Abstract
Filamentous fungi are often used as cell factories for recombinant protein production because of their ability to secrete large quantities of hydrolytic enzymes. However, even using strong transcriptional promoters, yields of nonfungal proteins are generally much lower than those of fungal proteins. Recent analyses revealed that expression of certain nonfungal secretory proteins induced the unfolded protein response (UPR), suggesting that they are recognized as proteins with folding defects in filamentous fungi. More recently, however, even highly expressed endogenous secretory proteins were found to evoke the UPR. These findings raise the question of whether the unfolded or misfolded state of proteins is selectively recognized by quality control mechanisms in filamentous fungi. In this study, a fungal secretory protein (1,2-α-D-mannosidase; MsdS) with a mutation that decreases its thermostability was expressed at different levels in Aspergillus oryzae. We found that, at moderate expression levels, wild-type MsdS was secreted to the medium, while the mutant was not. In the strain with a deletion for the hrdA gene, which is involved in the endoplasmic reticulum-associated degradation pathway, mutant MsdS had specifically increased levels in the intracellular fraction but was not secreted. When overexpressed, the mutant protein was secreted to the medium to a similar extent as the wild-type protein; however, the mutant underwent hyperglycosylation and induced the UPR. Deletion of α-amylase (the most abundant secretory protein in A. oryzae) alleviated the UPR induction by mutant MsdS overexpression. These findings suggest that misfolded MsdS and unfolded species of α-amylase might act synergistically for UPR induction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Filamentous fungi are the primary decomposers in many ecosystems because of their ability to secrete large quantities of hydrolytic enzymes. Therefore, these fungi have long been utilized for food processing and enzyme production (Ichishima 2016). Recombinant DNA technology exploits this property of hydrolytic enzyme secretion for the production of useful heterologous proteins. Use of strong transcriptional promoters achieves the production of recombinant proteins by fungi at levels of grams per liter of medium (Verdoes et al. 1995). In many cases, however, the yields of nonfungal proteins are limited to a few milligrams per liter through repression of transcription and/or translation of foreign genes, destabilization of transcripts, and degradation of polypeptides during and after secretion (Gouka et al. 1997). Several transcriptomic analyses revealed that overexpression of foreign proteins in filamentous fungi triggers the unfolded protein response (UPR), including upregulation of genes involved in protein folding in the endoplasmic reticulum (ER), glycosylation, vesicular trafficking, and ER-associated degradation (ERAD) (Sims et al. 2005; Guillemette et al. 2007; Ohno et al. 2011). Although it has not been proven that foreign proteins tend to be misfolded in filamentous fungi, study findings suggest that inefficient folding of foreign proteins and their removal from the ER might be bottlenecks in the production of foreign proteins by filamentous fungi. Interestingly, however, even major endogenous secretory proteins, which are assumed to be efficiently folded and transported through the secretory pathway, induce the UPR in the overexpressed state. For instance, introduction of multiple copies of the endogenous glucoamylase gene into Aspergillus niger evokes the UPR (Kwon et al. 2012). Even in the native (nonrecombinant) A. oryzae strain, which contains three copies of the α-amylase genes (amyA, amyB, and amyC), the UPR is upregulated when α-amylase expression is induced by the addition of maltose to the medium (Tanaka et al. 2015). These observations render it difficult to determine whether filamentous fungi, specifically, sense the unfolded/misfolded state of foreign proteins or just monitor crowdedness of nascent polypeptides in the ER lumen as general secretion stress. Various chemicals such as dithiothreitol (DTT) and tunicamycin are often used to analyze responses to unfolded proteins in organisms (Kaufman 1999; Travers et al. 2000; Sims et al. 2005; Guillemette et al. 2007). However, because these chemicals may affect most of the nascent polypeptides in the ER, it is difficult to evaluate the cellular response to specific misfolded secretory proteins and identify relationships between endogenous intact secretory proteins and misfolded proteins in the induction of UPR.
In the present study, we expressed a mutant version of the fungal secretory protein 1,2-α-D-mannosidase (MsdS), consisting of a single amino acid substitution known to decrease its thermostability (Tatara et al. 2005), in A. oryzae and compared its expression with strains of the organism expressing wild-type MsdS in order to analyze cellular responses to the expression of aberrant secretory proteins. By this strategy, we expected to circumvent flaws in the transcription and translation of target genes and thus can exclude outcomes provided by overexpression of MsdS regardless of whether it is mutated. We found that the mutant MsdS was a potential ERAD substrate and overexpression of the protein caused its overflow from the ERAD pathway to be secreted into the medium and the induction of UPR. We also demonstrated that this UPR induction was attenuated by the deletion of α-amylase genes and discussed the relationship between endogenous secretory proteins and misfolded proteins in UPR induction.
Materials and methods
Strains, media, and growth conditions
A. oryzae NS4 (niaD −, sC −), derived from A. oryzae RIB40 (ATCC42149), was used as the recipient strain for transformation (Yamada et al. 1997; Machida et al. 2005). Saccharomyces cerevisiae BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used to generate plasmids for disruption of the hrdA and pepE genes. For protein expression, approximately 1 × 106 conidiospores of fungal strains were cultivated in 50 ml of 1% yeast extract, 1% peptone, and 2% maltose (YPM) medium in a 250-ml Erlenmeyer flask at 30 °C for 24 h.
Construction of ΔhrdA, ΔpepE, and ΔamyABC strains
Gene disruption in A. oryzae was performed as previously described (Mizutani et al. 2008). The method developed by Colot et al. (2006), which utilized an in vivo recombination-mediated cloning in the yeast S. cerevisiae, was used to create plasmids for disruption of the hrdA and pepE genes. The 5′ and 3′ fragments of the hrdA gene were amplified by polymerase chain reaction (PCR) from A. oryzae NS4 genomic DNA with the primer pairs of hrdA-F1/hrdA-R1 and hrdA-F2/hrdA-R2, respectively (Table S1). S. cerevisiae BY4741 cells were transformed with these PCR products together with the circular plasmids pUSC (Yamada et al. 1997), which contains the sC gene from Aspergillus nidulans as a selective marker, and pYES2 (Thermo Fisher Scientific, Waltham, MA, USA), linearized with the restriction enzymes XbaI and HindIII. Yeast transformants were selected on SC-Ura plates. The plasmids were extracted and verified by restriction enzyme digestion and designated as pYES2-hrdA::sC. A. oryzae NS4 was transformed with pYES2-hrdA::sC and digested with SpeI and XbaI, as previously described (Gomi et al. 1987). Disruption of the hrdA gene was verified by southern blot analysis using the upstream region of hrdA as a probe, which was generated by PCR with the primers hrdA-F1 and hrdA-R1.
The same method was used to generate the pepE disrupting plasmid, pYES2-pepE::sC, except that the primers pepE-F1, pepE-R1, pepE-F2, and pepE-R2 (Table S1) were used to obtain the 5′ and 3′ fragments of the pepE gene. A. oryzae NS4 was transformed with the plasmid pYES2-pepE::sC, which was digested with NcoI. Transformants were analyzed by southern blot hybridization to verify disruption of the pepE gene.
Three plasmids, pΔamyA::loxPsC/Topo, pΔamyB::loxPsC/Topo, and pΔamyC::loxPsC/Topo, used for triple disruption of amyA, B, and C were constructed as follows. For amyA disruption, the amyA gene was PCR-amplified with the primer pair amyAFw and amyARv and the genomic DNA of A. oryzae RIB40 as the template, followed by ligation of the products into the pCR4Blunt-TOPO vector (Thermo Fisher Scientific, Waltham, MA, USA) to generate the plasmid pamyA/Topo. This plasmid was then used to amplify a DNA fragment using the primer pair ΔamyAInF Fw and ΔamyAInF Rv. This DNA fragment and the loxP-A. nidulans sC-loxP cassette excised from pUG6sC (Mizutani et al. 2012) with NotI were assembled using the In-Fusion kit (Takara Bio, Inc., Otsu, Japan) to generate pΔamyA::loxPsC/Topo. The plasmids pΔamyB::loxPsC/Topo and pΔamyC::loxPsC/Topo were obtained by essentially the same methods. The primers used to construct these plasmids are listed in Table S1.
A. oryzae ΔligD::ptrA, derived from NS4 (Mizutani et al. 2008), was transformed with pΔamyC::loxPsC/Topo digested with ApaLI and PmeI, as previously described (Gomi et al. 1987), to construct the amyC disruptant (ΔligD::ptrA ΔamyC::loxPsC). A. oryzae transformants were screened for sulfate prototrophy and purified by subculturing at least twice on CDE (1% glucose, 70 mM monosodium glutamate, 0.05% KCl, 0.2% KH2PO4, 0.05% MgSO4, and trace amounts of FeSO4, ZnSO4, CuSO4, MnSO4, Na2B4O7, and (NH4)6Mo7O24) agar plates (Mizutani et al. 2008). A single correct homologous integration resulting in the replacement of the resident amyC gene with ΔamyC::loxPsC was confirmed by colony PCR with the primer pair ΔamyC conf Fw and sC Rv and southern blot analysis with an amyC probe. The probe had been PCR-amplified with the primer pair amyCFw and amyC probe Rv and genomic DNA of A. oryzae RIB40 as the template.
The sC marker gene was removed from the ΔamyC::loxPsC strain by the Cre-loxP recombination with direct introduction of Cre recombinase to generate the ΔamyC strain (ΔligD::ptrA ΔamyC::loxP) (Mizutani et al. 2012). This strain was used for the second round of transformation with pΔamyB::loxPsC/Topo digested with ApaLI and PmeI to construct the amyB and C double disruptant (ΔamyB::loxPsC ΔamyC::loxP). The single correct homologous integration resulting in the replacement of the resident amyB gene with ΔamyB::loxPsC was confirmed by colony PCR with the primer pair ΔamyB conf Fw and sC Rv and southern blot analysis with the amyB probe. The probe had been amplified by PCR with the primer pair amyBFw and amyB probe Rv and A. oryzae RIB40 genomic DNA as the template. After removing the sC marker gene from the ΔamyB::loxPsC locus as described above, the resulting ΔamyBC strain (ΔligD::ptrA ΔamyB::loxP ΔamyC::loxP) was transformed with pΔamyA::loxPsC/Topo digested with ApaLI and PmeI to construct ΔamyABC (ΔligD::ptrA ΔamyA::loxPsC ΔamyB::loxP ΔamyC::loxP). The single correct homologous integration resulting in the replacement of the resident amyA gene with ΔamyA::loxPsC was confirmed by colony PCR with the primer pair ΔamyA conf Fw and sC Rv and southern blot analysis with the amyA probe. The probe had been amplified by PCR with the primer pair amyAFw and amyA probe Rv and A. oryzae RIB40 genomic DNA as the template. By sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), it was confirmed that the ΔamyABC strain did not produce α-amylase in media (Fig. S1).
Construction of strains expressing 1,2-α-D-mannosidase from Aspergillus tubingensis
The plasmids pNAN8142-msdS and pNAN8142-msdS-C443F were used to overexpress 1,2-α-D-mannosidase (MsdS) and its C443F variant in Aspergillus tubingensis (formerly designated as Aspergillus saitoi) (Tatara et al. 2005). It should be noted that a signal peptide sequence was exchanged with that of aspergillopepsin I from A. tubingensis, as a construct for efficient secretion (Ichishima et al. 1999). In order to produce moderate levels of the MsdS protein, the strong promoter P-No8142 was replaced with the enoA promoter from A. oryzae to generate penoA-msdS and penoA-msdS-C443F. These plasmids were digested, within the selective marker niaD gene with the restriction enzyme MfeI, to be targeted into the niaD − locus on the chromosome. The integration of one copy of plasmid DNA was verified by PCR and southern blot analysis (Figs. S2 and S3).
Isolation of total RNA and real-time quantitative reverse transcription PCR
Mycelia were obtained by filtration using Miracloth (Merck Millipore, Darmstadt, Germany), washed with sterilized water, and then immediately frozen in liquid nitrogen. The frozen mycelia were ground to a fine powder in a mortar and then used for isolation of total RNA with ISOGEN RNA extraction buffer (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions. Total RNA preparations treated with DNase I (Takara Bio, Inc.) were used as templates, and the first-strand complementary DNA (cDNA) was synthesized using the PrimeScript™ II First-Strand cDNA Synthesis Kit (Takara Bio, Inc.). The resulting single-stranded cDNA was subjected to real-time PCR analysis using SYBR® Green PCR Master Mix and a StepOnePlus™ Real-Time PCR system (Thermo Fisher Scientific). Relative quantification of messenger RNA (mRNA) was performed with the 2−ΔΔCT method using histone H4 mRNA as a reference. The nucleotide sequences of the primers used for this analysis are shown in Table S2.
Detection of MsdS proteins
The culture was separated into a medium fraction and mycelia by filtration with Miracloth. To analyze proteins secreted into the medium fraction, 2 μl of 6× SDS sample buffer (375 mM Tris-HCl, pH 6.8, 6% SDS, 30% glycerol, 15% 2-mercaptoethanol) was added to 10 μl of the culture medium and boiled for 3 min. Then, the samples were separated by SDS-PAGE using 10% polyacrylamide gels. For the intracellular fraction, the mycelia were protoplasted and then lysed in SDS sample buffer (62.5 mM Tris-HCl, pH 6.8, 1% SDS, 5% glycerol, 2.5% 2-mercaptoethanol). After boiling for 3 min, 10 μg of protein was separated by SDS-PAGE using 10% polyacrylamide gel. MsdS was detected by Coomassie brilliant blue R-250 staining or immunoblotting with anti-MsdS antiserum (kindly provided by Dr. Ichishima) (Tatara et al. 2003) and a Konica immunostaining HRP-1000 kit (Konica). To remove N-linked glycans, proteins equivalent to 10 μl of the medium fraction were treated with endoglycosidase H (Endo H; New England Biolabs Japan, Tokyo, Japan).
Results
Heterologous expression of MsdS in A. oryzae at two different expression levels
The Cys-443 residue of 1,2-α-D-mannosidase (MsdS) from A. tubingensis (formerly known as A. saitoi) does not contribute to disulfide bond formation; however, it is important for conformational stability (Tatara et al. 2005). In vitro study revealed that the substitution of this residue with another amino acid decreases its thermostability. Compared to wild-type MsdS, which was efficiently secreted from recombinant A. oryzae, production of some mutants including C443F MsdS (with substitution of Cys-443 to Phe) was greatly decreased, possibly because the mutant protein was recognized as misfolded and selected for degradation (Tatara et al. 2005). Thus, we decided to express the C443F mutant MsdS as a model protein to investigate the fates of misfolded secretory proteins in A. oryzae. Because cellular responses to the expression of aberrant secretory proteins may vary with their expression levels, two A. oryzae expression systems were utilized in this study. A modified No8 promoter (P-No8142), which is a hybrid of an A. niger DNA fragment with strong transcriptional activity and 12 copies of a cis element called region III from the A. oryzae agdA gene (α-glucosidase gene), was used to overexpress MsdS (Ozeki et al. 1996; Minetoki et al. 2003). Using this promoter, Ichishima et al. (1999) established an MsdS overexpression system in A. oryzae, which yielded >320 mg MsdS per liter of culture medium. For moderate expression, msdS cDNA was fused at the location immediately after the promoter region of enoA, which encodes the glycolytic enzyme enolase in A. oryzae. One copy of each construct was integrated into the niaD locus of A. oryzae NS4 strain in addition to an empty control vector (Figs. S2 and S3). We confirmed that msdS transcription was increased by approximately 10-fold to 12-fold with P-No8142 compared with P-enoA (Fig. 1). Along with elevated transcription levels, MsdS secretion was also increased when overexpressed with the P-No8142 promoter (Fig. 2a, lanes 2 and 5).
Quantitative RT-PCR analysis of msdS mRNA expressed with the promoters enoA and No8142. The fungal strains expressing wild-type MsdS from enoA (column 2) or No8142 promoter (column 5) and C443F MsdS from enoA (column 3) or No8142 promoter (column 6) were grown in YPM for 24 h, and the msdS gene expression levels were analyzed as described in the “Materials and methods” section. Fungal strains transformed with the empty vectors penoA (column 1) or pNAN8142 (column 4) were used as negative controls. Expression levels were normalized to histone H4. Data represent the fold changes relative to the strain containing penoA-msdS. Error bars indicate the standard deviations of three independent experiments
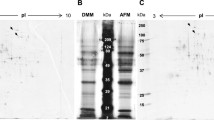
Western blot analysis of secreted and intracellular MsdS proteins expressed with the promoters enoA and No8142. a Aliquots (10 μl) of culture supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by western blot analysis using anti-MsdS antiserum (WB). Coomassie brilliant blue (CBB) staining was performed to detect α-amylase as loading controls. b Endoglycosidase H (Endo H) treatment of MsdS proteins. Culture supernatants were treated with or without Endo H and analyzed by western blotting using anti-MsdS antiserum. c Cell lysates (10 μg) were analyzed by western blotting with anti-MsdS antiserum. Total intracellular proteins were stained with CBB as loading controls
ERAD contributes to the degradation of mutant MsdS when expressed at a moderate level
Using the two expression systems described above, C443F MsdS and wild-type MsdS were expressed in A. oryzae. When the C443F mutant was expressed with the enoA promoter, the protein was not detected in the medium fraction, whereas a significant amount of the wild-type enzyme was secreted (Fig. 2a, lanes 2 and 3). We next investigated whether the mutant enzyme was accumulated inside the cells. To do so, cell lysates were prepared from protoplasts and then subjected to immunoblot analysis with anti-MsdS antibody. However, both wild-type and mutant MsdS were detected at only limited levels in the wild-type strain (Fig. 3a, b, lanes 2 and 3). Since we confirmed that the transcription levels of genes coding for the wild-type and mutant proteins were comparable (Fig. 1) and it was unlikely that a single codon exchange severely affected the translational efficiency, decreased levels of the mutant protein could be because of post-translational destabilization by the protein quality control system. Therefore, we next examined whether the mutant MsdS was subjected to the protein quality control mechanism. Vacuoles are known to be major sites of intracellular protein degradation. In the budding yeast S. cerevisiae, vacuolar proteinase A, which is coded by the PEP4 gene, is primarily required for activation of other vacuolar proteolytic enzymes. Thus, most vacuolar proteolytic activities were lost in pep4Δ cells (Jones et al. 1982). The aspartic proteinase PepE in A. oryzae is a potential ortholog of Pep4 and responsible for a major part of intracellular acid protease activity (Jin et al. 2007). We, therefore, decided to investigate whether PepE contributes to the degradation of the C443F mutant. To this end, we generated the ΔpepE strain and introduced the wild-type and mutant MsdS constructs as well as an empty control vector into the strain (Fig. S2b). As shown in Fig. 3a, the mutant MsdS was not detected in either the medium or cell fraction of the ΔpepE strain, suggesting that PepE was not the major limitation in the production of mutant MsdS. Interestingly, production of wild-type MsdS was significantly improved in both fractions by the deletion of pepE (Fig. 3a, lane 5). Although the cellular localization of PepE has not been identified, a portion of the wild-type MsdS may be sorted into the vacuoles for degradation (van den Hombergh et al. 1997). Moreover, some portions of PepE might also be secreted into the medium from the mycelia.
Effects of deletion of the pepE or hrdA genes on MsdS protein levels expressed with the enoA promoter. Extracellular (medium) and intracellular (cell lysate) fractions were prepared from the wild-type strain expressing wild type (a, b, lane 2) or C443F (a, b, lane 3) MsdS, the ΔpepE strain expressing wild type (a, lane 5) or C443F (a, lane 6), and the ΔhrdA strain expressing wild type (b, lane 5) or C443F (b, lane 6) grown in YPM for 24 h and analyzed by western blotting. Wild-type (a, b, lane 1), ΔpepE (a, lane 4), and ΔhrdA (b, lane 4) strains containing empty vectors (penoA) were used as negative controls
One of the major protein quality control systems in the protein secretory pathway is the ERAD pathway whereby misfolded proteins in the ER are retrotranslocated into the cytoplasm and degraded by the ubiquitin-proteasome system (Kostova and Wolf 2003; Nakatsukasa and Brodsky 2008). To test whether ERAD is responsible for the reduced production of mutant MsdS, we decided to disrupt the gene with a putative ortholog of Hrd1, a ubiquitin ligase required for ERAD in several organisms. A Basic Local Alignment Search Tool search revealed that A. oryzae bears a gene (gene ID AO090005000658) that codes for a translational product homologous to that in several organisms, including Aspergillus fumigatus (accession no. EDP48823.1 with an identity of 69%), Arabidopsis thaliana (accession no. NP_188230.1 with an identity of 31%), Drosophila melanogaster (accession no. NP_651894.3 with an identity of 26%), Caenorhabditis elegans (accession no. NP_505969.1 with an identity of 23%), Homo sapiens (accession no. NP_757385.1 with an identity of 25%), and S. cerevisiae (accession no. NP_014630.1 with an identity of 19%). Hereafter, this gene is referred to as hrdA for convenience. We introduced the wild-type and mutant MsdS constructs (penoA-msdS and penoA-msdS-C443F) into the ΔhrdA strain (Fig. S2c). Most of the wild-type MsdS was detected in the medium fraction of ΔhrdA cells, as observed in the cells of the wild-type strain, indicating that deletion of hrdA did not affect the secretion of the wild-type MsdS protein (Fig. 3b). On the other hand, levels of the mutant MsdS increased in the intracellular fraction of ΔhrdA cells; however, it was not secreted into the medium. These results suggested that the mutant MsdS was degraded by ERAD (Fig. 3b).
C443F MsdS was secreted but hyperglycosylated when overexpressed
In order to analyze the response to the overexpression of ERAD substrate in A. oryzae, both the wild-type and mutant MsdS were overexpressed from the No8142 promoter in the wild-type strain. Compared to the moderate expression from the enoA promoter, production of the wild-type MsdS was increased by >3-fold in the medium, as estimated by immunoblotting (Fig. 2a, lanes 2 and 5). Interestingly, migration of the C443F MsdS in the medium fraction was diffuse on SDS-PAGE (Fig. 2a, lane 6), possibly because of typical post-translational modification, known as hyperglycosylation in yeast but scarcely seen in filamentous fungi (Deshpande et al. 2008). To verify whether this was because of hyperglycosylation, the medium fraction of cells expressing wild-type or mutant MsdS were treated with Endo H and analyzed by immunoblotting. As shown in Fig. 2b, the smeared band of the overexpressed MsdS mutant converged to form a single band with a molecular weight identical to that of the wild-type MsdS, suggesting that the mutant MsdS was hyperglycosylated (Fig. 2b, lanes 6 and 8). Additionally, the amount of mutant MsdS was found to be similar to or slightly lower than that of the wild type (Fig. 2b, lanes 6 and 8), indicating that the efficiencies of secretion of these proteins were almost identical, although a portion of the mutant MsdS might be targeted to the ERAD. It should be noted that no mutant MsdS was observed even after Endo H treatment when expressed at a moderate level, further confirming that mutant MsdS was not present in the medium (Fig. 2b, lanes 3 and 4). Both wild-type and mutant MsdS were seen in the intracellular fraction when overexpressed. Interestingly, however, mutant MsdS did not undergo hyperglycosylation (Fig. 2c). Analogous to the N-glycosylation system in the yeast S. cerevisiae, hyperglycosylation seems to occur in the Golgi apparatus (Deshpande et al. 2008), suggesting that most intracellular mutant MsdS might be accumulated in the ER. Further analysis is required to verify the localization of the mutant MsdS.
Overexpression of C443F MsdS induces the UPR
Since C443F MsdS is a misfolded protein that undergoes ERAD, we next asked whether its overexpression could induce the UPR in A. oryzae. To this end, we expressed both wild-type and mutant MsdS in the wild-type strain with the enoA and No8142 promoters and analyzed the transcription of the ER chaperone genes bipA and pdiA, whose upregulation is a hallmark of the induction of the UPR (Punt et al. 1998; Guillemette et al. 2007; Richie et al. 2009; Tanaka et al. 2015). When C443F MsdS was expressed with the enoA promoter, no significant induction of bipA and pdiA transcription was observed compared to that observed for the control strain containing an empty vector or expression of wild-type MsdS from the same promoter (Fig. 4, columns 1–3). In contrast, overexpression of mutant MsdS, but not the wild-type MsdS, with the strong No8142 promoter significantly induced transcription of both bipA and pdiA (Fig. 4, columns 4–6). These results suggested that an intracellular accumulation of the mutant MsdS led to induction of the UPR. As shown in Fig. 2a, secretion of α-amylase to the medium was maintained at normal levels even when the mutant MsdS was overexpressed, implying that the ER stress by overexpression of the mutant MsdS did not affect the secretion of α-amylase.
Quantitative RT-PCR analysis of the bipA and pdiA genes in fungal cells expressing wild-type and C443F msdS from enoA or No8142 promoters. The fungal strains expressing wild-type MsdS from enoA (column 2) or No8142 promoters (column 5) and C443F MsdS from enoA (column 3) or No8142 promoters (column 6) were grown in YPM for 24 h, and expression levels of the bipA and pdiA genes were analyzed as described in the “Materials and methods” section. Strains transformed with empty vector penoA (column 1) or pNAN8142 (column 4) were used as negative controls. Expression levels were normalized to histone H4. Data represent the fold changes relative to the strain containing empty vector penoA. Error bars indicate the standard deviations of three independent experiments. Statistical analysis was conducted using a Student’s t test. *p < 0.05, **p < 0.01. ns not significant
We recently reported that A. oryzae upregulates the UPR under conditions inducing secretory production of endogenous amylolytic enzymes (Tanaka et al. 2015). Since α-amylases, encoded by three virtually identical genes (amyA, amyB, and amyC), are the most abundant secretory proteins of A. oryzae grown in YPM medium (Fig. 2a), we assumed that loading of α-amylases into the ER lumen might confer a synergistic effect with C443F MsdS on UPR induction. To test this hypothesis, we expressed both wild-type and mutant MsdS with the No8142 promoter in the ΔamyABC triple deletion strain and examined induction of the UPR by measuring mRNA levels of bipA and pdiA. First, we confirmed that the wild-type and mutant msdS genes were highly transcribed in both parent and triple mutant strains at similar levels (Fig. 5). Interestingly, the loss of α-amylase expression significantly attenuated the increase in bipA and pdiA transcription promoted by expression of mutant MsdS in the parent strain (Fig. 5). These results suggested that high-level expression of endogenous and misfolded secretory proteins cooperatively stimulated the UPR.
Effects of deletion of the α-amylase gene on induction of the UPR by overexpression of C443F MsdS. Quantitative RT-PCR was performed to analyze the mRNA levels of the msdS, bipA, and pdiA genes in parent and ΔamyA/B/C strains expressing wild-type and C443F msdS with the No8142 promoter. Expression levels were normalized to histone H4. Data represent the fold changes relative to the parent strain containing empty vector pNAN8142. Error bars indicate the standard deviations of three independent experiments. Statistical analysis was conducted using a Student’s t test. *p < 0.05, **p < 0.01. ns not significant
Discussion
In filamentous fungi, the expression of some nonfungal secretory proteins induces the UPR (Carvalho et al. 2011; Ohno et al. 2011). However, since UPR induction is dependent on the expression levels of heterologous proteins, it is unclear whether cells sense the folding state or concentration of these proteins in the ER to induce the UPR pathway. Furthermore, maltose-induced expression of even endogenous amylolytic enzymes, which are major secretory proteins, activates the UPR in A. oryzae (Tanaka et al. 2015). The results of this study showed that fungal cells actually recognize the folding state of aberrant secretory proteins to cope with this stress in various ways.
An in vitro study of MsdS revealed that the Cys-443 residue of MsdS contributes to its thermostability (Tatara et al. 2005). In the present study, we demonstrated that the substitution of this residue with phenylalanine leads to targeting of this protein to ERAD in A. oryzae when the protein is expressed at moderate levels. Under this condition, the mutant enzyme was not detected in the medium, whereas the wild type was secreted into the medium (Fig. 2a, b). Lack of secretion of the mutant enzyme might be caused by the ER quality control mechanism, including ERAD. Although levels of the mutant enzyme increased significantly in the ΔhrdA strain, it was retained inside the cells and not secreted (Fig. 3b). Similarly, for the ΔhrdC strain, deletion of hrdC, which codes for a putative ERAD component, increased intracellular accumulation of the glucoamylase-β-glucuronidase fusion protein without secretion into the medium (Carvalho et al. 2011). These findings suggest that other components of the ER quality control mechanism possibly contribute to this enzyme retention in cells. Since the overexpression of the mutant MsdS led to its secretion, the mechanism appears to be saturable (Fig. 2a). In S. cerevisiae, a mutant version of carboxypeptidase Y (G255R CPY), which is established as ERAD substrate, escapes the ERAD when it is overexpressed by a high copy number plasmid (Spear and Ng 2003). To further understand the mechanism of ER retention of unstable secretory proteins in filamentous fungi, other ERAD substrates need to be developed and tested for overexpressed protein overflow from the ERAD pathway.
Mutant MsdS that escaped from the ERAD pathway were hyperglycosylated and secreted to the medium (Fig. 2). The Endo H treatment removed this modification (Fig. 2b), indicating that elongation of the sugar chain might have occurred on the N-linked glycan(s) of misfolded MsdS. A specific sugar chain structure of the misfolded proteins that escaped from the ERAD could be recognized as a signal for hyperglycosylation. Otherwise, the post-ER quality control mechanism could detect immature folding of mutant proteins and modify the N-glycans. In A. oryzae, endogenous glucoamylase, which is coded by the glaB gene, is known to undergo hyperglycosylation (Tanaka et al. 2016). Since this modification is removable with Endo H treatment, N-linked high mannose-type glycans are possibly further modified on the GlaB protein as observed for the mutant MsdS. However, it is not yet known if the sugar chain structures are similar and native and misfolded proteins have the same molecular machineries for this modification. Additional experiments need to be conducted to elucidate the molecular mechanisms underlying hyperglycosylation and its role in filamentous fungi. This information may help in constructing host strains that prevent hyperglycosylation during recombinant protein production.
Overexpression of mutant MsdS, and not wild-type MsdS, caused induction of the UPR (Fig. 4), emphasizing that an aberrant tertiary structure is specifically recognized by the ER quality control mechanism. Interestingly, this response occurs synergistically with the high-level expression of α-amylase, the most abundant secretory proteins in A. oryzae, grown in a maltose-containing medium (Fig. 5). In S. cerevisiae, the ER chaperone Kar2 (BiP) associates with Ire1, a transmembrane ER stress sensor with serine/threonine kinase and endoribonuclease domains, and causes its inactivation under low ER stress conditions. Ire1 is activated via two steps (Kimata et al. 2007). Upon ER stress, such that the concentration of unfolded proteins increases in the ER lumen, Kar2 is released from Ire1 to bind to the unfolded proteins, leading to oligomerization of Ire1 (the first step) (Okamura et al. 2000; Kimata et al. 2003). In the second step, unfolded proteins directly associate with the core stress-sensing region (CSSR) of oligomerized Ire1 for further activation (Kimata et al. 2007). Previously, it has been shown that Kar2/BiP transiently associates with newly synthesized proteins and more stably with terminally misfolded proteins in the ER (Simons et al. 1995). We assume that a number of BipA (Kar2 ortholog) molecules may have been consumed by the import of newly synthesized α-amylase into the ER lumen and, thus, released from IreA (Ire1 ortholog) to activate IreA in A. oryzae. Since Kar2/BiP is known to bind more stably to mutant proteins with folding defects in yeast and mammalian cells (Hendershot et al. 1996; Suyama et al. 2014), the mutant MsdS, compared to the wild-type MsdS, could also bind more stably to BipA in A. oryzae, leading to the release of BipA from IreA. It might be also be possible that the mutant MsdS specifically binds to the CSSR of IreA. Further analysis is required to elucidate the complete mechanism underlying IreA activation by expression of α-amylase and the mutant MsdS.
UPR induction requires unconventional splicing of hacA mRNA by IreA in A. fumigatus (Feng et al. 2011). Although our analyses showed that the expression of α-amylases and mutant MsdS synergistically induced the UPR, we could not detect any obvious differences in the activation levels of hacA mRNA splicing, supposedly catalyzed by IreA, with overexpression of mutant MsdS (Fig. S4). In S. cerevisiae, with the depletion of inositol or treatment with a low concentration of DTT, the splicing of HAC1 mRNA is activated and then deactivated (Pincus et al. 2010). Similarly, in A. niger, the splicing of hacA mRNA is attenuated 2 h after the addition of DTT with a peak at 30 min (Guillemette et al. 2011). Inducible expression of tissue plasminogen activator fused with glucoamylase also activates splicing of hacA mRNA, which is then gradually attenuated (Mulder et al. 2004). Likewise, splicing of hacA mRNA might be activated at the earlier stage of cell growth by overexpression of mutant MsdS and α-amylase but attenuated after continuous incubation.
The protein quality control mechanism is thought to be one of the bottlenecks in efficient production of nonfungal proteins by filamentous fungi (Fleissner and Dersch 2010). The results of the present study showed that the ER quality control mechanism might contribute to the ER retention and degradation of mutant MsdS at a moderate protein expression level. Therefore, further studies are warranted to elucidate the mechanism underlying the ER retention of misfolded proteins for improved production of recombinant proteins.
References
Carvalho ND, Arentshorst M, Kooistra R, Stam H, Sagt CM, van den Hondel CA, Ram AF (2011) Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl Microbiol Biotechnol 89:357–373. doi:10.1007/s00253-010-2916-5
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi:10.1073/pnas.0601456103
Deshpande N, Wilkins MR, Packer N, Nevalainen H (2008) Protein glycosylation pathways in filamentous fungi. Glycobiology 18:626–637. doi:10.1093/glycob/cwn044
Feng X, Krishnan K, Richie DL, Aimanianda V, Hartl L, Grahl N, Powers-Fletcher MV, Zhang M, Fuller KK, Nierman WC, Lu LJ, Latge JP, Woollett L, Newman SL, Cramer RA Jr, Rhodes JC, Askew DS (2011) HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog 7:e1002330. doi:10.1371/journal.ppat.1002330
Fleissner A, Dersch P (2010) Expression and export: recombinant protein production systems for Aspergillus. Appl Microbiol Biotechnol 87:1255–1270. doi:10.1007/s00253-010-2672-6
Gomi K, Iimura Y, Hara S (1987) Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric Biol Chem 51:2549–2555. doi:10.1271/bbb1961.51.2549
Gouka RJ, Punt PJ, van den Hondel CA (1997) Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl Microbiol Biotechnol 47:1–11. doi:10.1007/s002530050880
Guillemette T, Ram AF, Carvalho ND, Joubert A, Simoneau P, Archer DB (2011) Methods for investigating the UPR in filamentous fungi. Methods Enzymol 490:1–29. doi:10.1016/B978-0-12-385114-7.00001-5
Guillemette T, van Peij N, Goosen T, Lanthaler K, Robson GD, van den Hondel CA, Stam H, Archer DB (2007) Genomic analysis of the secretion stress response in the enzyme-producing cell factory Aspergillus niger. BMC Genomics 8:158. doi:10.1186/1471-2164-8-158
Hendershot L, Wei J, Gaut J, Melnick J, Aviel S, Argon Y (1996) Inhibition of immunoglobulin folding and secretion by dominant negative BiP ATPase mutants. Proc Natl Acad Sci U S A 93:5269–5274
Ichishima E (2016) Development of enzyme technology for Aspergillus oryzae, A. sojae, and A. luchuensis, the national microorganisms of Japan. Biosci Biotechnol Biochem 80:1681–1692. doi:10.1080/09168451.2016.1177445
Ichishima E, Taya N, Ikeguchi M, Chiba Y, Nakamura M, Kawabata C, Inoue T, Takahashi K, Minetoki T, Ozeki K, Kumagai C, Gomi K, Yoshida T, Nakajima T (1999) Molecular and enzymic properties of recombinant 1,2-α-mannosidase from Aspergillus saitoi overexpressed in Aspergillus oryzae cells. Biochem J 339:589–597. doi:10.1042/bj3390589
Jin FJ, Watanabe T, Juvvadi PR, Maruyama J, Arioka M, Kitamoto K (2007) Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl Microbiol Biotechnol 76:1059–1068. doi:10.1007/s00253-007-1088-4
Jones EW, Zubenko GS, Parker RR (1982) PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics 102:665–677
Kaufman RJ (1999) Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13:1211–1233. doi:10.1101/gad.13.10.1211
Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K (2007) Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol 179:75–86. doi:10.1083/jcb.200704166
Kimata Y, Ishiwata-Kimata Y, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K (2003) Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell 14:2559–2569. doi:10.1091/mbc.E02-11-0708
Kostova Z, Wolf DH (2003) For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J 22:2309–2317. doi:10.1093/emboj/cdg227
Kwon MJ, Jorgensen TR, Nitsche BM, Arentshorst M, Park J, Ram AF, Meyer V (2012) The transcriptomic fingerprint of glucoamylase over-expression in Aspergillus niger. BMC Genomics 13:701. doi:10.1186/1471-2164-13-701
Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. doi:10.1038/nature04300
Minetoki T, Tsuboi H, Koda A, Ozeki K (2003) Development of high expression system with the improved promoter using the cis-acting element in Aspergillus species. J Biol Macromol 3:89–96
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol 45:878–889. doi:10.1016/j.fgb.2007.12.010
Mizutani O, Masaki K, Gomi K, Iefuji H (2012) Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl Environ Microbiol 78:4126–4133. doi:10.1128/AEM.00080-12
Mulder HJ, Saloheimo M, Penttilä M, Madrid SM (2004) The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol Gen Genomics 271:130–140. doi:10.1007/s00438-003-0965-5
Nakatsukasa K, Brodsky JL (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9:861–870. doi:10.1111/j.1600-0854.2008.00729.x
Ohno A, Maruyama J, Nemoto T, Arioka M, Kitamoto K (2011) A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl Microbiol Biotechnol 92:1197–1206. doi:10.1007/s00253-011-3487-9
Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K (2000) Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem Biophys Res Commun 279:445–450. doi:10.1006/bbrc.2000.3987
Ozeki K, Kanda A, Hamachi M, Nunokawa Y (1996) Construction of a promoter probe vector autonomously maintained in Aspergillus and characterization of promoter regions derived from A. niger and A. oryzae genomes. Biosci Biotechnol Biochem 60:383–389. doi:10.1271/bbb.60.383
Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P (2010) BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8:e1000415. doi:10.1371/journal.pbio.1000415
Punt PJ, van Gemeren IA, Drint-Kuijvenhoven J, Hessing JG, van Muijlwijk-Harteveld GM, Beijersbergen A, Verrips CT, van den Hondel CA (1998) Analysis of the role of the gene bipA, encoding the major endoplasmic reticulum chaperone protein in the secretion of homologous and heterologous proteins in black Aspergilli. Appl Microbiol Biotechnol 50:447–454. doi:10.1007/s002530051319
Richie DL, Hartl L, Aimanianda V, Winters MS, Fuller KK, Miley MD, White S, McCarthy JW, Latge JP, Feldmesser M, Rhodes JC, Askew DS (2009) A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog 5:e1000258. doi:10.1371/journal.ppat.1000258
Simons JF, Ferronovick S, Rose MD, Helenius A (1995) Bip/Kar2p serves as a molecular chaperone during carboxypeptidase-Y folding in yeast. J Cell Biol 130:41–49. doi:10.1083/Jcb.130.1.41
Sims AH, Gent ME, Lanthaler K, Dunn-Coleman NS, Oliver SG, Robson GD (2005) Transcriptome analysis of recombinant protein secretion by Aspergillus nidulans and the unfolded-protein response in vivo. Appl Environ Microbiol 71:2737–2747. doi:10.1128/AEM.71.5.2737-2747.2005
Spear ED, Ng DT (2003) Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol Biol Cell 14:2756–2767. doi:10.1091/mbc.E02-11-0717
Suyama K, Hori M, Gomi K, Shintani T (2014) Fusion of an intact secretory protein permits a misfolded protein to exit from the endoplasmic reticulum in yeast. Biosci Biotechnol Biochem 78:49–59. doi:10.1080/09168451.2014.877185
Tanaka M, Shintani T, Gomi K (2015) Unfolded protein response is required for Aspergillus oryzae growth under conditions inducing secretory hydrolytic enzyme production. Fungal Genet Biol 85:1–6. doi:10.1016/j.fgb.2015.10.003
Tanaka M, Yoshimura M, Ogawa M, Koyama Y, Shintani T, Gomi K (2016) The C2H2-type transcription factor, FlbC, is involved in the transcriptional regulation of Aspergillus oryzae glucoamylase and protease genes specifically expressed in solid-state culture. Appl Microbiol Biotechnol 100:5859–5868. doi:10.1007/s00253-016-7419-6
Tatara Y, Lee BR, Yoshida T, Takahashi K, Ichishima E (2003) Identification of catalytic residues of Ca2+-independent 1,2-α-D-mannosidase from Aspergillus saitoi by site-directed mutagenesis. J Biol Chem 278:25289–25294. doi:10.1074/jbc.M302621200
Tatara Y, Yoshida T, Ichishima E (2005) A single free cysteine residue and disulfide bond contribute to the thermostability of Aspergillus saitoi 1,2-α-mannosidase. Biosci Biotechnol Biochem 69:2101–2108. doi:10.1271/bbb.69.2101
Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258. doi:10.1016/S0092-8674(00)80835-1
van den Hombergh JPTW, van de Vondervoort PJI, Fraissinet-Tachet L, Visser J (1997) Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol 15:256–263. doi:10.1016/s0167-7799(97)01020-2
Verdoes JC, Punt PJ, van den Hondel CAMJJ (1995) Molecular genetic strain improvement for the overproduction of fungal proteins by filamentous fungi. Appl Microbiol Biotechnol 43:195–205. doi:10.1007/bf00172812
Yamada O, Lee BR, Gomi K (1997) Transformation system for Aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosci Biotechnol Biochem 61:1367–1369. doi:10.1271/bbb.61.1367
Acknowledgements
We thank Dr. Eiji Ichishima and Dr. Yota Tatara for providing the plasmids pNAN8142-msdS and pNAN8142-msdS-C443F and anti-MsdS antibody. This work was supported by the NISR Research Grant from the Noda Institute for Scientific Research, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any experiments performed using human participants or animals by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Jun-ichi Yokota and Daisuke Shiro contributed equally to this work.
Electronic supplementary material
ESM 1
(PDF 1742 kb)
Rights and permissions
About this article
Cite this article
Yokota, Ji., Shiro, D., Tanaka, M. et al. Cellular responses to the expression of unstable secretory proteins in the filamentous fungus Aspergillus oryzae . Appl Microbiol Biotechnol 101, 2437–2446 (2017). https://doi.org/10.1007/s00253-016-8086-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-8086-3