Abstract
Biological conversion of sulfide, acetate, and nitrate to, respectively, elemental sulfur (S0), carbon dioxide, and nitrogen-containing gas (such as N2) at NaCl concentration of 35–70 g/L was achieved in an expanded granular sludge bed (EGSB) reactor. A C/N ratio of 1:1 was noted to achieve high sulfide removal and S0 conversion rate at high salinity. The extracellular polymeric substance (EPS) quantities were increased with NaCl concentration, being 11.4-mg/g volatile-suspended solids at 70 mg/L NaCl. The denitrifying sulfide removal (DSR) consortium incorporated Thauera sp. and Halomonas sp. as the heterotrophs and Azoarcus sp. being the autotrophs at high salinity condition. Halomonas sp. correlates with the enhanced DSR performance at high salinity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
High-salinity wastewaters with excess nitrogen and sulfur compounds are produced in pharmaceutical, textile dyeing, and chemical industries (Liu et al. 2014; Teixeira et al. 2013). NaCl is usually one of the main salinity of these wastewaters with concentration of up to 70 g/L (Ng et al. 2015; Shi et al. 2014; Ng et al. 2014; Wang et al. 2009a). Biological processes are cost-effective and environmentally friendly for the removal of nitrogen or sulfur pollutants from waters (Li et al. 2009; Mahmood et al. 2009; Wang et al. 2009b). Certain chemolithotrophic denitrifiers, such as Thiobacillus denitrificans, can use reduced sulfur compounds as electron donors and oxidized nitrogen compounds as electron acceptors for denitrification reactions (Beristain-Cardoso et al. 2009; Lee et al. 2014; Wong and Lee 2011). The simultaneous removal of sulfide, nitrate, and acetate was achieved via denitrifying sulfide removal (DSR) process in the presence of both autotrophic and heterotrophic denitrifiers in the reactor (Reyes-Avila et al. 2004; Chen et al. 2008, 2009, 2010).
High salinity is inhibitory to activities of microorganisms and hence limits efficiency of biological wastewater treatment systems (Rinzema et al. 1988). Halophilic inocula were used to partially counteract these problems (Lefebvre et al. 2007; Morgan et al. 1990; Omil et al. 1995; Kargi and Uygur 1997). Liu et al. (2015) reported that the DSR sludge can tolerate 2–35 g/L of NaCl at a C/N ratio of 3:1 with high S0 conversion rate. The DSR reactors have to push to a higher NaCl concentration range, at least up to 70 g/L, to meet the practical needs.
This study for the first time operated using the DSR reactor up to 70 g/L of NaCl with acclimatization of halophilic inocula with denitrification activities. The performance of the DSR reactor and the associated extracellular polymeric substances (EPS) in the DSR sludge was investigated at different salinities. Shifts in microbial communities of the DSR process with high salinity wastewaters were reported.
Materials and methods
Reactor and synthetic wastewater
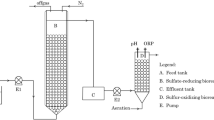
The utilized expanded granular sludge bed (EGSB) reactor in this study was modified by Liu et al (2015) and made by Plexiglas, which was 50 mm in diameter and 85 cm in height giving a working volume of 1.25 L. Waste sludge of 0.55 L was inoculated into the reactor to make the initial volatile suspended solid (VSS) concentration of 3.64 g/L. Reactor initial temperature was kept at 30±1 °C. Sludge was collected as inoculum from an anaerobic sludge thickener at the Nibuwan Municipal Wastewater Treatment Plant, Qingdao, China. The sludge was screened using a 0.2-mm Tyler mesh to remove most grids prior to use. Synthetic wastewater contained (per liter) the following: 200 mg S2−; 87.5 mg N-NO3 −; 75 mg C as acetate; and 1500 mg NaHCO3. The synthetic wastewater contained a sodium concentration of 985 mg/L with pH adjusted to 7.5. All the other details on the reactor setup as well as seed sludge are available in Liu et al. (2015).
Microbial population
Sludge samples were collected from the EGSB reactor and were immediately stored at −80 °C. Total genomic DNA was extracted in duplicate from each sample using the PowerSoil DNA Isolation Kit (MO BIO, Carlsbad, CA, USA) according to the manufacturer’s instructions. The extract was subsequently pooled to reduce sample variability. The quality of the DNA extracted was examined by 1 % (w/v) agarose gel electrophoresis, and the concentration was measured with a UV–Vis spectrophotometer (NanoDrop 2000, USA). The V3–V4 region of the 16S rRNA gene was amplified using bacterial primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′), with the reverse primer containing a 6-bp barcode used to tag each sample. PCR amplification was performed in triplicate for each sample using a reaction mixture (20 μL), which contains 4 μL of 5× PCR buffer, 10 ng of template DNA, 0.2 μM of each primer, 0.25 mM of each dNTP, and 1 U Fast Pfu polymerase (TransGen, China). The PCR procedure included an initial denaturation step at 95 °C for 2 min, subsequent 30 cycles of temperature gradient at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and finally an extension step at 72 °C for 5 min in a GeneAmp 9700 thermal cycler (ABI, USA). The triplicate amplicons were pooled together, electrophoresed on a 2 % (w/v) agarose gel, and recovered using an AxyPrep DNA Gel Extraction Kit (Axygen, USA).
The purified amplicon was quantified using a QuantiFluor-ST Fluorometer (Promega, USA), and then a composite sequencing library was constructed by combining equimolar ratios of amplicons from all samples. The resulting library for paired-end sequencing (2 × 250 bp) was analyzed on an Illumina MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China). Raw sequence data of this study have been deposited to the NCBI Sequence Read Archive with accession No. SRP063542.
Chemical analysis
After liquid samples were filtered with 0.45-μm membrane filters, the concentrations of nitrate, nitrite, sulfate, and thiosulfate in the samples were measured by ion chromatography (ICS-3000; Dionex, USA) equipped with a 4-mm IonOac AG4AAS4A-SC analytical column with carbonate/bicarbonate eluent (1.8 mmol/dm3 Na2CO3/1.7 mmol/L NaHCO3 at 1 cm3/min), and by sulfuric regeneration (25 mmol/L H2SO4 at 5 cm3/min). The sulfide concentration was determined using the methylene blue method (Truper and Schlegel 1964). Sulfide measured in this study includes all species in the liquid sample, i.e., dissolved H2S, HS−, and S2−. Other sulfur compounds apart from sulfide, sulfate, and thiosulfate were generally unstable and are neglected in this work. Hence, the daily average quantities of sulfide, sulfate, and thiosulfate in the influent and effluent streams were utilized to estimate the amount of S0 produced in the reactor (Chen et al. 2008). Acetate concentration was determined using an Agilent 6890 Gas Chromatograph (USA). The pH was measured using a pHS-25 meter (Shanghai, China).
The concentrations of suspended solids (SS) and VSS were measured according to Standard Methods (APHA 1998). Twenty milliliters of sludge from the reactor was centrifuged at 3200 rpm for 30 min. The supernatant was separated and kept as soluble EPS. The carbohydrate and protein in soluble EPS were also estimated. Carbohydrate measurements were acquired using the anthrone method with a glucose standard protein levels which were measured with the modified Lowry method using bovine serum albumin as a standard (Wu et al. 2009).
Results
Effects of salinity on DSR performance
The reactor was set up without NaCl addition for 26 days and was then operated at an incrementally increased influent NaCl concentration from 35 to 70 g/L (Table 1). Removals of nitrate, sulfide, and acetate were all nearly 100 % with no nitrite accumulation since day 2 (Fig. 1). About 120 mg-S/L of sulfate was noted in the effluent (Fig. 1b), with insoluble elemental sulfur (S0) solids being presented in the reactor. In this control test, about 40 % of fed sulfide was converted to S0 (Fig. 1c).
At 35 g/L of NaCl (stage II), the effluent sulfate was decreased to 90 mg-S/L, and the removal efficiencies of sulfide, acetate, and nitrate remained high (>95 %). The associated S0 conversion efficiency has experienced minor increase to 55 %. Little nitrite was accumulated in this stage.
With 50 g/L of influent NaCl (stage III(a)), the effluent sulfate was further decreased to 30 mg-S/L. Meanwhile, the corresponding nitrate removal efficiency was dropped to 87 %, although the removal efficiencies of sulfide and acetate were remained nearly perfect. The S0 conversion efficiency was increased to 85 %. About 1.5 mg-N/L of nitrite accumulation was observed in the effluent. After an increase in influent acetate from 75 to 225 mg C/L to make C/N ratio of 3:1 mol/mol at stage III(b), the effluent sulfate was decreased to 14 mg-S/L. The corresponding removal efficiencies of nitrate and acetate were at 87 and 100 %, respectively, while the corresponding sulfide removal was dropped to only 46.7 %. The S0 conversion efficiency maintained at 85 % at this stage. Effluent nitrite accumulation was decreased from 1.5 to 0.5 mg-N/L. In order to maintain high sulfide removal efficiency, influent acetate was decreased from 225 mg to 75 C/L to make C/N ratio of 1:1 mol/mol at stage III(c); the sulfide removal efficiency was increased back to 99 %. The corresponding removal efficiencies of nitrate and acetate were kept at 87 and 100 %, respectively. The effluent sulfate was increased from 15 to 30 mg-S/L. The S0 conversion efficiency maintained around 85 %, and the accumulation of nitrite was negligible.
At stage IV, the influent acetate concentration was maintained at 75 mg-C/L and the influent NaCl was increased to 70 g/L; removals of sulfide, nitrate, and acetate were at 87, 99, and 100 %, respectively. The corresponding accumulation of nitrite and sulfate was negligible, and S0 conversion efficiency was 100 %.
EPS at different NaCl concentrations
The compositions of the soluble EPS (proteins and polysaccharides) of the DSR sludge at different salinities were listed in Table 2. The quantities of soluble EPS increased with influent NaCl concentration and was 11.4 mg/g VSS at 70 kg/L NaCl. The proteins in EPS were increased markedly while polysaccharides in EPS which showed only minor increment. The increase in EPS quantities was proposed as a counteractive response of the biomass to the high sodium toxicity (Vyrides and Stuckey 2009).
Microbial communities at different NaCl concentrations
The phylogenetic classification of bacterial 16S rRNA sequence from the sludge samples at 0–70 g/L salinity was illustrated based on phylum in Fig. 2 and on genus in Fig. 3. The bacterial sequences affiliated with Proteobacteria were the most abundant, followed by the sequences affiliated with Firmicutes, Actinobacteria, and Chloroflexi, with Bacteroidetes being less dominated as influent NaCl concentration was increased.
At genus level, significant amounts of sequences could not be classified (18.2, 6.8, 2.7, and 3.8 % at 0, 35, 50, and 70 g/L salinity, respectively, indicating the presence of many taxa-unknown bacteria in the reactor (Fig. 3). Phylum sequences affiliated with Thauera, Halomonas, Azoarcus, Caldilineaceae_unclutured, Peptostreptococcaceae_incertae_sedis, Candidate_division_TM7_norank, Pseudomonas, Macellibacteroides, Blastocatella, Arcobacter, Anoxynatronum, Sulfurimonas, Citrobacter, Desulfuromusa, Sulfurovum were detected. The genera Thauera, Halomonas, and Pseudomonas were the hetetrophic denitrifiers. The dominant genus Thauera was 21.2 % after setup without NaCl and increased to 27.1 % with the influent NaCl concentration increasing to 35 g/L. Further increase of the influent NaCl concentrations to 50 and to 70 g/L reduced the abundance of Thauera to 8.3 and to 12.9 %, respectively. Abundance of Pseudomonas went a slight increase from 0.74 to 2.2 % and then to 3.6 % as NaCl concentration was increased from 0 to 35 g/L and then to 50 g/L. At 70 g/L NaCl, the abundance of Pseudomonas sharply decreased to 0.29 %, demonstrating its poor resistance to this level of salinity. Conversely, the abundance of Halomonas was significantly increased from 0.002 to 40.1 % at 70 g/L NaCl. The OTUs most closely reacted to Azoarcus, Arcobacter, Sulfurimonas, and Sulfurovum, the common autotrophic denitrifiers in DSR sludge, changed diversely with the influent NaCl concentration increasing from 0 to 70 g/L. The abundance of Azoarcus was increased from 0.60 to 39.5 % with NaCl concentration being increased from 0 to 50 g/L. Further increasing the influent NaCl concentrations to 70 g/L resulted in the decrease in Azoarcus abundance to 3.5 %. The maximum abundances of Arcobacter, Sulfurimonas, and Sulfurovum were increased over a range of 0–50 g/L NaCl. At 70 g/L NaCl, these stains were almost completely wiped out from the DSR reactor.
Discussions
The above experimental findings suggest that the DSR process can satisfactorily handle high-salinity wastewater with up to 70 g/L NaCl with the adaption of its microbial community with acclimation of halophilic inocula and production of excess EPS to resist salinity for the incorporated cells.
At 35–70 g/L NaCl (stages II, III(c), and IV), S0 conversion efficiency was enhanced to 100 % at the end, suggesting that high salinity was favorable for S0 conversion.
Arcobacter sp. is the dominant autotrophic denitrifier in the startup stage with no NaCl addition (Table 3). The genus Thauera was the main hetetrophic denitrifier at <35 g/L NaCl, while the genus Halomonas was the main hetetrophic denitrifier at >35 g/L NaCl. The genera Arcobacter, Sulfurovum, Azoarcus, and Sulfurimonas were the predominant autotrophic denitrifiers at NaCl concentration of 0–50 g/L, while at 70 g/L NaCl, only Azoarcus was left with an abundance of only 3.52 %. The high DSR performance achieved was proposed to depend on the balanced growth of both heterotrophic denitrifiers and autotrophic denitrifiers (Chen et al. 2008). The present study revealed an overwhelming growth of heterotrophic denitrifiers over authotrotrophic counterparts. The salinity likely depressed the activity of heterotrophic denitrifier but had minimal effects on the activity of autotrophic denitrifier, hence leading to increased S0 conversion at high salinity. Some species belonging to autotrophic denitrifiers, such as Sulfurimonas, can best grow on 95 mM NaCl (Bruckner et al. 2013) while the activity of heterotrophic denitrifier Halomonas sp. was inhibited at higher NaCl concentration (Mormile et al. 1999) .
Liu et al. (2015) noted that a C/N ratio of 3:1 is optimal for 2–35 g/L NaCl for maximum S0 conversion. In the present study, at higher salinity, the optimal C/N ratio is shifted to 1:1 (Fig. 1). At low salinity the strains adopted to fresh water environment may need excess carbon sources to survive at intermediate salinity (say, 35 g/L NaCl) environment (Liu et al. 2015). The incorporation of Halomonas sp. in the DSR sludge, which can remove nitrate effectively at high salinity conditions (Peyton et al. 2001), making the optimal C/N ratio of 1:1, the same as that proposed by Reyes-Avila et al. (2004) with no NaCl addition.
The results of this study indicated that the DSR process with Halomonas sp. could be used to treat salinity wastewater at NaCl concentration of 35–70 g/L.
References
APHA (1998) Standard methods for the examination of water and wastewater, 20th edition. American Public Health Association, Washington, DC
Beristain-Cardoso R, Texier AC, Sierra-Alvarea R, Razo-Flore EJ, Field A, Gomez J (2009) Effect of initial sulfide concentration on sulfide and phenol oxidation under denitrifying conditions. Chemosphere 74:200–205
Bruckner CG, Mammitzsch K, Jost G, Wendt J, Labrenz M, Jürgens K (2013) Chemolithoautotrophic denitrification of epsilonproteobacteria in marine pelagic redox gradients. Environ Microbiol 15(5):1505–1513
Chen C, Wang AJ, Ren NQ, Kan HJ, Lee DJ (2008) Biological breakdown of denitrifying sulfide removal process in high-rate expanded granular bed reactor. Appl Microbiol Biotechnol 81(4):765–770
Chen C, Wang AJ, Ren NQ, Lee DJ, Lai JY (2009) High-rate denitrifying sulfide removal process in expanded granular sludge bed reactor. Bioresour Technol 100(7):2316–2319
Chen C, Ren NQ, Wang AJ, Liu LH, Lee DJ (2010) Functional consortium for denitrifying sulfide removal process. Appl Microbiol Biotechnol 86(1):353–358
Kargi F, Uygur A (1997) Biological treatment of saline wastewater in a rotating biodisc contactor by using halophilic organisms. Bioprocess Eng 17(2):81–85
Lee DJ, Wong BT, Adav SS (2014) Azoarcus taiwanensis sp. nov., a denitrifying species isolated from a hot spring. Appl Microbiol Biotechnol 98(3):1301–1307
Lefebvre O, Quentin S, Torrijos M, Godon JJ, Delgenes JP, Moletta R (2007) Impact of increasing NaCl concentrations on the performance and community composition of two anaerobic reactors. Appl Microbiol Biotechnol 75(1):61–69
Li W, Zhao QL, Liu H (2009) Sulfide removal by simultaneous autotrophic and heterotrophic desulfurization-denitrification process. J Hazard Mater 162(2–3):848–853
Liu M, Liu T, Peng Y, Wang S, Xiao H (2014) Effect of salinity on N2O production during shortcut biological nitrogen removal from landfill leachate. J Biosci Bioeng 117(5):582–590
Liu C, Zhao C, Wang A, Guo Y, Lee DJ (2015) Denitrifying sulfide removal process on high-salinity wastewaters. Appl Microbiol Biotechnol 99(15):6463–6469
Mahmood Q, Hu BL, Cai J, Zheng P, Azim MR, Jilani G, Islam E (2009) Isolation of Ochrobactrum sp. QZ2 from sulfide and nitrite treatment system. J Hazard Mater 165(1-3):558–565
Morgan JW, Forster CF, Evison L (1990) A comparative study of the nature of biopolymers extracted from anaerobic and activated sludges. Water Res 24(6):743–750
Mormile MR, Romine MF, Garcia MT, Ventosa A, Bailey TJ, Peyton BM (1999) Halomonas campisalis sp. nov., a denitrifying, moderately haloalkaliphilic bacterium. Syst Appl Microbiol 22(4):551–8
Ng KK, Shi X, Ng HY (2015) Evaluation of system performance and microbial communities of a bioaugmented anaerobic membrane bioreactor treating pharmaceutical wastewater. Water Res 81(12):311–324
Ng KK, Shi X, Tang M, Ng H (2014) A novel application of anaerobic bio-entrapped membrane reactor for the treatment of chemical synthesis-based pharmaceutical wastewater. Sep Purif Technol 132(1-2):634–643
Omil F, Mendez R, Lema JM (1995) Anaerobic treatment of saline wastewaters under high sulphide and ammonia content. Bioresour Technol 54(3):269–278
Peyton BM, Mormile MR, Petersen JN (2001) Nitrate reduction with Halomonas campisalis: kinetics of denitrification at pH 9 and 12.5% NaCl. Water Res 35(17):4237–4242
Reyes-Avila JS, Razo-Flores E, Gomez J (2004) Simultaneous biological removal of nitrogen, carbon and sulfur by denitrification. Water Res 38(14–15):3313–3321
Rinzema A, Vanlier J, Lettinga G (1988) Sodium inhibition of acetoclastic methanogens in granular sludge from a UASB reactor. Enzyme Microb Technol 10(1):24–32
Shi X, Lefebvre O, Ng K, Ng H (2014) Sequential anaerobic-aerobic treatment of pharmaceutical wastewater with high salinity. Bioresour Technol 153(3):79–86
Teixeira C, Magalhaes C, Joye SB, Bordalo AA (2013) The role of salinity in shaping dissolved inorganic nitrogen and N2O dynamics in estuarine sediment waterInterface. Mar Pollut Bull 66(1-2):225–229
Truper H G, Schlegel H G (1964) Sulfur metabolism of Thiormodaceae. I. Quantitative measurements on growing cells of Chromatium okenii Antonie Van Leeuwenhoek 30: 225-238
Vyrides I, Stuckey DC (2009) Adaptation of anaerobic biomass to saline conditions: role of compatible solutes and extracellular polysaccharides. Enzyme Microb Tech 44(1):46–51
Wang AJ, Liu CS, Han HJ, Ren NQ, Lee DJ (2009a) Modeling denitrifying sulfide removal process using artificial neural networks. J Hazard Mater 168(2–3):1274–1279
Wang C, Chou W, Kuo Y, Chang F (2009b) Paired removal of color and COD from textile dyeing wastewater by simultaneous anodic and indirect cathodic oxidation. J Hazard Mater 169(1-3):16–22
Wong B, Lee DJ (2011) Denitrifying sulfide removal and carbon methanogenesis in a mesophilic, methanogenic culture. Bioresour Technol 102(12):6673–6679
Wu J, Zhou HM, Li HZ, Zhang PC, Jiang J (2009) Impacts of hydrodynamic shear force on nucleation of flocculent sludge in anaerobic reactor. Water Res 43(12):3029–3036
Acknowledgments
Funding
The research was supported by the National Natural Science Foundation of China under Grant No. 21307160, Natural Science Foundation of Shandong Province under Grant No. ZR2013EEQ030.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Liu, C., Zhao, D., Ma, W. et al. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp.. Appl Microbiol Biotechnol 100, 1421–1426 (2016). https://doi.org/10.1007/s00253-015-7039-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-7039-6