus WT and pdc mutant strains cultivated on various carbon sources including glucose, galactose, ethanol, and glycerol. The transcript levels of KmPDC1 were much higher in the presence of the fermentable carbon sources (glucose and galactose) than in the presence of nonfermentable carbon sources (ethanol and glycerol). It is noteworthy that the expression level of KmPDC1 in the presence of glucose as a carbon source was the highest as compared to the other carbon sources (Fig. 4a). The different PDC mRNA levels on different carbon sources have also been observed in the same fashion in S. cerevisiae (Kellermann et al. 1986), S. kluyveri (Moller et al. 2004), and K. lactis (Bianchi et al. 1996), indicating that the enhanced PDC1 expression under the influence of glucose is also well conserved among yeasts. In contrast, the transcript levels of KmPDC5 were extremely low compared to those of KmPDC1, and there was no detectable change in KmPDC5 expression on different carbon sources. It is also notable that the KmPDC5 expression level did not increase in the Kmpdc1Δ deletion strain (Fig. 4b); this finding is different from the induced expression of PDC5 upon the deletion of PDC1 in S. cerevisiae (Seeboth et al. 1990).
Quantitative real-time PCR (qRT-PCR) analysis of KmPDC1 and KmPDC5 expression in the presence of different carbon sources. Cells were grown in the YP medium containing different carbon sources, such as 2% glucose, 2% galactose, 3% ethanol, and 3% glycerol. The expression levels of KmPDC transcripts were normalized to the KmACT1 gene as a reference
Functional analysis of KmPDC1 and KmPDC5 in the Kmpdc1Δpdc5Δ double-deletion strain
No apparent defects of the Kmpdc5Δ mutant strain might be attributed to the negligible expression level of KmPDC5 transcript. On the other hand, as reflected by the divergence of the amino acid sequences critical for Pdc enzymatic activity in the K. marxianus Pdc5 protein (Fig. 1a), it can be speculated that KmPdc5p may lack Pdc enzymatic activity. To test these possibilities, we constructed the complementation vectors pKI-PDC1p-KmPDC1 and pKI-PDC1p-KmPDC5 containing the KmPDC1 and KmPDC5 expression cassettes under the control of the KmPDC1 promoter, KmPDC1(p)-PDC1 and KmPDC1(p)-PDC5. After linearization by digestion at SphI site, each of the PDC complementation vectors was integrated via single homologous recombination into the KmPDC1 promoter locus in the chromosome of the pdc1Δpdc5Δ double-deletion strain (Fig. 5a). The correct integration of the KmPDC1 and KmPDC5 expression cassettes was confirmed by PCR using suitable primer sets (Table S1, data not shown). Only the pdc1Δpdc5Δ strain complemented with KmPDC1 (pdc1Δpdc5Δ/PDC1) recovered its growth on YPD plate containing antimycin A, whereas the complementation strain with KmPDC5 (pdc1Δpdc5Δ/PDC5) could not recover from the growth defect in the presence of antimycin A (Fig. 5b). Moreover, we observed a noticeable change in colony morphology and size in the K. marxianus pdc1Δ and pdc1Δpdc5Δ strains on solid YPD media; these parameters returned to normal by reintroduction of the functional KmPDC1 gene, but not by the expression of KmPDC5 gene (Fig. 5c).
Complementation analysis of the KmPDC genes in the mutant strain Kmpdc1Δpdc5Δ. a Construction of the KmPDC1 and KmPDC5 complementation strains. The KmPDC1p-KmPDC1 and KmPDC1p-KmPDC5 expression cassettes were integrated, respectively, into the PDC1 promoter locus of the Kmpdc1Δ strain, generating the Kmpdc1Δpdc5Δ/PDC1 and Kmpdc1Δpdc5Δ/PDC5 complementation strains. b Growth analysis of the complementation strains on the YPD plate containing 5 μM antimycin A (YPD + Anti A). Yeast cells were precultured overnight in YPD, and 3 μl of the serially diluted cells corresponding to an OD600 of 1, 0.1, 0.01, and 0.001 were spotted. c Colony morphology of K. marxianus pdc mutant strains grown on YPD plate. (Left) Yeast cells were precultured overnight in YPD and diluted to OD600 0.001. 200 μl of the diluted yeast cells were spread on YPD plate and incubated at 37 °C for 5 days. (Right) The yeast cultures, containing 3.75 × 106 cells, were spotted on YPD plate and incubated 37 °C for 5 days
To examine the Pdc enzymatic activity in the K. marxianus pdc deletion mutant and complemented strains, total cell lysates were prepared from the WT S. cerevisiae and K. marxianus strains cultivated in YPD and subjected to the assay of Pdc enzymatic activity (Fig. 6a). Whereas the K. marxianus pdc1Δ mutant showed a dramatic decrease in Pdc activity, the pdc5Δ mutant did not manifest any detectable loss of enzymatic activity. The enzymatic activity of pdc1Δpdc5Δ double-mutant strain was almost identical to that of the single pdc1Δ mutant strain, suggesting that Pdc1p is responsible for most Pdc activity in K. marxianus under normal culture conditions. Indeed, the pdc1Δpdc5Δ mutant strain fully recovered the Pdc enzymatic activity after KmPDC1 complementation but not after KmPDC5 complementation. The level of Pdc activity has been regarded as one of the key differences in the carbon metabolism between Crabtree-positive and Crabtree-negative yeasts. It was reported that there is on average sixfold higher Pdc activity in cell-free extracts of Crabtree-positive yeasts (Van Urk et al. 1990). Nonetheless, we found that the Pdc activity in cell-free extracts of K. marxianus was comparable to that of S. cerevisiae (Fig. 6a). We next checked the transcription levels of the PDC genes expressed from the complementation constructs and found that the expression level of the KmPDC5 transcript, directed by the KmPDC1 promoter, was 40% of that of KmPDC1 (Fig. 6b). The observation that the Pdc activity was not detectable in the PDC5 complementation strain expressing a significant amount of the PDC5 transcript strongly indicates that K. marxianus Pdc5p does not retain the enzymatic activity that decarboxylates pyruvate.
Pdc activity analysis of the yeast cell lysates and the purified recombinant KmPdc proteins. a Pdc enzymatic activity of S. cerevisiae and K. marxianus strains. Total cell lysates were used for activity measurements. b Transcript level analysis of KmPDC1 and KmPDC5 in the complementation strains grown in the YPD medium. The expression levels of KmPDC genes were normalized to KmACT1 as a reference gene. c SDS-PAGE analysis of the purified KmPdc1 (Top) and KmPdc5 (Bottom) proteins expressed as maltose binding protein (MBP)-fused forms in E. coli. Lane 1, total cell lyaste; Lane 2, insoluble pellet from cell lysate; Lanes 3 and 4, soluble supernatant from total cell lysates; Lanes 5–7, PBS-washed flow through from MBP-trap; Lane 8, eluted KmPdc proteins by 20 mM maltose in PBS from MBP-trap. The arrow indicates MBP-KmPdc1 and MBP-KmPdc5 proteins, respectively. d Pdc activity of the purified MBP-KmPdc proteins
Pdc activity analysis of the purified recombinant KmPdc1 and KmPdc5 proteins
To validate our hypothesis that KmPdc5p does not possess Pdc activity, we purified KmPdc isozymes for in vitro activity analysis. Initially, the KmPdc1p and KmPdc5p were expressed with N-terminal His-tagged proteins in E. coli. However, only His-tagged KmPdc1p was recovered in soluble fraction from the transformed E. coli cell-free extracts, while His-tagged KmPdc5p was found mainly in inclusion bodies with much reduced protein level (data not shown). Such inclusion body formation of recombinant His-tagged S. cerevisiae Pdc5p in E. coli was also previously reported (Agarwal et al. 2013). Therefore, we expressed KmPdc1 and KmPdc5 proteins in E. coli as the maltose binding protein (MBP)-fused proteins containing TEV protease cleavage site. The MBP-fused KmPdc1 and KmPdc5 proteins were expressed in soluble form with the expected size of 103.4 and 104.9 kDa, respectively (Fig. 6c). The purified MBP-fusion proteins were subjected to Pdc activity analysis, revealing that KmPdc5p lacks the enzymatic activity in contrast to KmPdc1p with high activity (Fig. 6d). This is consistent with the result of complementation experiments (Fig. 5), in which the expression of KmPDC5 could not recover the intracellular Pdc enzymatic activity and growth defects of the pdc1Δpdc5Δ double-mutant strain.
Analysis of extracellular metabolites of K. marxianus pdc deletion strains
To examine the consumption and production of glucose and some metabolites, such as acetate, glycerol, and ethanol, metabolite profiling by HPLC analysis was performed on the culture supernatants of the K. marxianus pdc deletion strains cultivated in the YPD medium containing 2% glucose under aerobic conditions. During the shake flask cultivation, the ethanol production level was detected as less than 1 g/l even for the WT stain of K. marxianus KCTC 17555, which is a typical phenotype of Crabtree-negative yeast (Choo et al. 2016). Compared to the WT and Kmpdc5Δ single-deletion strains, the Kmpdc1Δ single- and Kmpdc1Δpdc5Δ double-deletion strains showed clearly retarded glucose consumption (Fig. 7, left panel). The profiles of acetate, glycerol, and ethanol in the culture supernatant of the single-deletion Kmpdc5Δ strain were found to be almost identical to those of the WT strain. In contrast, these metabolites were barely detectable in the culture supernatants of the Kmpdc1Δ single- and Kmpdc1Δpdc5Δ double-deletion strains (Fig. 7, right panel), while accumulation of pyruvate was observed (Fig. S4). These results further supported the notion that the lack of Pdc activity in the Kmpdc1Δ and Kmpdc1Δpdc5Δ strains blocks pyruvate conversion to aldehyde, thus leading to the shutdown of ethanol and acetate production.
Analysis of metabolite production and consumption in the K. marxianus Pdc-deficient strains. a WT, b pdc1Δ, c pdc5Δ, d pdc1Δpdc5Δ strains of K. marxianus KCTC 17555. Left-hand panels depict time course analysis of cell growth (OD600, □) and glucose concentration (g/l, ■). Right-hand panels show time course analysis of production and consumption of metabolites (ethanol, ●; acetate, ○; glycerol, ▲). Yeast cells were cultivated in 25 ml of the YP medium containing 2% of glucose in a 500-ml flask at 37 °C and 220 rpm
Discussion
Pyruvate, produced by glycolysis, is an important metabolite at the branch point of several metabolic pathways (Hua et al. 1999). Pyruvate can be converted to other carbohydrates such as ethanol, lactate, succinate, alanine, and some others that can be synthesized as polymer materials or drugs. Lately, pyruvate has been used as the key metabolic precursor of lactate, isobutanol, and 3-methyl-1-butanol, which have attracted attention as the second-generation biofuels that can serve as economical and renewable resources via metabolic engineering (Johnson and Beckham 2015). For such diverse applications, pyruvate is a valuable starting material in the biotechnology industry. The expression and activity of Pdc—as a key enzyme starting ethanol fermentation by converting pyruvate to acetaldehyde—has been studied in several yeast species with an industrial potential. Particularly, deletion of the PDC genes in yeast can lead to accumulation of the pyruvate pool, which can be converted to useful precursor biomaterials such as lactic acid or 2,3-butanediol via metabolic engineering (Kim and Hahn 2015; Porro et al. 1999). On the other hand, overexpression of PDCs has been a strategy useful for industrial fermentation processes such as a decarboxylation reaction for ethanol formation (Ishchuk et al. 2008) and biotransformation reactions involving aldehyde for the synthesis of addition products (Iding et al. 1998).
In the present study, we identified two K. marxianus genes encoding putative pyruvate decarboxylases, KmPDC1 and KmPDC5, in the genome of K. marxianus KCTC 17555 and carried out functional analysis to identify physiological roles of the two KmPDC genes. The presence of one K. marxianus gene encoding Pdc (YskPDC1a) was reported previously without any functional characterization (Holloway and Subden 1993). We showed here that the Kmpdc1Δ deletion mutant strain was able to grow quite well on glucose under aerobic conditions, although it has a slightly decreased growth rate as compared to the WT under aerobic conditions and cannot grow under oxygen-limited conditions (Fig. 3). Moreover, the K. marxianus pdc1Δ strains displayed a noticeable change in colony morphology and size on solid YPD media (Fig. 5c). The result suggests that the overall physiological changes due to the lack of Pdc enzymatic activity, e.g., possibly alteration of the cytosolic NAD+/NADH pool, decreased ATP production, and acidic pH conditions because of pyruvate accumulation, may cause the subtle growth retardation and aberrant colony morphology observed in the Kmpdc1Δ deletion strains. It can be also speculated that the growth of cells in the inner part of colonies might be suppressed by oxygen-limitation in the K. marxianus pdc1Δ strains, thus resulting in small and flat colonies.
In contrast to the KmPDC1 deletion, the deletion of KmPDC5 did not affect growth patterns and morphology of K. marxianus under any tested culture conditions, and KmPDC5 expression was not induced upon the loss of Pdc1p (Figs. 3 and 4), in contrast to the properties of S. cerevisiae PDC5 (Hohmann and Cederberg 1990). Moreover, the purified KmPdc5 protein did not show Pdc enzymatic activity, indicating that KmPdc5p is not a functional Pdc (Fig. 6). Even though KmPdc1 and KmPdc5 proteins have several domains that are well conserved among yeast Pdc proteins, we found that KmPdc5p shows notable divergence in the amino acid residues that are reported to be critical for the Pdc enzymatic activity in S. cerevisiae (Fig. 1). The amino acid residues including Pro-26, Asp-28, Glu-51, His-92, Cys-221, Ile-476, Glu-477, and Ile-480 are highly conserved in the Pdc1 homologs of various yeast species, but not conserved in KmPdc5p (Baburina et al. 1998; Lobell and Crout 1996; Mann et al. 2004). Such differences in the critical amino acid residues may explain the lack of Pdc enzymatic activity in KmPdc5p. Our data indeed strongly support the notion that KmPdc1p governs most of the Pdc activity in K. marxianus, while KmPdc5p does not contribute to the reaction of decarboxylation of pyruvate. This is quite contrast to S. cerevisiae Pdc proteins, in that ScPdc5p shows decarboxylation efficiency comparable to ScPdc1p (Agarwal et al. 2013). The possibility that KmPdc5p might have different functions in other metabolic pathways cannot be ruled out, considering that apart from the decarboxylation reaction, Pdc is known for its carboligation capabilities and has been exploited for the synthesis of commercially relevant compounds (Iding et al. 1998). S. cerevisiae WT strains are currently used in the whole-cell (R)-PAC biotransformation process (Rosche et al. 2002). Further study may be needed to characterize the carboligation activity of KmPdc1 and KmPdc5 proteins.
S. cerevisiae possesses five genes that are all showing strong sequence similarity to genes encoding TPP-dependent decarboxylases, including the three genes for Pdc isozymes (PDC1, PDC5, and PDC6), ARO10, and THI3 (Romagnoli et al. 2012). ScAro10p is shown as a 2-oxo-acid decarboxylase involved in production of higher alcohols, whereas ScThi3p is proposed to have a dual role in the metabolism of nutrients in yeast; utilization of thiamin and catabolism of leucine (Dickinson et al. 1997; Nosaka 2006). It is noteworthy that S. cerevisiae Thi3p, a protein of 609 amino acids, shows about 50% identity to all three ScPdc isozymes (Fig. S2, a). The BLAST search carried out with ScThi3p as a query against the K. marxianus KCTC 17555 genome also identified KmPdc1p (51.6% identity) and KmPdc5p (36.1% identity), respectively, as homologs of ScThi3p. Thus, it will be interesting to investigate whether KmPdc5p might be a functional homolog of ScThi3p. However, the phylogenetic tree analysis reveals the early divergence of KmPdc5p from the common ancestor of Pdc isozymes and ScThi3p, before the divergence from ScAro10p (Fig. S2, b). Despite of quite similar level of sequence identity to ScThi3p, the more conserved amino acid patterns are detected between KmPdc5p and ScPdc isozyames compared to those between KmPdc5p and ScThi3p (Fig. S3). Such peculiar sequence features of KmPdc5p strongly indicate that it might have evolved to possess unique enzyme activity, distinctive from the other TPP-dependent decarboxylases previously characterized in yeast.
It has long been regarded that Crabtree-negative yeasts, producing only marginal amounts of ethanol during aerobic growth, have much lower activities of the fermentative enzymes, such as Pdc and alcohol dehydrogenase (Adh), compared to Crabtree-positive yeasts. Indeed, a Crabtree-negative yeast Pichia stipitis was reported to have very low Adh and Pdc activities under aerobic conditions (Passoth et al. 1996). However, other Crabtree-negative yeasts, such as W. anomalus (Fredlund et al. 2004) and K. lactis (Kiers et al. 1998), were shown to have remarkably high activities of Pdc and Adh in spite of low ethanol production under aerobic conditions. It was recently proposed that the onset of Crabtree effect is mostly due to limited respiratory capacity in Crabtree-positive yeasts rather than due to overflow metabolism toward ethanol at the pyruvate branch point (Vemuri et al. 2007). In the present study, we observed that the intracellular Pdc activity of K. marxianus was comparable to that of S. cerevisiae (Fig. 6a), supporting the notion that the major part of pyruvate can enter the mitochondria to be metabolized via TCA cycle, regardless of Pdc activity, in Crabtree-negative yeasts with strong respiratory activity under aerobic conditions.
Another noticeable unique characteristic of Kmpdc1Δ single- and Kmpdc1Δpdc5Δ double-mutant strains was that they produced significantly smaller amounts of glycerol, acetate, and ethanol, as compared to the WT (Fig. 7). In the Crabtree-positive yeast S. cerevisiae, the purpose of NADH-consuming glycerol formation is to maintain the cytosolic redox balance. Inhibition of ethanol production reduces Adh-dependent NAD+ regeneration, resulting in increased glycerol production via the action of glycerol-3-phosphate dehydrogenase (GPD) as a compensation mechanism to maintain the redox balance in S. cerevisiae (Skory 2003). Therefore, deletion of genes GPD1 and GPD2 encoding GPD was found to be necessary to reduce byproduct formation in S. cerevisiae Pdc-deficient mutants (Ida et al. 2013; Kim and Hahn 2015). In this respect, significantly decreased glycerol production in the Kmpdc1Δ single- and Kmpdc1Δpdc5Δ double-deletion strains is quite different from the increased glycerol production in the pdc1Δpdc5Δ mutant of S. cerevisiae. Although we do not yet understand the molecular mechanism underlying the decreased glycerol production after KmPDC1 deletion, such a characteristic can be regarded as an advantageous property of Kmpdc1Δ to be used as a production host, along with its growth comparable to the growth of the WT on several carbon sources during aerobic cultivation.
The robust growth on glucose with reduced glycerol production of the Kmpdc1Δ strain constructed in the K. marxianus KCTC 17555 background is somewhat inconsistent with a recent report that the deletion of PDC1 in the K. marxianus YZJ051 strain decreases the growth rate, far below that of the parental YZJ051 strain (Zhang et al. 2017). The differences in features between these K. marxianus pdc1-null mutants may be partially attributed to the physiological and metabolic diversity within the K. marxianus species (Lane et al. 2011). K. marxianus YZJ051 is an engineered strain of K. marxianus NBRC1777 designed to carry a modified xylose assimilation pathway (Zhang et al. 2015). Although K. marxianus KCTC 17555, employed in this study, was shown to have low fermentation capacity (Fig. 7)—a typical phenotype of Crabtree-negative yeasts—K. marxianus NBRC1777 was reported as a strain selected for its high ethanol productivity (Hong et al. 2007). Moreover, the difference in culture medium used in the two studies might generate the different growth phenotypes. Altogether, our data strongly indicate that the single deletion of KmPDC1, which governs most of Pdc activity in K. marxianus, is sufficient to generate a starting host strain in the KCTC 17555 background for subsequent metabolic engineering aimed at production of high-value biomaterials from pyruvate without byproduct formation.
References
Agarwal PK, Uppada V, Noronha SB (2013) Comparison of pyruvate decarboxylases from Saccharomyces cerevisiae and Komagataella pastoris (Pichia pastoris). Appl Microbiol Biotechnol 97(21):9439–9449. https://doi.org/10.1007/s00253-013-4758-4
Baburina I, Dikdan G, Guo F, Tous GI, Root B, Jordan F (1998) Reactivity at the substrate activation site of yeast pyruvate decarboxylase: inhibition by distortion of domain interactions. Biochemistry 37(5):1245–1255. https://doi.org/10.1021/bi9709912
Bianchi MM, Tizzani L, Destruelle M, Frontali L, Wésolowski-Louvel M (1996) The ‘petite-negative’ yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol Microbiol 19(1):27–36. https://doi.org/10.1046/j.1365-2958.1996.346875.x
Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J (2003) Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 14(1):214–229. https://doi.org/10.1091/mbc.E02-08-0499
Choo JH, Han C, Kim JY, Kang HA (2014) Deletion of a KU80 homolog enhances homologous recombination in the thermotolerant yeast Kluyveromyces marxianus. Biotechnol Lett 36(10):2059–2067. https://doi.org/10.1007/s10529-014-1576-4
Choo JH, Hong CP, Lim JY, Seo JA, Kim YS, Lee DW, Park SG, Lee GW, Carroll E, Lee YW, Kang HA (2016) Whole-genome de novo sequencing, combined with RNA-Seq analysis, reveals unique genome and physiological features of the amylolytic yeast Saccharomycopsis fibuligera and its interspecies hybrid. Biotechnol Biofuels 9:246. https://doi.org/10.1186/s13068-016-0653-4
Dickinson JR, Lanterman MM, Danner DJ, Pearson BM, Sanz P, Harrison SJ, Hewlins MJ (1997) A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem 272(43):26871–26878
Dyda F, Furey W, Swaminathan S, Sax M, Farrenkopf B, Jordan F (1993) Catalytic centers in the thiamin diphosphate dependent enzyme pyruvate decarboxylase at 2.4-A resolution. Biochemistry 32(24):6165–6170
Erasmus DJ, van der Merwe GK, van Vuuren HJ (2003) Genome-wide expression analyses: metabolic adaptation of Saccharomyces cerevisiae to high sugar stress. FEMS Yeast Res 3(4):375–399
Fonseca GG, Heinzle E, Wittmann C, Gombert AK (2008) The yeast Kluyveromyces marxianus and its biotechnological potential. Appl Microbiol Biotechnol 79(3):339–354. https://doi.org/10.1007/s00253-008-1458-6
Franzblau SG, Sinclair NA (1983) Induction of pyruvate decarboxylase in Candida utilis. Mycopathologia 83(1):29–33
Fredlund E, Blank LM, Schnurer J, Sauer U, Passoth V (2004) Oxygen- and glucose-dependent regulation of central carbon metabolism in Pichia anomala. Appl Environ Microbiol 70(10):5905–5911. https://doi.org/10.1128/AEM.70.10.5905-5911.2004
Fredlund E, Beerlage C, Melin P, Schnurer J, Passoth V (2006) Oxygen and carbon source-regulated expression of PDC and ADH genes in the respiratory yeast Pichia anomala. Yeast 23(16):1137–1149. https://doi.org/10.1002/yea.1428
Gounaris AD, Turkenkopf I, Buckwald S, Young A (1971) Pyruvate decarboxylase. I. Protein dissociation into subunits under conditions in which thiamine pyrophosphate is released. J Biol Chem 246(5):1302–1309
Heo P, Yang TJ, Chung SC, Cheon Y, Kim JS, Park JB, Koo HM, Cho KM, Seo JH, Park JC, Kweon DH (2013) Simultaneous integration of multiple genes into the Kluyveromyces marxianus chromosome. J Biotechnol 167(3):323–325. https://doi.org/10.1016/j.jbiotec.2013.06.020
Hill J, Donald KA, Griffiths DE (1991) DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res 19(20):5791. https://doi.org/10.1093/nar/19.20.5791
Hohmann S, Cederberg H (1990) Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem 188(3):615–621. https://doi.org/10.1111/j.1432-1033.1990.tb15442.x
Holloway P, Subden RE (1993) The isolation and nucleotide sequence of the pyruvate decarboxylase gene from Kluyveromyces marxianus. Curr Genet 24(3):274–277. https://doi.org/10.1007/BF00351804
Hong J, Wang Y, Kumagai H, Tamaki H (2007) Construction of thermotolerant yeast expressing thermostable cellulase genes. J Biotechnol 130(2):114–123. https://doi.org/10.1016/j.jbiotec.2007.03.008
Hua Q, Yang C, Shimizu K (1999) Metabolic flux analysis for efficient pyruvate fermentation using vitamin-auxotrophic yeast of Torulopsis glabrata. J Biosci Bioeng 87(2):206–213. https://doi.org/10.1016/S1389-1723(99)89014-8
Ida Y, Hirasawa T, Furusawa C, Shimizu H (2013) Utilization of Saccharomyces cerevisiae recombinant strain incapable of both ethanol and glycerol biosynthesis for anaerobic bioproduction. Appl Microbiol Biotechnol 97(11):4811–4819. https://doi.org/10.1007/s00253-013-4760-x
Iding H, Siegert P, Mesch K, Pohl M (1998) Application of alpha-keto acid decarboxylases in biotransformations. Biochim Biophys Acta 1385(2):307–322. https://doi.org/10.1016/S0167-4838(98)00076-4
Ikushima S, Fujii T, Kobayashi O, Yoshida S, Yoshida A (2009) Genetic engineering of Candida utilis yeast for efficient production of L-lactic acid. Biosci Biotechnol Biochem 73(8):1818–1824. https://doi.org/10.1271/bbb.90186
Ishchuk OP, Voronovsky AY, Stasyk OV, Gayda GZ, Gonchar MV, Abbas CA, Sibirny AA (2008) Overexpression of pyruvate decarboxylase in the yeast Hansenula polymorpha results in increased ethanol yield in high-temperature fermentation of xylose. FEMS Yeast Res 8(7):1164–1174. https://doi.org/10.1111/j.1567-1364.2008.00429.x
Jeong H, Lee DH, Kim SH, Kim HJ, Lee K, Song JY, Kim BK, Sung BH, Park JC, Sohn JH, Koo HM, Kim JF (2012) Genome sequence of the thermotolerant yeast Kluyveromyces marxianus var. marxianus KCTC 17555. Eukaryot Cell 11(12):1584–1585. https://doi.org/10.1128/Ec.00260-12
Johnson CW, Beckham GT (2015) Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab Eng 28:240–247. https://doi.org/10.1016/j.ymben.2015.01.005
Kellermann E, Seeboth PG, Hollenberg CP (1986) Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Saccharomyces cerevisiae. Nucleic Acids Res 14(22):8963–8977. https://doi.org/10.1093/nar/14.22.8963
Kiers J, Zeeman AM, Luttik M, Thiele C, Castrillo JI, Steensma HY, van Dijken JP, Pronk JT (1998) Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast 14(5):459–469. https://doi.org/10.1002/(SICI)1097-0061(19980330)14:5<459::AID-YEA248>3.0.CO;2-O
Kim S, Hahn JS (2015) Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 31:94–101. https://doi.org/10.1016/j.ymben.2015.07.006
Konig S (1998) Subunit structure, function and organisation of pyruvate decarboxylases from various organisms. Biochim Biophys Acta 1385(2):271–286. https://doi.org/10.1016/S0167-4838(98)00074-0
Lane MM, Morrissey JP (2010) Kluyveromyces marxianus: a yeast emerging from its sister’s shadow. Fungal Biol Rev 24(1–2):17–26. https://doi.org/10.1016/j.fbr.2010.01.001
Lane MM, Burke N, Karreman R, Wolfe KH, O'Byrne CP, Morrissey JP (2011) Physiological and metabolic diversity in the yeast Kluyveromyces marxianus. Antonie Van Leeuwenhoek 100(4):507–519. https://doi.org/10.1007/s10482-011-9606-x
Lobell M, Crout DHG (1996) Pyruvate decarboxylase: a molecular modeling study of pyruvate decarboxylation and acyloin formation. J Am Chem Soc 118(8):1867–1873. https://doi.org/10.1021/Ja951830t
Lu P, Davis BP, Jeffries TW (1998) Cloning and characterization of two pyruvate decarboxylase genes from Pichia stipitis CBS 6054. Appl Environ Microbiol 64(1):94–97
Mann S, Melero CP, Hawksley D, Leeper FJ (2004) Inhibition of thiamin diphosphate dependent enzymes by 3-deazathiamin diphosphate. Org Biomol Chem 2(12):1732–1741. https://doi.org/10.1039/B403619k
Merico A, Sulo P, Piskur J, Compagno C (2007) Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J 274(4):976–989. https://doi.org/10.1111/j.1742-4658.2007.05645.x
Moller K, Langkjaer RB, Nielsen J, Piskur J, Olsson L (2004) Pyruvate decarboxylases from the petite-negative yeast Saccharomyces kluyveri. Mol Gen Genomics 270(6):558–568. https://doi.org/10.1007/s00438-003-0950-z
Nosaka K (2006) Recent progress in understanding thiamin biosynthesis and its genetic regulation in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 72(1):30–40. https://doi.org/10.1007/s00253-006-0464-9
Nosaka K, Esaki H, Onozuka M, Konno H, Hattori Y, Akaji K (2012) Facilitated recruitment of Pdc2p, a yeast transcriptional activator, in response to thiamin starvation. FEMS Microbiol Lett 330(2):140–147. https://doi.org/10.1111/j.1574-6968.2012.02543.x
Passoth V, Zimmermann M, Klinner U (1996) Peculiarities of the regulation of fermentation and respiration in the crabtree-negative, xylose-fermenting yeast Pichia stipitis. Appl Biochem Biotechnol 57-58:201–212
Porro D, Bianchi MM, Brambilla L, Menghini R, Bolzani D, Carrera V, Lievense J, Liu CL, Ranzi BM, Frontali L, Alberghina L (1999) Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl Environ Microbiol 65(9):4211–4215
Pronk JT, Yde Steensma H, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12(16):1607–1633. https://doi.org/10.1002/(SICI)1097-0061(199612)12:16<1607::AID-YEA70>3.0.CO;2-4
Romagnoli G, Luttik MA, Kotter P, Pronk JT, Daran JM (2012) Substrate specificity of thiamine pyrophosphate-dependent 2-oxo-acid decarboxylases in Saccharomyces cerevisiae. Appl Environ Microbiol 78(21):7538–7548. https://doi.org/10.1128/AEM.01675-12
Rosche B, Sandford V, Breuer M, Hauer B, Rogers PL (2002) Enhanced production of R-phenylacetylcarbinol (R-PAC) through enzymatic biotransformation. J Mol Catal B Enzym 19–20:109–115. https://doi.org/10.1016/S1381-1177(02)00157-1
Schaaff I, Green JB, Gozalbo D, Hohmann S (1989) A deletion of the PDC1 gene for pyruvate decarboxylase of yeast causes a different phenotype than previously isolated point mutations. Curr Genet 15(2):75–81. https://doi.org/10.1007/BF00435452
Seeboth PG, Bohnsack K, Hollenberg CP (1990) pdc1 0 mutants of Saccharomyces cerevisiae give evidence for an additional structural PDC gene: cloning of PDC5, a gene homologous to PDC1. J Bacteriol 172(2):678–685
Skory CD (2003) Induction of Rhizopus oryzae pyruvate decarboxylase genes. Curr Microbiol 47(1):59–64. https://doi.org/10.1007/s00284-002-3933-0
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
van Maris AJ, Geertman JM, Vermeulen A, Groothuizen MK, Winkler AA, Piper MD, van Dijken JP, Pronk JT (2004) Directed evolution of pyruvate decarboxylase-negative Saccharomyces cerevisiae, yielding a C2-independent, glucose-tolerant, and pyruvate-hyperproducing yeast. Appl Environ Microbiol 70(1):159–166. https://doi.org/10.1128/AEM.70.1.159-166.2004
Van Urk H, Voll WS, Scheffers WA, Van Dijken JP (1990) Transient-state analysis of metabolic fluxes in crabtree-positive and crabtree-negative yeasts. Appl Environ Microbiol 56(1):281–287
Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J (2007) Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104(7):2402–2407. https://doi.org/10.1073/pnas.0607469104
Xu QW, Vu H, Liu LP, Wang TC, Schaefer WH (2011) Metabolic profiles show specific mitochondrial toxicities in vitro in myotube cells. J Biomol NMR 49(3–4):207–219. https://doi.org/10.1007/s10858-011-9482-8
Zhang J, Zhang B, Wang D, Gao X, Sun L, Hong J (2015) Rapid ethanol production at elevated temperatures by engineered thermotolerant Kluyveromyces marxianus via the NADP(H)-preferring xylose reductase-xylitol dehydrogenase pathway. Metab Eng 31:140–152. https://doi.org/10.1016/j.ymben.2015.07.008
Zhang B, Zhu Y, Zhang J, Wang D, Sun L, Hong J (2017) Engineered Kluyveromyces marxianus for pyruvate production at elevated temperature with simultaneous consumption of xylose and glucose. Bioresour Technol 224:553–562. https://doi.org/10.1016/j.biortech.2016.11.110
Funding
This work was supported by the National Research Foundation of Korea (NRF), grant no. NRF-2017M3C1B5019295 (STEAM Research Project). Gyu Hun Sim was supported by Chung-Ang University Research Scholarship Grants in 2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 404 kb)
Rights and permissions
About this article
Cite this article
Choo, J.H., Han, C., Lee, D.W. et al. Molecular and functional characterization of two pyruvate decarboxylase genes, PDC1 and PDC5, in the thermotolerant yeast Kluyveromyces marxianus. Appl Microbiol Biotechnol 102, 3723–3737 (2018). https://doi.org/10.1007/s00253-018-8862-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-8862-3