Abstract
An agar-degrading bacterium, strain LGH, was isolated and identified as Cohnella sp. This strain had a capability of utilizing agar as a sole carbon source for growth and showed a strong agarolytic activity. A novel endo-type β-agarase gene agaW, encoding a primary translation product of 891 amino acids, including a 26 amino acid signal peptide, was cloned and identified from a genomic library of strain LGH. The AgaW belonged to the glycoside hydrolase (GH) GH50 family, with less than 39 % amino acid sequence similarity with any known protein, and hydrolyzed agarose into neoagarotetraose as the major end product and neoagarobiose as the minor end product through other neoagarooligosaccharide intermediates, such as neoagarohexaose.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Agar is an important matrix polysaccharide found in the cell walls of some red algae (Rhodophyceae), including Gelidium and Gracilaria, and is mainly composed of a hybrid structure of alternating 3-O-linked β-d-galactose and 4-O-linked α-L-galactose (Chi et al. 2012; Knutsen et al. 1994). The galactan backbones are often masked by ester sulfate groups, methyl groups, or pyruvic acid acetal groups, thereby increasing the number of possible agarobiose structures (Lahaye et al.1989; Usov 1998; van de Velde et al. 2002). Among the agar polysaccharides, agarose is mainly composed of two alternating heterogeneous galactan residues of 3-O-linked β-d-galactose and 4-O-linked 3,6-anhydro-α-L-galactose (Chi et al. 2012; Duckworth and Yaphe 1972). Because agarose is an uncharged polysaccharide, with well-known gelling and stabilizing characteristics, it has been commonly used in microbiological media and also as food additives used in pharmaceutical products and cosmetic applications (Araki 1959). More recently, biomass, such as terrestrial and marine biomass, has become a potential renewable resource to replace fossil resources for mankind in the near future. The red algae are good biomass resource candidates due to their easy cultivation in a broad range of oceans and rapid growth (Park et al. 2015). For efficient utilization of red algae, they must be completely hydrolyzed for saccharification of agar biomass.
The microbial enzymatic activities have shed light on the saccharification of agar biomass, especially of agarose biomass. Several distinct agar-degrading bacteria have been identified and characterized from seawater, marine sediments, and other environments (Chi et al. 2012; Fu and Kim 2010). In most agar-degrading bacteria, agarose is generally believed to be cleaved by agarases. Agarases are mainly classified into α-agarase (EC 3.2.1.158) and β-agarase (EC 3.2.1.81) according to their distinct cleavage pattern. α-Agarases cleave the α-1,3 linkages of agarose to produce agarooligosaccharides of series related to agarobiose, with 3,6-anhydro-l-galactose residues at their reducing ends, while β-agarases cleave β-1,4 linkages to produce neoagarooligosaccharides of series related to neoagarobiose, with d-galactose residues at their reducing ends (Duckworth and Turvey 1969). In some bacteria, neoagarobiose is then hydrolyzed into two monosaccharide units of 3,6-anhydro-l-galactose and d-galactose by a neoagarobiose hydrolase or a neoagarooligosaccharide hydrolase (Ha et al. 2012; Hehemann et al. 2012; Sugano et al. 1994a). In the agarolytic pathway in marine bacterium Vibrio sp. strain EJY3, 3,6-anhydro-l-galactose can be further catabolized to produce bioavailable 2-keto-3-deoxygalactonate through 3,6-anhydrogalactonate intermediate by an NADP+-dependent 3,6-anhydro-l-galactose dehydrogenase and an 3,6-anhydrogalactonate cycloisomerase, respectively (Yun et al. 2015). Escherichia coli harboring the genes of these two enzymes is capable of utilizing 3,6-anhydro-l-galactose as a sole carbon source for growth (Yun et al. 2015). These agarolytic enzymes can be used for the enzymatic saccharification of agarose biomass into bioavailable monosaccharides, which can be further converted into biofuels or industrial chemicals.
Currently, more than 60 agarases have been characterized and deposited in the CAZy database (http://www.cazy.org/Glycoside-Hydrolases.html). On the basis of amino acid sequence similarity, these agarases are at least classified into five distinct families of glycoside hydrolases (GHs) (Chi et al. 2012; Dong et al. 2006; Fu and Kim 2010; Henrissat 1991; Ma et al. 2007; Xie et al. 2013). Only two α-agarases have been reported in bacteria, including Alteromonas agarilytica GJ1B (Flament et al. 2007) and Thalassotalea agarivorans JAMB-A33 (Hatada et al. 2006), and belong to the GH96 family. In contrast, β-agarase-producing bacteria have been reported in several taxonomically diverse genera and are found in four distinct GH families of GH16, GH50, GH86, and GH118 (Chi et al. 2012; Dong et al. 2006; Fu and Kim 2010; Henrissat 1991; Ma et al. 2007; Xie et al. 2013). Of these, the GH16 family is the largest family, composed of more than 3880 functionally heterogeneous members, including endogalactosidases, endoglucanases, κ-carrageenases, lichenases, and xyloglucanases etc. (http://www.cazy.org/Glycoside-Hydrolases.html). The GH50, GH86, and GH118 families, however, consist solely of β-agarases and presently have only 217, 49, and 8 members, respectively (http://www.cazy.org/Glycoside-Hydrolases.html).
So far, most of the agar-degrading bacteria are aquatic isolates (Chi et al. 2012; Fu and Kim 2010). Agarolytic activity has also been rarely observed in terrestrial bacteria, such as Paenibacillus (Hosoda et al. 2003), Pseudomonas (Song et al. 2015), and Streptomyces (Temuujin et al. 2012). Here, we reported the isolation of a Gram-positive soil bacterium, Cohnella sp. LGH, which can degrade agar and utilize it as a sole carbon source for growth. A novel endo-type β-agarase gene agaW was cloned from Cohnella sp. LGH and expressed in Escherichia coli. The AgaW belonged to the GH50 family, with less than 39 % amino acid sequence similarity with any known protein and had a capability of hydrolyzing agarose into neoagarobiose and neoagarotetraose as two end products through other neoagarooligosaccharide intermediates, such as neoagarohexaose. The biochemical properties of the recombinant AgaW were also characterized in this investigation.
Materials and methods
Chemicals
Agarose and a series of neoagarooligosaccharides were purchased from Qingdio BZ Oligo Biotech Co., Ltd., China. Other chemical reagents used in this study were all of analytical grade and obtained from Shanghai Sangon Biological Engineering Technology & Service Co., Ltd., China. All enzymes and kits necessary for DNA manipulation were purchased from Takara Biotechnology (Dalian) Co., Ltd., China. The ultra-15 centrifugal filter unit with ultracel-5 regenerated cellulose membrane (5-kDa cutoff size) was purchased from Amicon.
Isolation and identification of an agar-degrading bacterium
Soil samples, collected from a farmland in Nanjing, China, were diluted in distilled water and spread onto mineral salt medium containing 0.1 % NaCl, 0.1 % NH4NO3, 0.15 % K2HPO4, 0.05 % KH2PO4, 0.01 % CaCl2, 0.01 % MgSO4, and 1.5 % agar (pH 7.0–7.5) (Sambrook and Russell 2001). After incubation at 30 °C for 72 h, grown colonies that formed obvious depressions on agar plates were isolated and purified, and each colony was detected by Lugol’s iodine solution (5 % iodine and 10 % KI) staining to confirm the agarolytic activity by clearing zones. One Gram-positive promising colony, designated LGH, was selected for further investigation. This strain was then identified by morphological, physiological, and biochemical characteristics and 16S rRNA gene sequence analysis and deposited in China General Microbiological Culture Collection Center (CGMCC) under accession number CGMCC 10018. Alignment of the 16S rRNA gene sequence was performed by ClustalX 1.8.3 (Thompson et al. 1997) with default settings. Phylogenesis was analyzed by MEGA version 5.2 Software. Distances were calculated using the Kimura two-parameter distance model. Unrooted trees were built by the neighbor-joining method. The dataset was bootstrapped 1000 times (Weisburg et al. 1991).
Six polysaccharide utilization assays
Strain LGH was grown at 30 °C in LB medium plus 2 % sucrose with shaking overnight (optical density at 600 nm (OD600) of 1.0). Cells were centrifuged for 5 min at 12,000×g at 4 °C, washed three times with distilled water, and then transferred at 2 % (v/v) to mineral salt medium (Sambrook and Russell 2001) plus 0.2 % each polysaccharide as a sole carbon source. Each flask was incubated at 30 °C with shaking for 5 days, and the OD600 was recorded as a representation of microbial growth using a UV-2000 UV–vis spectrophotometer (Shimadzu, Kyoto, Japan). In this assay, the six polysaccharides were carrageenan, chitin, carboxymethyl cellulose (CMC), agarose, soluble starch, and xylan.
Cloning of the agarase gene and sequence analysis
Genomic DNA of strain LGH was extracted by the method of high-salt precipitation (Miller et al. 1988), and the restriction enzyme Sau3AI was employed to obtain randomly digested chromosomal fragments. These 4- to 8-kb fragments were recovered by a DNA purification kit (TaKaRa) and ligated into pUC118 previously digested with BamHI and treated with calf intestinal alkaline phosphatase (TaKaRa). The ligation product was then transformed into E. coli DH5α cells. The resulting bacterial suspension was spread onto LB agar medium plus 100 μg ml−1 of ampicillin and incubated at 37 °C for 24 h or more. Grown colonies were picked and purified from those having formed obvious depressions on agar plates. The agarolytic activity was further tested by Lugol’s iodine solution analysis as described above.
The inserted fragment in the transformant was sequenced at Takara Biotechnology Co. Ltd. Nucleotide and deduced amino acid sequence analyses were performed using Omiga software. BlastN and BlastP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and pairwise sequence alignment (http://www.ebi.ac.uk/Tools/psa/) were used for the nucleotide sequence and deduced amino acid identity and similarity searches, respectively.
Expression and purification of the recombinant AgaW
The agaW gene fragment without the 78-bp signal peptide-encoding region was amplified by PCR using LGH genomic DNA as a template, and the primer pairs P F to which was added a KpnI site (underlined) (5′-CGG GGT ACC GCC ACC CCG TTC CCT ACT TTG AAC TTC-3′) and P R to which was added a HindIII site (underlined) (5′-CCC AAG CTT CTT TGA TAT TAG CAA ATG ATC CAT TAT AAA-3′). The amplified fragment of 2595 bp was then ligated into pET29a, and the resulting plasmid pET29a-agaW was transformed into E. coli BL21 (DE3) pLysS cells. An obtained transformant clone was grown at 37 °C on LB plus 50 μg ml−1 of kanamycin to an OD600 of 0.5, at which 0.1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce the gene expression. After induction at 15 °C for 18 h, cells were harvested, resuspended in 10 mM potassium phosphate buffer (pH 7.5) and disrupted by sonication. After centrifugation at 15,000×g for 30 min, the supernatant was collected and further purified by a 2-ml volume of NTA-Ni2+ agarose (Sangon) at 4 °C. His-tagged target protein was allowed to bind to the resin in 10 mM potassium phosphate buffer (pH 7.5) plus 0.5 M sodium chloride and 10 mM imidazole and then was eluted by 10 mM potassium phosphate buffer (pH 7.5) plus 0.5 M sodium chloride and 500 mM imidazole. The purified enzyme was concentrated with an ultra-15 centrifugal filter unit and stored in 10 mM potassium phosphate buffer (pH 7.5) plus 1 mM 2-mercaptoethanol and 10 % glycerol at 4 °C. The protein concentration was quantified by the protein assay kit (Shanghai Sangon Biological Engineering Technology & Service Co., Ltd., China) with bovine serum albumin as a standard.
Determination of molecular mass and isoelectric point of the recombinant AgaW
The molecular mass of denatured recombinant AgaW was estimated by 0.1 % sodium dodecyl sulfate-10 % polyacrylamide gel electrophoresis (SDS-PAGE) using a broad-range molecular weight protein standard. Proteins were visualized after Coomassie brilliant blue R-250 staining. The exact native molecular mass of recombinant AgaW was determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDITOF-MS). The isoelectric point (pI) of recombinant AgaW was estimated by gel isoelectric focusing (IEF) using a precast Ampholine PAGplate (Amersham Bioscience, Uppsala, Sweden) and IEF standards (Amersham Bioscience).
Determination of the agarolytic activity of the recombinant AgaW
The agarolytic activity was measured by the 3,5-dinitrosalicylic acid (DNS) method as previously reported (Chi et al. 2014; Temuujin et al. 2011), with slight modification. Briefly, 10 μl of the enzyme solution was mixed with 390 μl of 10 mM potassium phosphate buffer (pH 7.0) plus 0.5 % agarose and incubated at 50 °C for 10 min. The reaction mixture was mixed with 400 μl of DNS reagent solution containing (per liter) 6.5 g of DNS, 325 ml of 2 M NaOH, and 45 ml of glycerol and heated in a boiling water bath for 5 min. After cooling to room temperature, the absorbance of the mixture was measured spectrophotometrically at 540 nm to determine the produced reducing sugar. As blank control, 10 μl of the distilled water was used instead of the enzyme solution; other steps were the same as above. One activity unit (U) was defined as the amount of enzyme required to produce 1 μmol of reducing oligosaccharides or monosaccharides per minute at 50 °C. For the preparation of the standard curve, galactose was used as the reference reducing monosaccharide.
Characterization of biochemical properties of the recombinant AgaW
The optimum temperature for the agarolytic activity was determined in 10 mM potassium phosphate buffer (pH 7.0) at temperatures ranging from 30 to 70 °C at an interval of 5 °C. The optimum pH was determined at 50 °C in 10 mM buffer solutions at pH ranging from 3.0 to 12.0 at an interval of 1.0: Na2HPO4 citrate buffer of pH 3.0 to 5.0, potassium phosphate buffer of pH 6.0–7.0, boric acid-borax buffer of pH 8.0 to 9.0, and glycine-NaOH buffer of pH 10.0 to 12.0. The temperature stability of recombinant AgaW was determined in 10 mM potassium phosphate buffer (pH 7.0) at 50 °C after preincubation of enzyme at temperatures ranging from 30 to 70 °C for 1 h. The pH stability was determined also in 10 mM potassium phosphate buffer (pH 7.0) at 50 °C after preincubation of enzyme at pH ranging from 3.0 to 12.0 at 30 °C for 1 h. The effects of potential inhibitors or activators on the agarolytic activity were determined at 50 °C in 10 mM potassium phosphate buffer (pH 7.0) containing various metal ions and chemical reagents at a final concentration of 10 mM. The relative activity was defined as a percentage of the activity obtained in the absence of an additive.
The kinetic parameters of the recombinant AgaW were determined in 10 mM potassium phosphate buffer (pH 7.0) by addition of enzyme at a final concentration of 62.5 μg ml−1 with agarose as substrate at a final concentration of 0.5 to 10 mg ml−1. The reaction was performed at 50 °C for 10 min, and the agarolytic activity was measured as described above. At least three independent determinations were performed for each kinetic constant. The substrate-free assay system was also used as blank simultaneously. Kinetic parameters were calculated from nonlinear regression data analysis against various substrate concentrations using SigmaPlot Version 8.0 software (SPSS Inc.).
The substrate specificity of the recombinant AgaW was measured by using two artificial chromogenic substrates, p-nitrophenyl-α-d-galactopyranoside, and p-nitrophenyl-β-d-galactopyranoside, respectively. Briefly, 20 μg of the recombinant AgaW was incubated in 700 μl of 10 mM potassium phosphate buffer (pH 7.0) with each substrate at a final concentration of 3 mg ml−1 at 50 °C for 2 h. The reaction was stopped by addition of 500 μl of 1 M Na2CO3. The activity of the enzyme was measured spectrophotometrically at 420 nm by evaluating the release of p-nitrophenol from the hydrolysis of the artificial chromogenic substrate.
Measurement of viscosity
The kinematic viscosity of the recombinant AgaW was determined using the Ubbelohde viscometer (Shanghai Sangon Biological Engineering Technology & Service Co., Ltd., China). Ten micrograms of the recombinant AgaW was mixed in 15 ml of 10 mM potassium phosphate buffer (pH 7.0) with 0.5 % agarose as substrate. The reaction was incubated at 50 °C at various reaction times ranging from 0 to 120 min, and the efflux time was measured at each time. The experiment was repeated three times.
Structural analysis of the enzymatic product
The recombinant AgaW was incubated at 50 °C in 10 mM potassium phosphate buffer (pH 7.0) with 0.5 % agarose, neoagarobiose, neoagarotetraose, or neoagarohexaose as substrate, respectively. Aliquots were applied to a Silica Gel 60 thin-layer chromatography (TLC) plate and developed with an n-butanol-acetic acid-water solution (1:2:1, by volume) for analysis of the hydrolytic products of agarose or an n-butanol-ethanol-water solution (3:3:1, by volume) for analysis of the hydrolytic products of neoagarobiose, neoagarotetraose, and neoagarohexaose. The oligosaccharide spots that developed were visualized by spraying with 10 % (by volume) H2SO4 in ethanol and heating at 100 °C for 10 min. For mass analysis, the spots corresponding to the hydrolyzed products were scraped out and dissolved in methanol for drying. The molecular mass distribution of the products was determined using an Agilent 6410B triple-quadrupole mass spectrometer (Agilent, USA) equipped with electrospray ionization (ESI) under negative-ion ionization conditions. The ESI-MS conditions were optimized as follows: drying gas temperature, 350 °C; drying gas flow (nitrogen), 10 L min−1; nebulizer gas pressure (nitrogen), 30 psi; and capillary voltage, 4000 V. Negative ions were acquired in full scan mode in the range of m/z 100–1000 molecular mass units for identification within a 1-s scan time interval.
For nuclear magnetic resonance (NMR) analysis, adequate enzyme was incubated at 50 °C in 10 mM potassium phosphate buffer (pH 7.0) with 0.5 % agarose for at least 24 h. The hydrolyzate was centrifuged at 15,000×g for 15 min, and the supernatant was filtered through an ultra-15 centrifugal filter unit. The filtrate was lyophilized, then collected as the oligosaccharide products, and finally dissolved in D2O. Carbon-13 nuclear magnetic resonance (13C NMR) spectrum was recorded using the NMR spectrometer (400 MHz, DRX-400; Bruker, USA).
Nucleotide sequence accession numbers
The nucleotide sequences of the 16S rDNA and agaW genes of Cohnella sp. strain LGH were deposited in the GenBank database under accession numbers KP099053 and KR296705, respectively.
Results
Isolation and identification of an agar-degrading bacterium
One promising colony, with a capability of forming obvious depression on the mineral salt medium agar plate, was picked and purified through the isolation procedure. This strain, designated LGH, was capable of utilizing agar as a sole carbon source for growth and showed strong agarolytic activity through Lugol’s iodine solution staining (Fig. 1a). LGH was Gram-positive, aerobic, rod shaped, and motile by means of peritrichous flagella (Fig. 1b) strain. Catalase, gelatinase, oxidase, and milk coagulation were positive. Nitrate reduction and urease were negative. Polysaccharides such as agarose, carrageenan, soluble starch, and xylan were assimilated, but chitin and CMC were not.
Identification of an agar-degrading strain LGH. a Detection of agarolytic activity on an agar plate by Lugol’s iodine solution staining. b A typical field of Cohnella sp. strain LGH (×40,000). The scale bar represents 1.0 μm. c Phylogenetic tree illustrating the 16S rDNA gene similarity of LGH to strains exhibiting highest sequence similarity (RDP analysis and FASTA)
The 16S rRNA gene sequence of strain LGH showed 99 % identity to C. phaseoli strain GSPC1 and 97 % identities to other typical strains of the genus Cohnella, such as C. luojiensis strain HY-22R, C. boryungensis strain BR-29, C. hongkongensis strain HKU3, and C. cellulosilytica strain FCN3-3. Based on the results of morphological and physiological characteristics, and phylogenetic tree analysis based on 16S rRNA gene sequence (Fig. 1c), LGH was identified as a Cohnella sp. strain.
Cloning and sequence analysis of agaW
In order to investigate the agar-degrading molecular mechanism, the genomic library of Cohnella sp. LGH was constructed for identifying the open reading frame (ORF) corresponding to the agarase gene. Several positive transformants, having formed obvious depressions on agar plates, were screened from approximately 15,000 transformants. These clones showed strong agarolytic activities through Lugol’s iodine solution staining, while the negative control, E. coli DH5α harboring only pUC118, did not at all (data not shown). The sequences analysis (Microbial Genome Annotation Tools Glimmer 3.0 (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi)) showed that the inserted fragments in these transformants all contained a putative ORF. This ORF, designated agaW, was subcloned into the linear vector pMD18-T (TaKaRa) and the resulting plasmid was transformed into E. coli DH5α. Its encoding protein AgaW was then confirmed to be the target agarase.
The agaW was 2676 nucleotides in length, with TTG (not ATG) start and TAA stop codons, and encoded a protein of 891 amino acids. Promoter prediction (http://www.fruitfly.org/seq_tools/promoter.html) revealed that there was a promoter-like region located at position 145 to 96 bp upstream from the start codon. A potential ribosome-binding site, GGGAGG, similar to that found in E. coli, was found 10 bp upstream from the start codon. Signal peptide prediction (SignalP 4.1 server, http://www.cbs.dtu.dk/services/SignalP/) revealed that there was a potential signal sequence of 26 amino acid residues at its amino-terminal end (Figure S1). Comparative sequence analyses using the BLAST program showed that the deduced amino acid sequence of agaW had no obviously overall sequence similarity to any characterized agarase, only sharing at most 26.7 % identity and 38.5 % similarity to a GH50 agarase HZ2 (GenBank accession No. ADY17919) from Agarivorans sp. strain HZ105 (Lin et al. 2012), 26.6 % identity and 38.3 % similarity to a GH50 agarase AgaB (GenBank accession No. BAA04744) from Vibrio sp. strain JT0107 (Sugano et al. 1994b), 24.8 % identity and 36.9 % similarity to a GH50 agarase AgaA (GenBank accession No. BAA03541) also from strain JT0107 (Sugano et al. 1993), and 25.3 % identity and 36.0 % similarity to a GH50 agarase Aga41A (GenBank accession No. ADM25828) from Vibrio sp. strain CN41 (Liao et al. 2011). Although no typical conserved domains were detected in the deduced amino acid sequence of agaW, a partially conserved catalytic module of GH50 family, extending for at least 375 amino acid residues (Ekborg et al. 2006; Fu and Kim 2010), was present at positions approximately 278 to 704 in its carboxy-terminal end (Figure S1). On the other hand, four regions at positions 207 to 257, 258 to 301, 362 to 726, and 430 to 602 (Figure S1) were well conserved, respectively, with 58.8, 77.3, 59.2, and 54.9 % similarities to the homologous regions, annotated as domain 2, domain 3 (containing two regions), and a conserved domain of the GH42 family, of a known crystal structure GH50 agarase Aga50D (GenBank accession No. ABD81904) from Saccharophagus degradans 2–40 (Pluvinage et al. 2013; Weiner et al. 2008). Therefore, AgaW appeared to be classified into the GH50 family.
Expression and purification of the recombinant AgaW
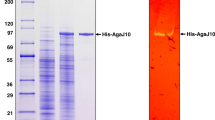
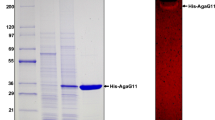
The agaW gene fragment, encoding a mature form of the protein without the signal peptide sequence (Leu1-Ala26), was expressed as an intracellular protein in E. coli BL21 (DE3) pLysS with or without IPTG, and total proteins were analyzed by SDS-PAGE. Only one induced protein corresponding to 97 kDa was observed (Fig. 2), which closed well to the calculated recombinant AgaW molecular mass of 97,979 Da. The His-tagged recombinant enzyme was then purified from the crude extract using NTA-Ni2+ agarose affinity chromatography. The SDS-PAGE analysis showed that only one single 97-kDa protein band was observed with 0.5 mg loading quantity of protein sample (Fig. 2).
SDS-PAGE analysis of recombinant AgaW protein produced in E. coli BL21 (DE3) pLysS harboring plasmid pET29a-agaW. Lane 1, protein molecular marker with the sizes 200, 116, 97.2, 66.4, 44.3, 29, and 20.1 kDa; lane 2, total proteins of E. coli BL21 (DE3) pLysS harboring plasmid pET29a-agaW without IPTG induction; lane 3, total proteins of E. coli BL21 (DE3) pLysS harboring plasmid pET29a-agaW with IPTG induction at 15 °C for 18 h; lane 4, 0.5 mg of purified recombinant AgaW protein after being purified by NTA-Ni2+ agarose
Biochemical characterization of the recombinant AgaW
The exact native molecular mass of the recombinant AgaW was determined to be 97.684 Da by MALDITOF-MS. The molecular mass of the denatured recombinant AgaW was determined to be 97 kDa by SDS-PAGE. These results indicated that the recombinant AgaW was a monomeric protein. The pI of the recombinant AgaW was estimated to be 5.1 by gel IEF.
The recombinant AgaW exhibited agarolytic activity at wide temperatures ranging from 30 to 70 °C, with its maximum activity at 50 °C. Although it sustained more than 80 % of maximum activity at temperatures ranging from 40 to 55 °C, its relative activity began to drop rapidly at temperatures beyond 55 °C, with only 20 % of its maximum activity at 70 °C (Fig. 3a). The enzyme was thermostable at temperatures below 50 °C and retained at least 95 % of initial activity at 50 °C over a 60-min period, but its stability dropped dramatically at temperatures beyond 50 °C, with only 50 % of its initial activity at 55 °C (Fig. 3a). The recombinant AgaW also exhibited a wide pH profile, showing maximum agarolytic activity at pH 7.0 and retaining more than 90 % of maximum activity at pH ranging from 6.0 to 10.0 (Fig. 3b). The enzyme was stable at pH ranging from 6.0 to 10.0, and retained at least 90 % of initial activity over a 60-min period, but its stability dropped obviously at pH beyond 5.0 and 10.0, with only 38 % of its initial activity at pH 12.0, and close to inactivation at pH 3.0 (Fig. 3b). The agarolytic activity of the recombinant AgaW was completely inhibited by Cu2+ and surfactant SDS and partially inhibited by Mn2+, Ba2+, Zn2+, Fe3+, and EDTA, with 86.8 ± 0.02, 67.06 ± 0.02, 45.6 ± 0.04, 12.48 ± 0.02, and 89.88 ± 0.06 % of relative activity, respectively. On the other hand, the enzymatic activity was able to be enhanced by Ca2+, Na+, K+, Mg2+, and DTT, with 144.1 ± 0.01, 130.26 ± 0.1, 121.61 ± 0.03, 120.73 ± 0.02, and 142.42 ± 0.15 % of relative activity, respectively. No significant activation or inhibition was observed by urea. The K m, V max, k cat, and k cat/K m values of the recombinant AgaW for agarose as substrate were 3.43 × 10−4 M (3.43 mg ml−1), 387.11 U mg−1, 2.5 × 104 s−1, and 7.3 × 107 M−1 s−1, respectively.
Effect of temperature and pH on agarolytic activity of the recombinant AgaW. a Effect of temperature on agarolytic activity of the recombinant AgaW. Filled circle with solid line, optimum temperature; open circle with dash line, thermostability. b Effect of pH on agarolytic activity of the recombinant AgaW. Filled triangle with solid line, optimum pH; open triangle with dash line, pH stability
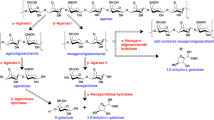
Mode of action
The agarose hydrolytic products by the recombinant AgaW were separated and detected by TLC with the standard compounds neoagarohexaose, neoagarotetraose, and neoagarobiose (Fig. 4a). Short-chain oligosaccharides were not detectable until the first 15 min. One spot, sharing the same traveling distance as neoagarotetraose, was clearly detected after 10 min. Its amount gradually increased in a time-dependent manner and finally became the major product. This product had two strong quasimolecule ions at m/z of 629.0 and m/z of 665.5, represented [M-H]− and [M + Cl]−, respectively, corresponding to (neo)agarotetraose (Fig. 4e). Another spot, sharing the same traveling distance as neoagarohexaose, was obviously detected after 10 min as well, but gradually decreased after 60 min, and finally disappeared. This product had a weak quasimolecule ion at m/z of 935.0, represented [M-H]− and a strong quasimolecule ion at m/z of 971.5, represented [M + Cl]−, corresponding to (neo)agarohexaose (Fig. 4f). With the decrease of (neo)agarohexaose, one spot, sharing the same traveling distance as neoagarobiose, began to present and remarkably increased in a time-dependent manner. This product had a strong quasimolecule ion at m/z of 359.5, represented [M + Cl]−, corresponding to (neo)agarobiose (Fig. 4d).
Analysis of the mode of action of the recombinant AgaW. a The agarolytic products were taken out at the indicated intervals and analyzed by TLC. S, neoagarobiose, neoagarotetraose, and neoagarohexaose. b The recombinant AgaW hydrolyzed neoagarohexaose into neoagarobiose and neoagarotetraose as two end products. c Changes in the viscosity of the recombinant AgaW reaction mixture. d Mass spectrum of one spot corresponding to neoagarobiose. e Mass spectrum of one spot corresponding to neoagarotetraose. f Mass spectrum of one spot corresponding to neoagarohexaose
The anomeric configuration of agarolytic products was examined by 13C NMR (Fig. 5). The chemical shifts of the anomeric carbon signals in the spectrum were identical to those of a typical pattern for neoagarooligosaccharides (Fu et al. 2008; Rochas et al. 1986; Temuujin et al. 2012).The resonances at 92.360 and 96.182 ppm were signals for the α- and β-anomeric forms of the galactose unit at the reducing end of the neoagarooligosaccharides, respectively. No resonance was detected at 90.8 ppm, which was the characteristic signal of the reducing end of the hydrated form of 3,6-anhydro-l-galactose of the agarooligosaccharides (Fu et al. 2008; Rochas et al. 1994; Temuujin et al. 2012). Thus, the dimer, tetramer, and hexamer agarolytic products were identified as neoagarobiose, neoagarotetraose, and neoagarohexaose, respectively. These results also demonstrated that the recombinant AgaW was a β-agarase.
TLC analysis showed that only two spots corresponding to neoagarobiose and neoagarotetraose were detected from the reaction products even after incubation of long time, such as 1440 min (Fig. 4a), suggesting that the two products cannot be recognized by the recombinant AgaW. When we incubated excess enzyme with the standard neoagarobiose, neoagarotetraose, and neoagarohexaose as substrates, respectively, neoagarobiose and neoagarotetraose were not hydrolyzed as expected, while neoagarohexaose were completely hydrolyzed into neoagarobiose and neoagarotetraose (Fig. 4b). This observation clearly demonstrated that the recombinant AgaW hydrolyzed agarose into neoagarotetraose and neoagarobiose as two end products through other neoagarooligosaccharide intermediates, such as neoagarohexaose.
To investigate the hydrolysis pattern of the recombinant AgaW, the viscosity changes of the enzymic reaction system was measured. During the period of incubation, the viscosity began to drop rapidly within the first 10 min, then continued to drop at a very slow rate from 10 to 20 min, and finally became constant (Fig. 4c). At the same time, TLC analysis showed that short-chain oligosaccharides were not detectable within the first 10 min (Fig. 4a), in spite of the dramatically dropped viscosity during this time. This result suggested that the rapid drop of viscosity was caused by random endocleavage of agarose and accumulation of long-chain neoagaroligosaccharides. This hydrolysis pattern indicated that the recombinant AgaW had an endolytic activity.
Discussion
In present study, a novel β-agarase AgaW was identified and characterized from Cohnella sp. LGH and classified into the GH50 family according to the analysis of its homologous regions and the partially conserved catalytic module. Biochemical characterization of a few GH50 agarases reveals that they have exolytic or both endolytic and exolytic activities, and the vast majority of them produce neoagarobiose as a major or end product from neooligosaccharide intermediates, such as neoagarotetraose and neoagarohexaose (Fu et al. 2008; Lee et al. 2006; Ohta et al. 2005; Sugano et al. 1993) or direct from agarose (Kim et al. 2010). Just only one GH50 agarase, AgaACN41, from Vibrio sp. strain CN41, produces neoagarotetraose as the sole end product (Liao et al. 2011). However, AgaW just presented an endolytic activity and produced neoagarotetraose as the major end product and neoagarobiose as the minor end product, which were uncommon in all characterized GH50 agarases.
Recently, two artificial chromogenic substrates, p-nitrophenyl-α-d-galactopyranoside and p-nitrophenyl-β-D-galactopyranoside, have been reported to be utilized for preliminary determination of the mode of action of agarases. Two GH16 β-agarases AgaG1 from Alteromonas sp. GNUM-1 (Chi et al. 2014) and AgaH71 from Pseudoalteromonas hodoensis H7 (Park et al. 2015) exhibited strong hydrolytic activity toward p-nitrophenyl-β-d-galactopyranoside but negligible activity toward p-nitrophenyl-α-d-galactopyranoside, indicating that they recognized the β-linkage but not α-linkage. The similar recognition mode was believed to be present in the hydrolysis of agarose as well. This hypothesis was further confirmed by 13C NMR analysis of the anomeric configuration of their agarolytic products (Chi et al. 2014; Park et al. 2015). In present study, we also utilized p-nitrophenyl-α-d-galactopyranoside and p-nitrophenyl-β-d-galactopyranoside to determine the mode of action of AgaW firstly. Unexpectedly, AgaW exhibited a strong hydrolytic activity toward both of two artificial substrates (Fig. 6), indicating that it recognized not only the β-linkage but also the α-linkage. On the other hand, the structure information of the agarolytic products provided by 13C NMR demonstrated that AgaW was only able to hydrolyze the β-1,4 linkages of agarose, but not the α-1,3 linkages. Thus, our results indicated that the linkage recognition mode of agarases toward two artificial chromogenic substrates was not wholly identical to that toward agarose.
Neoagarooligosaccharides as the major agarolytic products have potentially high economic values due to their physiological and biological activities without any toxicity. Using β-agarases to produce the neoagarooligosaccharides is becoming a tendency for its high efficiency and nonpollution. However, industrial application requires agarases with high activities and stabilities at temperatures higher than the gelling temperature of agar (1.5 % agar solution solidifies at 43 °C), which limits the use of agarases. AgaW exhibited a wide temperatures and pH stability profile, with an optimum temperature of 50 °C, which was higher than the agarose gelling temperature. Therefore, AgaW was believed to have potential applications to produce neoagarooligosaccharides in the industry.
Cohnella sp. strain LGH had an ability to utilize agarose as a sole carbon source for growth, which suggested that the agarolytic products of neoagarobiose and neoagarotetraose were able to be further catabolized in this bacterium. This hypothesis was further confirmed by the growth of strain LGH in liquid mineral salt medium using the neoagarobiose and neoagarotetraose mixture as the sole carbon source (data not shown). The result suggested that there must be other enzyme(s), such as a neoagarobiose hydrolase or a neoagarooligosaccharide hydrolase, to catalyze the further steps in the agarose catabolism as well.
References
Araki C (1959) Seaweed polysaccharides. In: Wolfrom ML (ed) Carbohydrate chemistry of substances of biological interest. Pergamon Press, London, pp. 15–30
Chi WJ, Chang YK, Hong SK (2012) Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol 94:917–930
Chi WJ, Park da Y, Seo YB, Chang YK, Lee SY, Hong SK (2014) Cloning, expression, and biochemical characterization of a novel GH16 β-agarase AgaG1 from Alteromonas sp. GNUM-1. Appl Microbiol Biotechnol 98:4545–4555
Dong J, Hashikawa S, Konishi T, Tamaru Y, Araki T (2006) Cloning of the novel gene encoding beta-agarase C from a marine bacterium, Vibrio sp. strain PO-303, and characterization of the gene product. Appl Environ Microbiol 72:6399–6401
Duckworth M, Turvey JR (1969) The action of a bacterial agarase on agarose, porphyran and alkali-treated porphyran. Biochem J 113:687–692
Duckworth M, Yaphe W (1972) The relationship between structures and biological properties of agars. In: Nisizawa K (ed) Proceedings of the 7th international seaweed symposium. Halstead Press, New York, pp. 15–22
Ekborg NA, Taylor LE, Longmire AG, Henrissat B, Weiner RM, Hutcheson SW (2006) Genomic and proteomic analyses of the agarolytic system expressed by Saccharophagus degradans 2–40. Appl Environ Microbiol 72:3396–3405
Flament D, Barbeyron T, Jam M, Potin P, Czjzek M, Kloareg B, Michel G (2007) Alpha-agarases define a new family of glycoside hydrolases, distinct from beta-agarase families. Appl Environ Microbiol 73:4691–4694
Fu XT, Kim SM (2010) Agarase: review of major sources, categories, purification method, enzyme characteristics and applications. Mar Drugs 8:200–218
Fu XT, Lin H, Kim SM (2008) Purification and characterization of a novel beta-agarase, AgaA34, from Agarivorans albus YKW-34. Appl Microbiol Biotechnol 78:265–273
Ha SC, Lee S, Lee J, Kim HT, Ko HJ, Kim KH, Choi IG (2012) Crystal structure of a key enzyme in the agarolytic pathway, α-neoagarobiose hydrolase from Saccharophagus degradans 2–40. Biochem Biophys Res Commun 412:238–244
Hatada Y, Ohta Y, Horikoshi K (2006) Hyperproduction and application of alpha-agarase to enzymatic enhancement of antioxidant activity of porphyran. J Agric Food Chem 54:9895–9900
Hehemann JH, Smyth L, Yadav A, Vocadlo DJ, Boraston AB (2012) Analysis of keystone enzyme in agar hydrolysis provides insight into the degradation of a polysaccharide from red seaweeds. J Biol Chem 287:13985–13995
Henrissat B (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280:309–316
Hosoda A, Sakai M, Kanazawa S (2003) Isolation and characterization of agar-degrading Paenibacillus spp. associated with the rhizosphere of spinach. Biosci Biotechnol Biochem 67:1048–1055
Kim HT, Lee S, Lee D, Kim HS, Bang WG, Kim KH, Choi IG (2010) Overexpression and molecular characterization of Aga50D from Saccharophagus degradans 2–40: an exo-type beta-agarase producing neoagarobiose. Appl Microbiol Biotechnol 86:227–234
Knutsen SH, Myslabodski DE, Larsen B, Usov AI (1994) A modified system of nomenclature for red algal galactans. Bot Mar 37:163–169
Lahaye M, Yaphe W, Viet MTP, Rochas C (1989) 13C NMR spectroscopic investigation of methylated and charged agarose oligosaccharides and polysaccharides. Carbohydrate Res 190:249–265
Lee DG, Park GT, Kim NY, Lee EJ, Jang MK, Shin YG, Park GS, Kim TM, Lee JH, Lee JH, Kim SJ, Lee SH (2006) Cloning, expression, and characterization of a glycoside hydrolase family 50 β-agarase from a marine Agarivorans isolate. Biotechnol Lett 28:1925–1932
Liao L, Xu XW, Jiang XW, Cao Y, Yi N, Huo YY, Wu YH, Zhu XF, Zhang XQ, Wu M (2011) Cloning, expression, and characterization of a new beta-agarase from Vibrio sp. strain CN41. Appl Environ Microbiol 77:7077–7079
Lin B, Lu G, Zheng Y, Xie W, Li S, Hu Z (2012) Gene cloning, expression and characterization of a neoagarotetraose-producing β-agarase from the marine bacterium Agarivorans sp. HZ105. World J Microbiol Biotechnol 28:1691–1697
Ma C, Lu X, Shi C, Li J, Gu Y, Ma Y, Chu Y, Han F, Gong Q, Yu W (2007) Molecular cloning and characterization of a novel beta-agarase, AgaB, from marine Pseudoalteromonas sp. CY24. J Biol Chem 282:3747–3754
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Ohta Y, Hatada Y, Ito S, Horikoshi K (2005) High-level expression of a neoagarobiose-producing β-agarase gene from Agarivorans sp. JAMB-AII in Bacillus subtilis and enzymic properties of the recombinant enzyme. Biotechnol Appl Biochem 41:183–191
Park da Y, Chi WJ, Park JS, Chang YK, Hong SK (2015) Cloning, expression, and biochemical characterization of a GH16 β-agarase AgaH71 from Pseudoalteromonas hodoensis H7. Appl Biochem Biotechnol 175:733–747
Pluvinage B, Hehemann JH, Boraston AB (2013) Substrate recognition and hydrolysis by a family 50 exo-β-agarase, Aga50D, from the marine bacterium Saccharophagus degradans. J Biol Chem 288:28078–28088
Rochas C, Lahaye M, Yaphe W, Viet MTP (1986) 13C-N.M.R.-spectroscopic investigation of agarose oligomers. Carbohydr Res 148:199–207
Rochas C, Potin P, Kloareg B (1994) NMR spectroscopic investigation of agarose oligomers produced by an alpha-agarase. Carbohydr Res 253:69–77
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Song T, Zhang W, Wei C, Jiang T, Xu H, Cao Y, Cao Y, Qiao D (2015) Isolation and characterization of agar-degrading endophytic bacteria from plants. Curr Microbiol 70:275–281
Sugano Y, Matsumoto T, Kodama H, Noma M (1993) Cloning and sequencing of agaA, a unique agarose 0107 gene from a marine bacterium, Vibrio sp. strain JT0107. Appl Environ Microbiol 59:3750–3756
Sugano Y, Kodama H, Terada I, Yamazaki Y, Noma M (1994a) Purification and characterization of a novel enzyme, α-neoagrooligosaccharide hydrolase (α-NAOS hydrolase), from a marine bacterium, Vibrio sp. strain JT0107. J Bacteriol 176:6812–6818
Sugano Y, Matsumoto T, Noma M (1994b) Sequence analysis of the agaB gene encoding a new beta-agarase from Vibrio sp. strain JT0107. Biochim Biophys Acta 1218:105–108
Temuujin U, ChiWJ LSY, Chang YK, Hong SK (2011) Overexpression and biochemical characterization of DagA from Streptomyces coelicolorA3(2): an endo-type β-agarase producing neoagarotetraose and neoagarohexaose. Appl Microbiol Biotechnol 92:749–759
Temuujin U, Chi WJ, Chang YK, Hong SK (2012) Identification and biochemical characterization of Sco3487 from Streptomyces coelicolor A3(2), an exo- and endo-type β-agarase-producing neoagarobiose. J Bacteriol 194:142–149
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Usov AI (1998) Structural analysis of red seaweed galactans of agar and carrageenan groups. Food Hydrocoll 12:301–308
van de Velde F, Knutsen SH, Usov AI, Rollema HS, Cerezo AS (2002) 1H and 13C high resolution NMR spectroscopy of carrageenans: application in research and industry. Trends Food Sci Technol 13:73–92
Weiner RM, Taylor 2nd LE, Henrissat B, Hauser L, Land M, Coutinho PM, Rancurel C, Saunders EH, Longmire AG, Zhang H, Bayer EA, Gilbert HJ, Larimer F, Zhulin IB, Ekborg NA, Lamed R, Richardson PM, Borovok I, Hutcheson S (2008) Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2–40 T. PLoS Genet 4:e1000087
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Xie W, Lin B, Zhou Z, Lu G, Lun J, Xia C, Li S, Hu Z (2013) Characterization of a novel β-agarase from an agar-degrading bacterium Catenovulum sp. X3. Appl Microbiol Biotechnol 97:4907–4915
Yun EJ, Lee S, Kim HT, Pelton JG, Kim S, Ko HJ, Choi IG, Kim KH (2015) The novel catabolic pathway of 3,6-anhydro-L-galactose, the main component of red macroalgae, in a marine bacterium. Environ Microbiol 17:1677–1688
Acknowledgments
This study was funded by the Fundamental Research Funds for the Central Universities of The People’s Republic of China (Project nos. KYZ201409 and KJQN201517) and the National Natural Science Foundation (Project nos. 31470551, 41401347, and 41401254).
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
This article does not contain any studies with human participants performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 217 kb)
Rights and permissions
About this article
Cite this article
Li, G., Sun, M., Wu, J. et al. Identification and biochemical characterization of a novel endo-type β-agarase AgaW from Cohnella sp. strain LGH. Appl Microbiol Biotechnol 99, 10019–10029 (2015). https://doi.org/10.1007/s00253-015-6869-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6869-6