Abstract
Gluconobacter (G.) oxydans is able to incompletely oxidize various sugars and polyols for the production of biotechnologically important compound. Recently, we have shown that the organism produces and accumulates mannitol as compatible solute under osmotic stress conditions. The present study describes the role of two cytoplasmic mannitol dehydrogenases for osmotolerance of G. oxydans. It was shown that Gox1432 is a NADP+-dependent mannitol dehydrogenase (EC 1.1.1.138), while Gox0849 uses NAD+ as cofactor (EC 1.1.1.67). The corresponding genes were deleted and the mutants were analyzed for growth under osmotic stress and non-stress conditions. A severe growth defect was detected for Δgox1432 when grown in high osmotic media, while the deletion of gox0849 had no effect when cells were exposed to 450 mM sucrose in the medium. Furthermore, the intracellular mannitol content was reduced in the mutant lacking the NADP+-dependent enzyme Gox1432 in comparison to the parental strain and the Δgox0849 mutant under stress conditions. In addition, transcriptional analysis revealed that Gox1432 is more important for mannitol production in G. oxydans than Gox0849 as the transcript abundance of gene gox1432 was 30-fold higher than of gox0849. In accordance, the activity of the NADH-dependent enzyme Gox0849 in the cell cytoplasm was 10-fold lower in comparison to the NADPH-dependent mannitol dehydrogenase Gox1432. Overexpression of gox1432 in the corresponding deletion mutant restored growth of the cells under osmotic stress, further strengthening the importance of the NADP+-dependent mannitol dehydrogenase for osmotolerance in G. oxydans. These findings provide detailed insights into the molecular mechanism of mannitol-mediated osmoprotection in G. oxydans and are helpful engineering strains with improved osmotolerance for biotechnological applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms are often exposed to fluctuations of the external osmolarity and have to cope with osmotically induced water fluxes across the cytoplasmic membrane. As a consequence, they must be able to make physiological adjustments to ensure adequate levels of cellular hydration for the protection of cell function and turgor. In prokaryotes, synthesis and accumulation of compatible solutes are among some common strategies to cope with reduced water activity. Small organic compounds such as glycine, betaine, trehalose, ectoine, glutamate, glutamine, mannitol and proline are well known to be compatible with cellular metabolic functions and stabilize the positive turgor pressure inside the cell (da Costa et al. 1998). Mannitol is the most abundant polyol in nature and is used as an osmoprotectant mostly in eukaryotes such as plants and fungi (Empadinhas and da Costa 2008). In a previous study, it was shown that the industrially important microorganism, Gluconobacter oxydans, copes with osmotic stress by synthesizing mannitol inside the cell (Zahid et al. 2015). The organism accumulates mannitol as a compatible solute in media with high concentrations of sucrose, glucose, or fructose (Table S1). Additionally, mannitol improved the growth of the bacterium when present in low concentrations in high osmotic glucose media. However, until now, the enzymes involved in stress-mediated mannitol production in G. oxydans were unknown. There are two potential pathways present in bacteria for the synthesis of mannitol. One includes a mannitol-1-phosphate dehydrogenase and a mannitol-1-phosphatase which convert fructose-6-phosphate to mannitol. There are also reports on a bifunctional enzyme for mannitol biosynthesis in Acinetobacter baylyi that combines mannitol-1-phosphate dehydrogenase and phosphatase activities in a single polypeptide chain (Sand et al. 2015). The second pathway is based on a mannitol-2-dehydrogenase reducing fructose directly to mannitol and is frequently found in lactic acid bacteria (Saha 2004). Both, mannitol-1-phosphate dehydrogenase and mannitol-2-dehydrogenase belong to the class of oxidoreductase and require NAD(P)+ or NAD(P)H as cofactors.
In the present study, we identified two genes gox1432 and gox0849 that potentially encode mannitol dehydrogenases in G. oxydans 621H. Both mannitol dehydrogenase-encoding genes were deleted and the effect on osmoprotection in knockout mutants was analyzed. In addition, both enzymes were overproduced and their catalytic efficiency was investigated. We found that the NADP+-dependent mannitol dehydrogenase (Gox1432) was crucial for cell survival under osmotic stress. These findings provide first insight into the mechanism of osmoprotection in G. oxydans.
Materials and methods
Chemicals
All chemicals, antibiotics, and media components were purchased from Carl Roth GmbH (Karlsruhe, Germany) and Sigma-Aldrich (Munich-Germany). NADPH, NADH, and NAD+ were purchased from Carl Roth GmbH (Karlsruhe, Germany), while NADP+ was from Merck (Darmstadt, Germany). Restriction endonucleases, T4 DNA ligase, Taq DNA polymerase, and other PCR reagents were obtained from Thermo Fisher Scientific Biosciences GmbH (Schwäbisch-Gmünd, Germany). Phusion DNA Polymerase was bought from Biozym Scientific GmbH (Oldendorf, Germany). pASK-IBA3 and pASK-IBA5 plasmids were obtained from IBA GmbH (Goettingen, Germany). Oligonucleotides were synthesized by Eurofins MWG Operon (Ebersberg, Germany).
Microorganisms and culture conditions
High-efficiency competent Escherichia coli (NEB 5-α) cells were obtained from NEB New England Biolabs Inc. (Frankfurt am Main, Germany) and grown in Lysogeny broth at 37 °C (Miller 1972). For the overproduction of Gox1432 and Gox0849, E. coli transformants were grown in maximal induction medium (Mott et al. 1985) containing ampicillin (100 μg ml−1). G. oxydans 621H ΔhsdR (DSM 2343; Table 1) was obtained from S. Bringer-Meyer (Research Center Juelich GmbH, Germany) and cultured in yeast extract (0.6 % w/v) mannitol (2 % w/v) medium with cefoxitin (50 μg ml−1) in baffled flasks at 30 °C. Strains containing pBBR.p264.ST derivatives were grown in the presence of kanamycin (50 μg ml−1). To study the effect of osmotic stress, cultures were grown in YGP medium containing yeast extract (0.6 % w/v), glucose (0.9 % w/v), and 100 mM sterile phosphate buffer (pH 6.8–7.0). The osmolarity of the medium was increased by the addition of sucrose (450 mM). Optical density (OD) was measured photometrically at 600 nm. All experiments were performed in triplicates using two biological replicates.
Standard molecular biology techniques and cloning
All routine molecular biology techniques were performed according to Sambrook et al. (1989). Genomic DNA of G. oxydans was isolated using Gen Jet Genomic DNA purification kit from Thermo Fisher Scientific (Schwäbisch-Gmünd, Germany) and was used as template for gene amplification. For cloning of gox1432 and gox0849, primers containing Eco1l restriction sites (Table 1) were designed, respectively, using Primer D’signer 1.1 software (www.iba-lifesciences.com). The PCR products and the expression vectors pASK-IBA 3 and pASK-IBA 5 were digested with Eco1l, purified, and ligated to generate the plasmids pASK.3_gox1432.ST and pASK.5_gox0849.ST, respectively. Both constructs contained an ampicillin resistance cassette, plasmid-encoded ribosomal binding sites, and a C-terminal Strep-tag II (pASK.3_gox1432.ST) or a N-terminal Strep-tag II (pASK.5_gox0849.ST). The plasmids were transferred into E. coli (NEB 5-α) according to manufacturer’s instructions. Positive clones were selected by colony PCR and the correct insertion of the PCR products was checked by sequencing (StarSEQ GmbH, Mainz, Germany). For the generation of G. oxydans strains carrying in-frame deletion mutations for gox1432 and/or gox0849, the codAB markerless in-frame deletion system was used (Kostner et al. 2013). For the generation of the Δgox1432 mutant 1 kb of the up- and downstream regions were amplified using primers up_gox1432_Fw/up_gox1432_Rev and do_gox1432_Fw/do _gox1432_Rev (Table 1). The amplified fragments were used as template for a fusion PCR reaction using primers up_gox1432_Fw and do_gox1432_Rev. After digestion using Kpn1 and Xba1 and ligation in the corresponding sites of pKOS6b, the resulting vector pKos6bΔ1432 was transferred into competent E. coli (NEB 5-α). Finally, pKos6bΔ1432 was transferred from E. coli NEB 5-α into G. oxydans with the help of E. coli ECG 18 by triparental mating (Condon et al. 1991). The markerless deletion of gox1432 was generated by plating G. oxydans pKos6bΔ1432 on 5-fluorocytosine containing agar plates as described by Kostner et al. (2013). The construction of the Δgox0849 mutant and the double mutant (Δgox1432 Δgox0849) was performed analogously. For the complementation of gox1432 deletion mutants, the vector pBBR.p264.ST was used (Kallnik et al. 2010). Gene gox1432 including its native ribosomal binding site was amplified using the pBBR.p264.gox1432_Fw and pBBR.p264.gox1432_rev primer pair. The resulting PCR product was digested with EcoRV/Asc1 and cloned into the EcoRV/Asc1 sites of the pBBR.p264.ST vector. The resulting plasmid, pBBR.p264.gox1432.ST, was transformed into E. coli (NEB 5-α) and used for conjugal transfer of the plasmid into the G. oxydans Δgox1432 strain. All clones and mutants were verified by sequencing (StarSeq, Mainz, Germany).
Reverse transcription quantitative PCR
Reverse transcription quantitative PCR (RT-qPCR) was performed to quantify the relative abundance of transcripts from gox1432 and gox0849 in G. oxydans using gox1709 as a reference gene (Rauch et al. 2010). Total RNA from osmotically stressed and non-stressed cultures (OD600 of 0.4–0.6) was isolated by the TRI reagent extraction method (Chomczynski 1993). One hundred milliliter cultures were harvested by centrifugation at 10,000 rpm and 4 °C. The cell pellet was dissolved in 5 ml TRI reagent and lysed by freeze-thaw treatment at −80 °C for 15 min. Total RNA was extracted according to manufacturer’s instructions (Sigma-Aldrich, Munich, Germany) and purified using the SurePrep RNA Cleanup and Concentration Kit (Thermo Fisher Scientific, Schwerte, Germany), including a DNaseI treatment. RNA concentration and purity was analyzed spectrophotometrically by the 260/280 nm ratio as well as by denaturing agarose gel electrophoresis. Primers for RT-qPCR were generated using primer3 software (http://primer3.ut.ee/) (Table 1). Each PCR reaction contained 250–300 ng RNA and the transcript abundance was quantified using a SYBR Green RT-PCR Kit (Qiagen, Hilden, Germany) with a master cycler gradient from Eppendorf (Hamburg, Germany). A relative value (ΔCt) for the target cDNA concentration in each reaction was determined by subtracting Ct values of the genes of interest from the reference gene and the fold change was expressed as 2(ΔCt). The efficiency of the primer pairs RT_Gox1432_Fw/RT_Gox1432_rev and RT_Gox849_Fw/RT_Gox0849_rev was 96 and 100 %, respectively, and was checked by qPCR using serial dilutions of the template DNA (Stahlberg et al. 2003).
Overproduction and purification of proteins
For the overproduction of Gox1432 and Gox0849, E. coli DH5α cells containing plasmid pASK.3_gox 1432.ST or pASK.5_gox0849.ST were grown in 100 ml maximal induction medium (Mott et al. 1985) with ampicillin (100 μg ml−1) at 37 °C. The cultures were induced with 200 ng ml−1 anhydrotetracyclin at an optical density of 0.5 and were incubated overnight at 16 °C. Cells were harvested by centrifugation (10,000 rpm, 4 °C, 15 min) and resuspended in 15 ml buffer W (100 mM Tris-Cl, 100 mM NaCl, pH 8.0). After addition of lysozyme, DNaseI and protease inhibitor cocktail (Sigma-Aldrich, Munich, Germany) cells were lysed by sonication (Branson Sonifier Cell Disruptor with Branson Ultrasonics converter, Danbury USA). The proteins were purified by Strep-tactin Superflow® affinity chromatography as described by the manufacturer (IBA GmbH, Göttingen, Germany) and quantified by Bradford method (Bradford 1976). SDS-PAGE and western blots were performed as described by Meyer et al. (2015) according to the methods of Laemmli (1970) and Towbin et al. (1979), respectively. Native PAGE was done on 4–20 % gradient gels (BioRad, Munich, Germany) as described by Rauch et al. (2010).
Enzyme assays
The activities of purified enzymes were measured photometrically by recording the rate of change in absorption of NAD(P)H/NAD(P)+ at 340 nm (ε = 6.22 mM−1 cm−1). For the analysis of substrate reduction (d-fructose, l-sorbose, or 5-keto d-gluconate), the 1 ml reaction mixtures contained 50 μmol potassium phosphate, pH 7.0, 0.125 μmol NADH or NADPH, and 100 μmol of substrates. For the quantification of polyol oxidation, 50 mM Tris-Cl, pH 9.0 (for Gox0849) or 50 mM sodium carbonate (for Gox1432), pH 10 were used that were supplemented with 0.125 μmol NAD+ or NADP+ and 100 μmol of substrates. The reactions were started by the addition of substrates. One unit corresponded to the conversion of 1.0 μmol of pyridine dinucleotide per minute. For the determination of activities of mannitol dehydrogenases in the cytoplasm, 100–250 ml stressed or non-stressed cultures were harvested in the mid log phase by centrifugation at 10,000 rpm for 15 min and 4 °C. Cells were resuspended in 10 ml cell suspension buffer containing 100 mM potassium phosphate buffer, pH 7.0, 5 mM MgSO4·7H2O and 0.75 mM dithiothreitol. Cells were lyzed by sonication and the cell-free extract was centrifuged at 15,000 rpm for 15 min followed by ultracentrifugation at 45,000 rpm for 1 h. The clear supernatant (cytoplasmic fraction) was used to measure the activities of NADP+ and NAD+-dependent mannitol dehydrogenases at pH 7.0 as described above. Protein concentration was measured according to Bradford (1976).
Determination of glucose consumption and gluconate production
The rate of glucose consumption and gluconate formation was analyzed for the double deletion mutant (Δgox1432 Δgox0849) in comparison to the wild type. Both cultures were grown in 70-ml YGP media containing 1000 mM glucose in 250-ml baffled flasks. The flasks were inoculated by the addition of 5 ml of an overnight culture and were incubated at 30 °C and a stirring rate of 180 rpm. Samples (0.7 ml) were taken at regular intervals and were centrifuged at 12,000 rpm for 10 min. The supernatant was analyzed on a Aminex-HPX87H column (BioRad, Munich, Germany, 300 mm × 7.8 mm) using 5 mM H2SO4 as eluent at 65 °C and a flow rate of 0.3 mL min−1. Glucose and gluconate concentrations were quantified according to standard calibration curves. Analysis of intracellular mannitol content was performed as described by Zahid et al. (2015).
Results
Identification and characterization of mannitol dehydrogenases in G. oxydans
Two mannitol dehydrogenases are present in G. oxydans that vary with respect to their cofactor specificity (Adachi et al. 1999). One enzyme was described as a NADP+-dependent mannitol dehydrogenase (EC 1.1.1.138) and the second mannitol dehydrogenase showed a preference for NAD+ (EC 1.1.1.67). The amino acid sequences of these enzymes are not available, and the respective gene loci could not be identified. A search using the NCBI non-redundant BLAST database revealed that gox1432 and gox0849 encode putative mannitol dehydrogenases in G. oxydans. Both proteins were homologous to several putative mannitol dehydrogenases from acetic acid bacteria. However, these enzymes have not been characterized with respect to their catalytic activities. A BLAST analysis using the SwissProt database indicated several biochemically analyzed homologues of the mannitol dehydrogenase family with identities of 32–40 %, similarities of 50–57 %, and coverage of 84–95 %. Among them was a mannitol-2-dehydrogenase from Rhodobacter sphaeroides (Schneider et al. 1993) and Pseudomonas fluorescens (Kavanagh et al. 2002) and a d-arabinitol-4 dehydrogenase from Klebsiella pneumanie (Heuel et al. 1998). Gox1432 and Gox0849 contained Rossmann fold NAD+/NAD(P)+ binding domains (Rossmann et al. 1974) and mannitol dehydrogenase signature sequence motifs (PDOC00751). Gox1432 and Gox0849 were annotated as NADP-d-sorbitol dehydrogenase and NADPH-dependent l-sorbose reductase, respectively (Prust et al. 2005). Because of the unknown substrate spectrum of the predicted mannitol dehydrogenases, the corresponding genes were cloned into pASK-IBA vectors and heterologously overexpressed in E. coli. The proteins were purified from crude extract to apparent homogeneity using Strep-tactin affinity chromatography obtaining 1–2 mg of protein from 1 l E. coli cultures.
SDS-PAGE and western blot analysis revealed molecular masses of 54.9 kDa for Gox1432 and 55.7 kDa for Gox0849, which are in accordance with the expected mass of 54.6 and 55.0 kDa for the recombinant tagged proteins, respectively (Fig. 1). Similarly, Adachi et al. (1999) reported that the NADP+-dependent mannitol dehydrogenase from Gluconobacter suboxydans IFO 12528 was a monomeric protein with an apparent molecular mass of 50 kDa. Gel filtration chromatography showed that the NAD+-dependent mannitol dehydrogenase had a native molecular mass of 130 kDa and was composed of two identical subunits (Adachi et al. 1999). In agreement, native PAGE revealed that Gox1432 formed monomers, while Gox0849 had a homodimeric structure (data not shown). In addition, very faint protein bands were detected for Gox1432 at 100 kDa and for Gox0849 at 160 kDa representing low amounts of homodimers and homotrimers, respectively.
SDS-PAGE and western blot of purified proteins. Lanes 1, 3, 5, 7: SDS-PAGE (12 % gel) of molecular mass marker (page ruler Prestained Protein Ladder, Thermo Fisher Scientific, Schwerte, Germany). Lane 2: SDS-PAGE of purified Gox1432 (silver staining). Lane 4: western blot of purified Gox1432. Lane 6: SDS-PAGE of purified Gox0849 (silver staining). Lane 8: western blot of purified Gox0849. Molecular mass of the purified proteins was determined by plotting the mobility versus molecular mass of the standard marker proteins
For further characterization, the activity of Gox1432 from G. oxydans 621H was determined using different sugars and polyols in the presence of either NAD(P) + or NAD(P)H. It was found that Gox1432 oxidized the polyols d-mannitol, d-sorbitol, d-arabitol, and d-glycerol and was only active with NADP+ as an electron acceptor. Furthermore, the enzyme was able to reduce d-fructose, l-sorbose, and 5-keto d-gluconate in the presence of NADPH as an electron donor. The optimum pH for substrate reduction and oxidation was 7.0 and 10, respectively. Gox0849 also showed cofactor preference, but could only use NADH for fructose reduction. No other tested sugars were reduced. Furthermore, only d-mannitol was oxidized by Gox0849 using NAD+ as an electron acceptor. The optimum pH for reduction of d-fructose and oxidation of d-mannitol was 7.0 and 9.0, respectively. The kinetic parameters of both proteins were measured for different substrates and pH values (Table 2). At a physiological pH of 7, Gox1432 and Gox0849 reduced most rapidly d-fructose to d-mannitol with catalytic efficiencies of 2.8 × 103 s−1 M−1 and 2.9 × 103 s−1 M−1, respectively. In contrast, the rate of mannitol oxidation at pH 7 was 4-fold lower for Gox1432 and 3-fold lower for Gox0849. These data suggest that although the enzymes are able to oxidize mannitol, their primarily function in vivo is the reduction of fructose. High oxidative activities of the enzymes were obtained only at non-physiological pH values of 9–10 with K cat/K m of 7.8 × 103 s−1 M−1 and 6.4 × 103 for Gox1432 and Gox0849, respectively.
Transcript abundance of genes encoding mannitol dehydrogenases in G. oxydans
The transcript abundance of gox1432 and gox0849 was determined by RT-qPCR in cells grown under unstressed (YGP) and osmotic stress conditions (YGP with 450 mM sucrose). Gene gox1709 coding for the gluconokinase was used as an internal reference as it is constitutively expressed (Rauch et al. 2010). As shown in Fig. 2, the expression of both genes was not regulated as their quantification cycle (Ct) values did not differ significantly under control and osmotic stress conditions. However, the transcript abundance of gox1432 was 3.5-fold higher in comparison to the reference gene indicating its importance for cellular metabolism. In contrast, the amount of transcripts from gox0849 was about 8-fold lower than the reference gene.
Transcript abundance of the genes encoding mannitol dehydrogenases (gox1432 and gox0849) in comparison to the house keeping gene gox1709. Dark gray bars indicate transcript abundance of indicated genes under non-stress conditions; light gray bars indicate transcript abundance of the corresponding genes under osmotic stress conditions. ΔCt values were determined by subtracting average Ct values of the genes of interest from the reference gene gox1709
Characterization of mannitol dehydrogenase deletion mutants
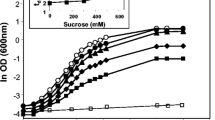
To elucidate the function of the two mannitol dehydrogenases in osmotolerance of G. oxydans, the deletion mutants Δgox1432 and Δgox0849 and the corresponding double mutant were generated. Growth, enzymatic activities, and intracellular mannitol content of all strains were monitored in YGP medium, and 450 mM sucrose was used as an osmoticum as it is not metabolized by G. oxydans and does not interfere with glucose metabolism. Growth parameters of all mutants were similar to the wild type under non-osmotic stress conditions with doubling times of 2 h and final optical densities of 2.0 (not shown). Hence, the experiments indicated that gox1432 and gox0849 were not essential for growth under standard conditions with glucose as substrate. In YGP medium supplemented with 450 mM sucrose, growth of the wild type and the Δgox0849 mutant was almost identical with doubling times of 3.2 and 3.3 h, respectively (Fig. 3a). Also the final OD600 was similar with 0.91 for Δgox0849 versus 0.96 for the wild type, indicating the minor importance of mannitol dehydrogenase Gox0849 for cells growth under osmotic stress conditions. In contrast, growth of G. oxydans Δgox1432 was impaired in highly osmotic media. The doubling time was extended to 6 h and the final optical density was severely reduced by 45 % in comparison to the parental strain (Fig. 3a). To examine whether a plasmid-borne allele could complement the genomic deletion of gox1432, the mutant was transformed with a broad host range plasmid pBBR1-MCS with the strong promoter p264 in front of gene gox1432. The complementation led to slightly improved growth parameters (t d = 3.1 h, final OD600 = 1.4) in comparison to the wild type (Fig. 3b). These results confirmed the importance of gox1432 for cell survival and growth under osmotic stress.
Growth of wild-type and mutant strains of G. oxydans on YPG supplemented with 450 mM sucrose. a Growth profile of the wild-type (filled circle), mutant strain Δgox0849 (open triangle), mutant strain Δgox1432 (open square), double mutant Δgox1432 Δgox0849 (filled triangle). b Wild-type (filled circle), Δgox1432 complemented strain (inverted open triangle), double mutant Δgox1432 Δgox0849 with 5 mM mannitol (inverted filled triangle), growth of non-stressed wild type grown on YGP (open circle)
The growth defect of cells under osmotic stress was even more pronounced when both mannitol dehydrogenase-encoding genes (gox1432, gox0849) were deleted. The double mutant (Δgox1432 Δgox0849) showed an extensive lag phase of 8–10 h when grown on YGP + 450 mM sucrose medium followed by poor growth with doubling times of 6 h and a final optical density of 0.4 (Fig. 3a). However, when the double mutant was cultured in YGP + 450 mM sucrose medium in the presence of 5 mM mannitol, growth improved significantly with doubling times of 2.1 h and a final OD600 of 2.0. These values were in the same range as for the wild type without osmotic stress (t d = 2 h, final OD600 = 2.0). Hence, low concentrations of mannitol could alleviate osmotic stress from the cells imposed by sucrose (Zahid et al. 2015). The results indicate that the growth retardation observed for the Δgox1432 mutant and the double mutant under osmotic stress was due to defects in mannitol synthesis. Furthermore, it became evident that gox1432 encoding the NADP+-dependent mannitol dehydrogenase was the major player for osmoprotection of G. oxydans. However, gox0849, encoding the NAD+-dependent mannitol dehydrogenase, is important for survival of the cell under osmotic stress when gox1432 is absent. The slow growth of the double mutant after a long lag phase points towards a third unknown enzyme that is able to produce low amounts of mannitol in the presence of 450 mM sucrose as osmoticum.
Intracellular mannitol formation and activity of cytoplasmic mannitol dehydrogenase
The single and double mannitol dehydrogenase deletion mutants were also analyzed for the presence of intracellular mannitol under osmotic stress induced by 450 mM sucrose (Fig. 4). It was found that the deletion of gox0849 did not significantly alter the intracellular mannitol accumulation (1.7 ± 0.1 μmol mg protein−1) in comparison to the wild type (2.0 ± 0.1 μmol mg protein−1). In contrast, the Δgox1432 mutant and the double mutant revealed a 60 to 70 % reduction of intracellular mannitol. These values correlated with the findings from the growth experiments and revealed that the deletion of mannitol dehydrogenases Gox1432 retarded cell growth under osmotic stress due to a decrease in intracellular mannitol production. Again, it was found that the double deletion mutant had some residual potential for mannitol synthesis (0.4 ± 0.1 μmol mannitol mg protein−1). This finding indicated the presence of a third intracellular mannitol dehydrogenase with limited activity.
Effect of deletion of mannitol dehydrogenases on intracellular mannitol formation. The wild-type and the mannitol dehydrogenase deletion mutants were cultivated in YGP medium containing 450 mM sucrose. Analysis of intracellular mannitol content was performed as described by Zahid et al. (2015)
To confirm these results, the cytoplasmic fractions of the wild type, the single mutant, and the double mutant strains were prepared from stressed and non-stressed cells. The intracellular pH of acetic acid bacteria is in the range of 7.0 (Sugisawa et al. 1991). Hence, it is tempting to speculate that Gox1432 and Gox0849 function as fructose reductases in the cytoplasm of G. oxydans as shown for the purified enzymes at pH 7. Therefore, all cytoplasmic fractions were assayed for d-fructose reduction activity (Fig. 5). The maximum reduction rate for d-fructose was 3.0 ± 0.4 μmol min−1 mg of cytoplasmic protein−1 as catalyzed by the NADP+-dependent mannitol dehydrogenase Gox1432 in the wild-type strain under osmotic stress. In contrast, the activity was only 1.25 ± 0.2 μmol min−1 mg protein−1 in the cytoplasm of wild-type cultures grown in the absence of sucrose. Similar rates for NADPH-dependent fructose reduction were found in the cytoplasm of the Δgox0849 mutant. Under stress and non-stress conditions the NADH-specific reduction rate of d-fructose was only 0.3 ± 0.03 μmol min−1 mg of cytoplasmic protein−1 in the wild type. NADH-dependent reduction of fructose in G. oxydans Δgox0849 was 2-fold lower compared to the wild type. In the Δgox1432 mutant, only NAD+-specific mannitol dehydrogenase activity was detected with an activity of 0.3 ± 0.04 μmol min−1 mg of cytoplasmic protein−1. In the double mutant NADPH-dependent d-fructose reduction was completely absent, but the cytoplasm was still reactive with NADH to reduce d-fructose with a specific activity of 0.16 ± 0.05 U mg protein−1 (Fig. 5). This indicated the presence of another intracellular NAD+-specific mannitol dehydrogenase. In addition, the cytoplasmic fractions of all strains were tested for d-mannitol oxidation at pH 7. It was found that the ratio of NAD+ and NADP+-dependent mannitol oxidation in stressed and unstressed cultures were similar to the rates of fructose reduction but the specific activities were 10-fold lower (data not shown).
Activities of mannitol dehydrogenases in the cytoplasmic fractions of the wild-type and mutant strains of G. oxydans grown on YGP medium (non-stressed) or YGP medium + 450 mM sucrose (stressed). Dark and light gray bars represent the activity of the NADP+ and the NAD+-dependent mannitol dehydrogenase, respectively
One important biotechnological application of G. oxydans is the synthesis of gluconate that is used in pharmaceutical and food industries (Roblin et al. 2006). Therefore, the wild-type and the double mutants were tested for their efficiency with respect to gluconate production in YP media containing 1 M glucose as substrate and as osmoticum. It is known, that only low amounts of glucose are transported into the cells for biomass production. Most glucose is oxidized in the periplasm by the membrane-bound glucose dehydrogenase to form δ-gluconolactone, which is then hydrolyzed to gluconate. We observed a severe effect on growth, gluconate production, and pH values of the cultures during glucose-induced osmotic stress (Fig. 6a, b). After a long lag phase, the double deletion mutant reached a final OD600 of 0.2 and had a doubling time of approximately 10 h. Consequently, the gluconate production rate of the double mutant strain was almost 4-fold lower compared to the wild-type strain in the exponential growth phase. At the end of the experiment (t = 30 h), the osmotically stressed wild-type cells produced about 350 mM gluconate in contrast to the double mutant that formed only 100 mM gluconate. Accordingly, the drop of the pH values during growth of the cultures was different. The pH of the double mutant decreased from 6.5 to 3.5 within 30 h. In the wild type, the same decrease was observed after 14 h. Thus, the deletion of mannitol dehydrogenase-encoding genes had a direct influence on cell division and cellular catalytic efficiencies under osmotic stress. Furthermore, it was found that the deletion mutant accumulated only 0.4 ± 0.2 μmol mannitol mg cell protein−1 while the mannitol concentration was 2.0 ± 0.4 μmol mg protein−1 in the wild type with glucose as osmoticum (Table S1). In summary, the results demonstrate the necessity of mannitol dehydrogenases for G. oxydans to tolerate and survive high osmotic conditions for effective product formation.
Effect of deletion of mannitol dehydrogenases on growth and gluconate formation. a Correlation between gluconate formation (filled and open squares) and decrease in pH (filled and open triangles) of the wild-type G. oxydans (filled symbols) and Δgox1432 Δgox0849 double mutant (open symbols) on yeast-glucose (1000 mM) media with 100 mM K-phosphate buffer (pH 6.8). b Growth profile of the wild-type (filled circle) and the Δgox1432 Δgox0849 double mutant (open circle)
Discussion
Microorganisms thrive in different ecological niches depending on local biotic and abiotic factors. Water activity is one of the most important abiotic parameters that directly influence growth of microbial species. Therefore, microorganisms adapt to cope with reduced water activity either by the uptake of extracellular ions (K+) or by synthesis and accumulation of small organic molecules referred to as osmolytes or compatible solutes (da Costa et al. 1998; Empadinhas and da Costa 2009; Wood 2015). Numerous low weight organic molecules are known to be compatible solutes such as amino acids (glutamate, proline, glutamine, ectoine), sugars (trehalose, sucrose), phosphodiesters (di-myo-inositol phosphate), glyceric acid derivatives (mannosylglycerate, glucosylglycerate), and polyols (sorbitol, mannitol) (Wood 2015; Empadinhas and da Costa 2009). These molecules maintain a positive intracellular turgor pressure and secure the hydration of the cell cytoplasm to attenuate external water fluctuations (Cayley et al. 1992; Cayley and Record 2004).
Recently, we have reported the role of d-mannitol as an osmoprotectant in G. oxydans 621H (Zahid et al. 2015). Mannitol functions as an osmolyte in plants and fungi (Jennings 1984; Grant 2004; Empadinhas and da Costa 2009) as well as in different bacteria like Pseudomonas fluorescence (Kets et al. 1996) and A. baylyi (Sand et al. 2013) and in hetero-fermentative lactic acid bacteria (Wisselink et al. 2002). Other short-chain polyols such as glycerol, arabitol, and sorbitol are also known to act as compatible solutes in different microorganisms under osmotic as well as cold and oxidative stress conditions (Sootsuwan et al. 2013; de Barros and Celligoi, 2006; Efiuvwevwere et al. 1999; Loos et al. 1994; Hocking 1988). In G. oxydans, mannitol was accumulated inside the cells during growth under osmotic stress conditions. Moreover, external mannitol had a positive effect on growth and physiology of cells when added in small amounts to osmotically stressed cultures (Zahid et al. 2015). Hence, it seems obvious that the osmotolerance of G. oxydans can be improved by increasing the intracellular mannitol production. Therefore, it was of great interest to unravel the pathway of mannitol synthesis in this organism.
Here, we show that mannitol dehydrogenases are responsible for osmotolerance of G. oxydans 621H. This bacterium naturally lives in sugar-rich environments such as fruits, flowers, nectars, wine, and honey syrups (De Muynck et al. 2007). Due to the abundance of nutrients in its environment, G. oxydans performs an overflow type of metabolism where most of the sugars, polyols, and alcohols are incompletely oxidized to keto-sugars and acids (De Ley et al. 1984; Deppenmeier et al. 2002; Deppenmeier and Ehrenreich 2009). Due to high product and low growth yield, G. oxydans is used for several biotechnological processes in textile, food, pharmaceutical, and fodder industries to produce valuable substances such as d-gluconate, dihydroxyacetone, l-sorbose (vitamin C production), and 6-amino-6-deoxy-l-sorbose (Miglitol production) (Hommel and Ahnert 1999; Hancock 2009; Macauley et al. 2001; Claret et al. 1994; De Muynck et al., 2007). G. oxydans can thrive in concentrated sugar solutions, for example, up to 30 % (w/v) glucose, but its growth and productivity declines by increased osmolarity of medium (Luchterhand et al. 2015; Zahid et al. 2015). Therefore, there is a need to explore the physiology of G. oxydans under osmotic stress to understand and improve its bio-catalytic efficiency.
Since mannitol cannot be synthesized by the primary metabolism of G. oxydans (Zahid et al. 2015), it can be argued that this compatible solute is produced as a secondary metabolite inside the cell. Fructose is probably the direct precursor which could be formed from glucose by the catalytic activities of a glucokinase, a glucose-6-phosphate isomerase and a phosphofructose phosphatase (Richhardt et al. 2012; Rauch et al. 2010). In addition, the production of fructose 6-phosphate is also possible by using the oxidative pentose phosphate pathway with subsequent dephosphorylation. Finally, fructose would be reduced to mannitol by a mannitol dehydrogenase reaction (Fig. 7). A second pathway for mannitol synthesis is found in fungi which includes the direct production of fructose from glucose by the catalytic activity of a glucose isomerase. However, such an enzyme is not encoded on the chromosome of G. oxydans 621H.
Four types of polyol dehydrogenases are known to exist in Gluconobacter species which use mannitol (oxidative direction) or fructose (reductive direction) as substrates. These enzymes can be distinguished by their molecular mass, cofactor specificity, and cellular localization: Type 1 comprise membrane-bound and PQQ-dependent polyol dehydrogenases with broad substrate spectrum which are involved in incomplete oxidation of a range of substrates. It has been shown that the enzyme from G. oxydans 621H oxidizes mannitol, glycerol and gluconate forming fructose, dihydroxyacetone, and 5-ketogluconate, respectively. The enzyme is also responsible for the oxidation of d-sorbitol to produce l-sorbose, an important intermediate in vitamin C production (Matsushita et al. 2003; Hoshino et al. 2003). Type 2 proteins are NADP+-dependent mannitol dehydrogenases with limited substrate spectrum and molecular masses of about 50 kDa per single subunit that convert only mannitol or fructose (Adachi et al. 1999). Type 3 NADP+ mannitol dehydrogenases catalyze the reversible oxidation of different polyols such as d-sorbitol, mannitol, and arabitol (Klasen 1994; Shibata et al. 2000; Shinjoh et al. 2002). Blast analysis and multiple alignments indicated that Gox1432 belongs to this group of mannitol dehydrogenases (data not shown). Class 4 comprise NAD+ dependent mannitol dehydrogenases which were found in cell extracts from G. oxydans LMG 1489 (Parmentier et al. 2005) and G. suboxydans IFO12528 (Adachi et al. 1999). Here, we describe the first purified enzyme of this class that is encoded by gene gox0849.
Gox1432 and Gox0849 reduced d-fructose to d-mannitol with comparable catalytic efficiencies at pH 7.0. However, neither was able to reduce fructose-6-phosphate to mannitol-1-phosphate. Hence, it is tempting to speculate that there is a phosphatase that is able to dephosphorylate fructose-6-phosphate in the course of mannitol synthesis. Efficient conversion of d-mannitol to d-fructose by Gox1432 and Gox0849 was only observed at non-physiological alkaline conditions. These data indicate that the enzymes work as reductases in vivo and can be classified as d-fructose reductases rather than d-mannitol dehydrogenases. Strong evidence was presented that Gox1432 is more important for mannitol production in G. oxydans than Gox0849, since its transcript abundance was much higher than the expression level of gox0849. This was in agreement with the activity of the NADH-dependent enzyme in the cell cytoplasm being 10-fold lower in comparison to the NADPH-dependent d-fructose reductase Gox1432. The importance of Gox1432 on cell physiology and compatible solute accumulation was further highlighted by analyzing the corresponding deletion strain. Under osmotic stress condition the growth rate and the final optical density of the Δgox1432 strain was reduced by 37 and 45 %, respectively, compared to the wild type. Furthermore, deletion of gox1432 resulted in a complete loss of NADPH-dependent d-fructose reduction in extracts of cells grown under stress and non-stress conditions. As expected, NADH-dependent d-fructose reduction was unaltered. These results indicated that Gox1432 is solely responsible for intracellular NADPH mediated d-fructose reduction. Similar effects were observed in Zymomonas mobilis, when the gfo gene (encoding the glucose-fructose oxidoreductase) was deleted which is responsible for the synthesis of the osmolyte d-sorbitol (Sootsuwan et al. 2013). The cells became osmosensitive and the growth rates were reduced from 0.18 to 0.08 h−1 in sugar-rich media (Loos et al. 1994; de Barros and Celligoi 2006). The growth defect of Z. mobilis Δgfo was completely relieved either by complementation of the gfo gene or by the addition of the osmolyte under osmotic stress conditions (Sootsuwan et al. 2013). In almost the same manner, the addition of low amounts of mannitol or the complementation with gox1432 led to a recovery of normal growth parameters of the Δgox1432 mutant.
In contrast to the Δgox1432 strain, growth of the gox0849 deletion mutant was comparable to wild-type cells in high osmotic media. Interestingly, the deletion of gox0849 did not result in a complete loss of NADH-dependent d-fructose reduction, indicating the presence of another yet unknown NADH-specific d-fructose reductase with low activity. Evidence in favor of this assumption came from the analysis of the double deletion mutant strain (Δgox1432 Δgox0849) that revealed the same residual activity of NADH-dependent d-fructose reduction under stress and non-stress conditions. However, the unknown mannitol dehydrogenase could not completely restore normal growth of the double deletion mutant since the cultures revealed a long lag phase and reached only 50 % of the final optical density compared to the parental strain.
Sugisawa et al. (1991) deleted the orthologous gene of gox1432 in Gluconobacter melanogenus N44-1 and observed that the mutant strain was unable to grow in media containing 500 mM l-sorbose. Likewise, it was shown that in G. suboxydans IFO 3291, the ortholog of Gox1432 is mainly responsible for l-sorbose assimilation (Shinjoh et al. 2002). Furthermore, the enzyme also catalyzes 5-ketogluconate reduction as shown for G. oxydans DSM 3503 (Klasen 1994). Analogous to these results, we found that G. oxydans 621H Δgox1432 was not able to grow with l-sorbose and 5-ketogluconate as carbon and energy source, respectively (data not shown). Therefore, Gox1432 is the major factor for intracellular reduction of partially oxidized products that are formed by the membrane-bound polyol dehydrogenase (Gox0854–0855) (Fig. 7). When the major substrates (d-sorbitol or gluconate) are consumed, incompletely oxidized products (l-sorbose or 5-ketogluconate) are taken up by the cells and the reduced forms are channeled in the central metabolism. l-Sorbose is reduced to d-sorbitol that is converted into d-fructose and finally phosphorylated to fructose-6-phosphate (Richhardt et al. 2012). 5-Ketogluconate is reduced to gluconate and phosphorylated to 6-phosphogluconate by Gox1709. Both intermediates are then channeled into the oxidative pentose phosphate pathway. As shown here, Gox1432 has another important function because it plays a central role in the synthesis of mannitol as osmoprotectant in osmotically stressed G. oxydans (Fig. 7). Thus, we suggest that Gox1432 is the key player for survival in the sugar-rich natural habitats of the organism. This information is especially useful to optimize the strategies of engineering G. oxydans strains with improved osmotolerance for biotechnological applications.
References
Adachi O, Toyama H, Matsushita K (1999) Crystalline NADP-dependent D-mannitol dehydrogenase from Gluconobacter suboxydans. Biosci Biotechnol Biochem 63:402–407
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cayley S, Record MT Jr (2004) Large changes in cytoplasmic biopolymer concentration with osmolality indicate that macromolecular crowding may regulate protein–DNA interactions and growth rate in osmotically stressed Escherichia coli K-12. J Mol Recognit 17:488–496
Cayley S, Lewis BA, Record MT Jr (1992) Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J Bacteriol 174:1586–1595
Chomczynski P (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15(3):532–537
Claret C, Salmon JM, Romieu C, Bories A (1994) Physiology of Gluconobacter oxydans during dihydroxyacetone production from glycerol. Appl Microbiol Biotechnol 41:359–365
Condon C, FitzGerald RJ, O’Gara F (1991) Conjugation and heterologous gene expression in Gluconobacter oxydans ssp. suboxydans. FEMS Microbiol Lett 80:173–178
da Costa MS, Santos H, Galinski EA (1998) An overview of the role and diversity of compatible solutes in Bacteria and Archaea. Adv Biochem Eng Biotechnol 61:117–153
de Barros M, Celligoi MAPC (2006) Synthesis of sorbitol by Zymomonas mobilis under high osmotic pressure. Braz J Microbiol 37:324–328
De Ley J, Gillis M, Swings J (1984) The genus Gluconobacter. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams and Wilkins, Baltimore, pp. 267–278
De Muynck C, Pereira CSS, Naessens M, Parmentier S, Soetaert W, Vandamme EJ (2007) The genus Gluconobacter oxydans: comprehensive overview of biochemistry and biotechnological applications. Crit Rev Biotechnol 27:147–171
Deppenmeier U, Ehrenreich A (2009) Physiology of acetic acid bacteria in light of the genome sequence of Gluconobacter oxydans. J Mol Microbiol Biotechnol 16:69–80
Deppenmeier U, Hoffmeister M, Prust C (2002) Biochemistry and biotechnological applications of Gluconobacter strains. Appl Microbiol Biotechnol 5(9):1513–1533
Efiuvwevwere BJO, Gorris LGM, Smid EJ, Kets EPW (1999) Mannitol-enhanced survival of Lactococcus lactis subjected to drying. Appl Microbiol Biotechnol 51:100–104
Empadinhas N, da Costa MS (2008) Osmoadaptation mechanisms in prokaryotes: distribution of compatible solutes. Int Microbiol 11(3):151–161
Empadinhas N, da Costa MS (2009) Diversity, distribution and biosynthesis of compatible solutes in prokaryotes. Contrib Sci 5:95–105
Grant WD (2004) Life at low water activity. Philos Trans R Soc Lond Ser B Biol Sci 359:1249–1266
Hancock RD (2009) Recent patents on vitamin C: opportunities for crop improvement and single-step biological manufacture. Recent Pat Food Nutr Agric 1(1):39–49
Heuel H, Garakani AS, Turgut S, Lengeler JW (1998) Genes for D-arabinitol and ribitol catabolism from Klebsiella pneumoniae. Microbiology 144:1631–1639
Hocking AD (1988) Strategies for microbial growth at reduced water activities. Microbiol Sci 5:280–284
Hommel RK, Ahnert P (1999) Acetobacter. In: Encyclopedia of food microbiology, Vol. 1. Academic Press, pp 352–359
Hoshino T, Sugisawa T, Shinjoh M, Tomiyama N, Miyazaki T (2003) Membrane-bound D-sorbitol dehydrogenase of Gluconobacter suboxydans IFO 3255—enzymatic and genetic characterization. Biochim Biophys Acta 1647:278–288
Jennings DH (1984) Polyol metabolism in fungi. Adv Microb Physiol 25:149–193
Kallnik V, Meyer M, Deppenmeier U, Schweiger P (2010) Construction of expression vectors for protein production in Gluconobacter oxydans. J Biotechnol 150:460–465
Kavanagh KL, Klimacek M, Nidetzky B, Wilson DK (2002) Crystal structure of Pseudomonas fluorescens mannitol 2-dehydrogenase binary and ternary complexes: specificity and catalytic mechanism. J Biol Chem 277(45):43433–43442
Kets EPW, Galinski EA, De Witt M, De Bont JAM, Heipieper H (1996) Mannitol, a novel compatible solute in Pseudomonas putida S12. J Bacteriol 178:6665–6670
Klasen R (1994) Molecularbiologische Untersuchungen zur 5-Ketoglukonatbildung bei Gluconobacter oxydans. Thesis, Heinrich-Heine University of Duesseldorf, Germany
Kostner D, Peters B, Mientus M, Liebl W, Ehrenreich A (2013) Importance of codB for new codA-based markerless gene deletion in Gluconobacter strains. Appl Microbiol Biotechnol 97(18):8341–8349
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Loos H, Krämer RH, Sprenger GA (1994) Sorbitol promotes growth of Zymomonas mobilis in environments with high concentrations of sugar: evidence for a physiological function of glucose-fructose oxidoreductase in osmoprotection. J Bacteriol 176:7688–7693
Luchterhand B, Fischöder T, Grimm AR, Wewetzer S, Wunderlich M, Schlepütz T, Büchs J (2015) Quantifying the sensitivity of G. oxydans ATCC 621H and DSM 3504 to osmotic stress triggered by soluble buffers. J Ind Microbiol Biotechnol 42:585–600
Macauley S, McNeil B, Harvey LM (2001) The genus Gluconobacter and its applications in biotechnology. Crit Rev Biotechnol 21(1):1–25
Matsushita K, Fujii Y, Ano Y, Toyama H, Shinjoh M, Tomiyama N, Miyazaki T, Sugisawa T, Hoshino T, Adachi O (2003) 5-Keto-D-gluconate production is catalyzed by a quinoprotein glycerol dehydrogenase, major polyol dehydrogenase, in Gluconobacter species. Appl Environ Microbiol 69(4):1959–1966
Meyer M, Schweiger P, Deppenmeier U (2015) Succinic semialdehyde reductase Gox1801 from Gluconobacter oxydans in comparison to other succinic semialdehyde-reducing enzymes. Appl Microbiol Biotechnol 99(9):3929–3939
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor, Cold Spring Harbor Laboratory
Mott JF, Grant RA, Ho YS, Platt T (1985) Maximizing gene expression from plasmid vectors containing the λ PL promotor: strategies of overproducing termination factor ρ. Proc Natl Acad Sci U S A 82:88–92
Parmentier S, Beauprez J, Arnaut F, Soetaert W, Vandamme EJ (2005) Gluconobacter oxydans NAD-dependent, D-fructose reducing, polyol dehydrogenases activity: screening, medium optimisation and application for enzymatic polyol production. Biotechnol Lett 27:305–311
Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U (2005) Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat Biotechnol 23:195–200
Rauch B, Pahlke J, Schweiger P, Deppenmeier U (2010) Characterization of enzymes involved in the central metabolism of Gluconobacter oxydans. Appl Microbiol Biotechnol 88:711–718
Richhardt J, Bringer S, Bott M (2012) Mutational analysis of the pentose phosphate and Entner-Doudoroff pathways in Gluconobacter oxydans reveals improved growth of a Δedd Δeda mutant on mannitol. Appl Environ Microbiol 78:6975–6986
Roblin L, Urban M, Flicoteau D, Martin C, Pradeau D (2006) Topical treatment of experimental hydrofluoric acid skin burns by 2.5 % calcium gluconate. J Burn Care Res 27(6):889–894
Rossmann MG, Moras D, Olsen KW (1974) Chemical and biological evolution of a nucleotide-binding protein. Nature 250:194–199
Saha BC (2004) Purification and characterization of a novel mannitol dehydrogenase from Lactobacillus intermedius. Biotechnol Prog 20:537–542
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor, Cold Spring Harbor Laboratory
Sand M, Mingote AI, Santos H, Müller V, Averhoff B (2013) Mannitol, a compatible solute synthesized by Acinetobacter baylyi in a two-step mechanism including a salt-induced and salt-dependent mannitol-1-phosphate dehydrogenase. Environ Microbiol 15:2187–2197
Sand M, Rodrigues M, González JM, de Crécy-Lagard V, Santos H, Müller V, Averhoff B (2015) Mannitol-1-phosphate dehydrogenases/phosphatases: a family of novel bifunctional enzymes for bacterial adaptation to osmotic stress. Environ Microbiol 17:711–719
Schneider KH, Giffhorn F, Kaplan S (1993) Cloning, nucleotide sequence and characterization of the mannitol dehydrogenase gene from Rhodobacter sphaeroides. J Gen Microbiol 139:2475–2484
Shibata T, Ichikawa C, Matsuura M, Takata Y, Noguchi Y, Saito Y, Yamashita M (2000) Cloning of a gene for D-sorbitol dehydrogenase from Gluconobacter oxydans G624 and expression of the gene in Pseudomonas putida IFO3738. J Biosci Bioeng 89:463–468
Shinjoh M, Tazoe M, Hoshino T (2002) NADPH-dependent L-sorbose reductase is responsible for L-sorbose assimilation in Gluconobacter suboxydans IFO 3291. J Bacteriol 184:861–863
Sootsuwan K, Thanonkeo P, Keeratirakha N, Thanonkeo S, Jaisil P, Yamada M (2013) Sorbitol required for cell growth and ethanol production by Zymomonas mobilis under heat, ethanol, and osmotic stresses. Biotechnol Biofuels 6:180–193
Stahlberg A, Aman P, Ridell B, Mostad P, Kubista M (2003) Quantitative real-time PCR method for detection of B-lymphocyte monoclonality by comparison of κ and λ immunoglobulin light chain expression. Clin Chem 49(1):51–59
Sugisawa T, Hoshino T, Fujiwara A (1991) Purification and properties of NADPH-linked L-sorbose reductase from Gluconobacter melanogenus N44-1. Agric Biol Chem 55(8):2043–2049
Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354
Wisselink HW, Weusthuisa RA, Egginka G, Hugenholtza J, Grobbena GJ (2002) Mannitol production by lactic acid bacteria: a review. Int Dairy J 12:151–161
Wood JM (2015) Bacterial responses to osmotic challenges. J Gen Physiol 145(5):381–388
Zahid N, Schweiger P, Galinski E, Deppenmeier U (2015) Identification of mannitol as compatible solute in Gluconobacter oxydans. Appl Microbiol Biotechnol 99(13):5511–5521
Acknowledgments
Special thanks to the Higher Education Commission of Pakistan and their German collaborators DAAD for the financial support (scholarship for NZ). We thank Elisabeth Schwab and Marlene Hecker for the technical assistance as well as Sarah Refai, Anna Siemen, and Jana Hiltner for the critical proof reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical statement
The authors affirm that the said publication complies with obligations and norms of the ethical standards of research.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 10 kb)
Rights and permissions
About this article
Cite this article
Zahid, N., Deppenmeier, U. Role of mannitol dehydrogenases in osmoprotection of Gluconobacter oxydans . Appl Microbiol Biotechnol 100, 9967–9978 (2016). https://doi.org/10.1007/s00253-016-7680-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7680-8