Abstract
Ammonia oxidation is performed by both ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB). Few studies compared the adaptability of AOA and AOB for oxygenated/hypoxic alternant conditions in water-level-fluctuating zones. Here, using qPCR and 454 high-throughput sequencing of functional amoA genes of AOA and AOB, we examined the changes of abundances, diversities, and community structures of AOA and AOB in periodically flooded soils compared to the non-flooded soils in Three Gorges Reservoir. The increased AOA operational taxonomic unit (OTU) numbers and the higher ratios of abundance (AOA:AOB) in the periodically flooded soils suggested AOA have better adaptability for oxygenated/hypoxic alternant conditions in the water-level-fluctuating zones in the Three Gorges Reservoir and probably responsible for the ammonia oxidation there. Canonical correspondence analysis (CCA) showed that oxidation-reduction potential (ORP) had the most significant effect on the community distribution of AOA (p < 0.01). Pearson analysis also indicated that ORP was the most important factor influencing the abundances and diversities of ammonia-oxidizing microbes. ORP was significantly negatively correlated with AOA OTU numbers (p < 0.05), ratio of OTU numbers (AOA:AOB) (p < 0.01), and ratio of amoA gene abundances (AOA:AOB) (p < 0.05). ORP was also significantly positively correlated with AOB abundance (p < 0.05).
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrification, a two-step process in the oxidation of ammonia to nitrate, begins with the aerobic oxidation of ammonia to nitrite (Gorman-Lewis et al. 2014). Betaproteobacteria and Gammaproteobacteria were long thought to be the only two clades to perform ammonia oxidation (Prosser and Nicol 2008). However, the theory of ammonia oxidation changed significantly since the first ammonia-oxidizing archaea (AOA) Nitrosopumilus maritimus was discovered (Konneke et al. 2005). Since then, several other strains of AOA were enriched or isolated from various ecosystems (de la Torre et al. 2008; Hatzenpichler et al. 2008; Blainey et al. 2011; Laura et al. 2011). So far, a series of environmental factors such as ammonia concentration (Verhamme et al. 2011), oxygen concentration (Molina et al. 2010), and pH (Gubry-Rangin et al. 2011) seemed to influence the relative contributions of AOA and ammonia-oxidizing bacteria (AOB) and lead to the niche segregation of the two kinds of ammonia-oxidizing microbes.
Ammonia as an electron donor and oxygen as an electron acceptor were necessary for AOA and AOB to perform ammonia oxidation. However, the affinities of AOA and AOB for the two substrates were quite different. The half-saturation constant (K m = 133 nM total ammonium) of Nitrosopumilus maritimus SCM1 is much lower than that of Nitrosomonas europaea (K m = 553 μM) (Martens-Habbena et al. 2009). The other strains of AOA enriched from soils, such as Candidatus Nitrososphaera sp. JG1 and Candidatus Nitrosoarchaeum koreensis MY1, also have very low half-saturation constant for ammonia (2.15 and 0.69 μM) (Jung et al. 2011; Kim et al. 2012). That is the reason why AOA has competitive advantages over AOB in the oligotrophic environments like open seas or in soils with relative low ammonia concentrations (Beman et al. 2008; Verhamme et al. 2011). As for oxygen, AOA also have competetive advantages over AOB. The K m values for oxygen of Nitrosopumilus maritimus SCM1, Candidatus Nitrososphaera sp. JG1, and Candidatus Nitrosoarchaeum koreensis MY1 were 3.90, 4.67, and 10.38 μM, separately (Martens-Habbena et al. 2009; Jung et al. 2011; Kim et al. 2012). AOB have relative lower affinities for oxygen. The K m value for oxygen of Nitrosomonas oligotropha NL7 and Nitrosomonas europaea C-31 is 76.3 and 183.3 μM, respectively (Park and Noguera 2007; Park et al. 2010). The fact that AOA could live better in hypoxic environments was also demonstrated by a series of ecological studies. Nitrosopumilus maritimus SCM1 that dominated the transcriptome in 80-m depths suggested a substantial role for archaeal nitrification in the upper OMZ (Stewart et al. 2012). In the Arabian Sea, both thaumarchaeotal 16S rDNA and amoA gene abundances and gene expression had peaks in the upper OMZ transition zone (200 m) where oxygen concentrations were around 5 μM (Pitcher et al. 2011). In the Black Sea, thaumarchaeotal amoA expression could explain nearly 80 % of the nitrite variations in the lower oxic zone (Lam et al. 2007). In soil ecosystem, the AOA amoA genes and the ratio of AOA/AOB increased along with the soil depth (Leininger et al. 2006).
The Three Gorges Reservoir was completely built in 2009 in southwestern part of China to produce electric power. The area of the zone is 348.93 km2, and a total of 22 cities or counties were included. It is being operated between a high water level of 175 m and a low water level of 145 m. As a result, the water-level-fluctuating zone of the reservoir has an annual fluctuation up to 30 m and the soils in the zones are half-year inundated and half-year exposed (Fang et al. 2011). All the nitrogen transformation processes (including nitrification, anammox, denitrification, DAMO) and the fluxes of nitrogen cycle in these zones might be quite specific since the physicochemical properties (moisture, oxygen concentration, ammonia, nitrite, nitrate, etc.) of the soil would change a lot even during a single year. To date, no survey focusing on the ammonia-oxidizing microorganisms in the water-level-fluctuating zone in Three Gorges Reservoir has been performed. So, the primary objectives of the present study are as follows: (1) to investigate the abundances, diversities, and communities of AOA and AOB in the water-level-fluctuating zones in the Three Gorges Reservoir, finding the most important environmental factors affecting the abundances, diversities, and communities of ammonia-oxidizing microbes; (2) to explore whether AOA or AOB has better adaptability to live in the water-level-fluctuating zones in the Three Gorges Reservoir and is responsible for the ammonia oxidation there.
Materials and methods
Sampling sites description, soil collection, and physicochemical properties analysis
The soil samples were collected from the water-level-fluctuating zones in Three Gorges Reservoir. Sampling sites from two counties (Kaixian and Yunyang) (Fig. 1) were located in the main stream of Yangtze River, with Baijia creek from Kaixian county and Yanglu lake from Yunyang county. Three Gorges Reservoir has a high water level of 175 m from October to April and has a low water level of 145 m in the rest of the year, so the water-lever-fluctuating zones about 30 m were formed in these areas (Zhang et al. 2012). The four periodically flooded soil samples (PXH3 and PXH9 from Baijia creek; PXH5 and PXH11 from Yanglu lake, Table 1) from water-level-fluctuating zones were collected in April in the year 2010 and 2011 immediately after the water level went down to 145 m, and the soils were exposed to air. The four soil samples (PXH3, PXH5, PXH9, and PXH11) were flooded by the impoundment for more than 6 months with a water depth about 30 m. The other four non-flooded soil samples (PXH2 and PXH8 from Baijia creek; PXH4 and PXH10 from Yanglu lake, Table 1) over the 175-m water-level line were collected as control. At each sampling site, five soil samples (5 cm diameter and 5 cm depth) were collected within an area of 20 m2 and were mixed together. Each of the eight samples was sectioned into two subsamples: one was stored at 4 °C for the soil physicochemical analysis, and the other one was frozen at −80 °C for the molecular analysis. The latitude and longitude, pH, temperature, oxidation-reduction potential (ORP), conductivity, and moisture content of all the eight samples were measured in situ. Total nitrogen (TN), total phosphorus (TP), organic matters (OM), and the inorganic nitrogen contents (including ammonium (NH4 +-N), nitrite (NO2 −-N) and nitrate (NO3 −-N)) were measured as previously described (Zhang et al. 2012; Liu et al. 2013).
DNA extraction and clone library construction
DNA extraction was carried out as previously described (Hu et al. 2013). The functional genes (amoA genes) of bacteria and archaea were amplified using the amoA-1F/amoA2-R (Rotthauwe et al. 1997) and CamoA-19f/CamoA-616r (Pester et al. 2012) primer pairs, respectively. Four hundred fifty-four high-throughput sequencing was used to ensure the good coverage of clone libraries. To distinguish amoA amplicons that originated from different samples, barcode oligonucleotides were ligated to each end of the two primer pairs. The protocols used to amplify the two functional genes were previously described (Shen et al. 2008; Pester et al. 2012). For each sample, triplicate PCR products were obtained and mixed together. The length of the PCR products was determined by electrophoresis in a 1.5 % agarose gel. The PCR products were purified using the UNIQ-10 purification kit (Sangon Biotech, Shanghai) before sequencing on the GS FLX Titanium platform.
Bioinformatic analysis
Bioinformatic analysis was performed with the Mothur software package (http://www.mothur.org.) (Schloss et al. 2009). Firstly, the achieved sequences were screened for their barcodes and primers and only sequences with exact matches were reserved for further analysis. Secondly, sequences were trimmed and archaeal amoA sequences with length of more than 400 nt and bacterial amoA sequences with length of more than 450 nt were included. Thirdly, the datasets of AOA and AOB were subsampled to equalize differences in sampling intensity. Then, both of the functional genes were aligned against the AOA amoA and AOB amoA databases that were established before (Hu et al. 2014a, b). Then, the results of alignment were screened for the chimeras and the sequences with chimeras were removed. After all the procedures above, we used the high-quality sequences reserved to calculate the operational taxonomic units (OTUs) of AOA and AOB. The high-quality sequences were filtered and calculated using the default setting for each parameter in the Mothur package. The cutoff for both functional genes was set at 15 %, as previously recommended (Pester et al. 2012). Finally, the diversity indices such as ACE, Chao, Shannon, and Simpson diversity and coverage were calculated.
Statistical analysis
BLASTN search was used to examine the representative sequences for all the OTUs of AOA and AOB. Sequences that were not AOA and AOB amoA genes were deleted, along with their OTUs. The ecological distributions of AOA and AOB communities and their correlations with environmental factors were determined using Canoco software 4.5 as previously introduced (Hu et al. 2014a, b). Principal components analysis (PCA), canonical correspondence analysis (CCA), and redundancy analysis (RDA) analysis were carried out using all the OTUs of AOA and AOB as previously introduced (Shen et al. 2013). Pearson correlation analysis (significance level α = 0.05) was used to examine the correlations between the diversities and abundances of AOA and AOB and environmental factors. The representative sequences of all the OTUs for both AOA and AOB and were imported into MEGA 4.1 to construct alignment files in combination with the sequences of known sequences of AOA and AOB. The representative sequences of emerging OTUs of AOA in the periodically flooded soils combined with some environmental archaeal amoA sequences were also used to construct alignment files. Phylogenetic trees were constructed as previously described (Shen et al. 2014).
Real-time quantitative PCR (qPCR)
The two primer sets mentioned above were used to determine the copy numbers of AOA and AOB amoA genes in the soil samples. qPCR was performed as previously described (Hu et al. 2014a, b).
Deposited 454 read accession numbers
Sequences were submitted to the Sequence Read Archive (SRA) at NCBI under the accession numbers SRR1046086 and SRR1047950.
Results
Physicochemical properties of the soil samples
The physicochemical properties of the collected soil samples, including pH, temperature (T), oxidation-reduction potential (ORP), conductivity, moisture content, organic matter content (OM), ammonium content, nitrite content, nitrate content, total nitrogen content (TN), and total phosphorus content (TP) are presented in Table 1. All of the soils had nearly neutral pH values (i.e., 6.60–7.23). The ORP of non-flooded soils such as PXH2 and PXH4 from Baijia creek and PXH8 and PXH10 from Yanglu lake were positive. On the contrary, periodically flooded soils from the rest of the four sampling sites had negative ORP. The conductivity, moisture content, TN, OM, and ammonium content of periodically flooded soils were always higher than the non-flooded soils. The temperature and nitrate content of periodically flooded soils were relative lower than that of the non-flooded soils. The TP went down after the soils were inundated except for the group of PXH10 and PXH11. All the soil samples had low nitrite content as expected.
Abundance of AOA and AOB
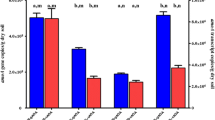
In this study, the functional genes (i.e., amoA gene) of AOA and AOB from the 8 soil samples were quantified. The number of AOA amoA genes of these sites ranged from 8.56 × 107 to 1.12 × 108 in the non-flooded soils and from 3.12 × 107 to 9.67 × 107 in the periodically flooded soils. The AOA amoA gene copy mumbers were always lower in the periodically flooded soils compared with the corresponding non-flooded soils. The number of AOB amoA genes ranged from 9.98 × 106 to 7.70 × 107 in the non-flooded soils and ranged from 5.50 × 105 to 2.37 × 106 in the periodically flooded soils (Fig. 2). Similar to AOA, the AOB amoA gene copy mumbers were always lower in the periodically flooded soils compared with the corresponding non-flooded soils. AOA dominated numerically in all of the 8 soils, with the ratios of AOA:AOB varying from 2.4 to 8.6 in the non-flooded soils and from 14.1 to 56.7 in the periodically flooded soils.
Diversities, phylogenetic analyses, and community structures of AOA and AOB
After all the quality control procedures, normalized and high-quality sequences of AOA and AOB were used for the diversity analysis. For AOA, a total of 7942 high-quality sequences were obtained for the 8 soil samples, with an average of 993 sequences per soil samples (Table S1). Using the 15 % cutoff that was recommended in a previous study (Pester et al. 2012), a total of 90 OTUs were obtained for the archaeal amoA gene. The AOA OTU numbers of each soil sample ranged from 25 to 54, with PXH8 displayed the lowest diversity (25 OTUs) and PXH11 displayed the highest diversity (54 OTUs). The AOA OTU numbers obtained in the periodically flooded soils (PXH3, PXH5, PXH9, PXH11) were always higher than the OTU numbers obtained from the corresponding non-flooded soils (PXH2, PXH4, PXH8, PXH10) (Fig. 3, Table S1). The coverage of the archaeal amoA clone libraries ranged from 98.3 to 99.4 % (Table S1). According to the previously reported classification of archaeal amoA genes (Pester et al. 2012), the representative sequences of the 90 OTUs were grouped into 4 different clusters as shown in Fig. 4a. Among all the 90 OTUs, Nitrosopumilus and Nitrosotalea clusters contained only one OTU. OTU 57 belonging to Nitrosotalea cluster was retrieved from periodically flooded soil sample PXH3. OTU 80 assigned to Nitrosopumilus cluster was retrieved from non-flooded soil sample PXH2. There was no OTU affiliated with thermal AOA cluster. The rest of the 88 OTUs were all affiliated with Nitrososphaera and Nitrososphaera sister cluster. Among the 7942 sequences, 7935 sequences were affiliated with Nitrososphaera and Nitrososphaera sister cluster, accounted for 99.91 % of all the sequences obtained. All the 8 soil samples were composed dominantly by sequences belonging to Nitrososphaera and Nitrososphaera sister cluster.
Neighbor-joining phylogenetic tree showing the phylogenetic affiliations of the representative sequences of each AOA (a) and AOB (b) OTU recovered from water-level-fluctuating zones in Three Gorges Reservoir. The numbers at the nodes are percentages that indicate the levels of bootstrap support from 1000 replicates. The scale bar represents 0.1 nucleic acid substitutions per nucleotide position
As for AOB, after all the quality control procedures same as for AOA, a total of 16,037 sequences were retrieved from all the 8 soil samples. For each sample, 2005 sequences were obtained on the average (Table S2). Using the 15 % cutoff that was recommended in a previous study (Purkhold et al. 2000), 16,037 sequences were assigned to 71 OTUs. The AOB OTUs of each sample ranged from 13 to 32, with PXH8 from non-flooded soil showed the highest diversity (32 OTUs) and PXH5 from periodically flooded soil displayed the lowest diversity (13 OTUs) (Fig. 3). The diversity of AOA was higher than that of AOB in each sampling sites, except in PXH8. Opposite to AOA, The AOB OTU numbers obtained in the periodically flooded soils (PXH5, PXH9, PXH11) were always lower than the OTU numbers obtained from the corresponding non-flooded soils (PXH4, PXH8, PXH10), except in PXH2 and PXH3. The diversity of PXH3 from periodically flooded soil was a little higher than PXH2 (Fig. 3, Table S2). The coverage of the bacterial amoA clone libraries ranged from 99.2 to 99.9 % (Table S2). The representative sequences of the 71 OTUs were grouped into two clusters, with 3 OTUs clustered into the Nitrosomonas cluster and the remaining 68 OTUs clustered into the Nitrosospira cluster (Fig. 4b). Only 11 sequences were affiliated with the Nitrosomonas cluster. All the 8 AOB community structures were primarily composed of Nitrosospira-related sequences.
The newly emerging AOA species in the periodically flooded soils
The diversity of AOA in the periodically flooded soils (PXH3, PXH5, PXH9, PXH11) was higher than that in the corresponding non-flooded soils (PXH2, PXH4, PXH8, PXH10) (Fig. 3, Table S1). A total of 23 OTUs (based on 15 % cutoff) were newly emerged in the periodically flooded soils compared to the non-flooded soils. The 23 OTUs contained 142 sequences and were clustered into three clusters (Figure S3). Nitrososphaera cluster contained 8 OTUs and 69 sequences, Nitrososphaera sister cluster contained 14 OTUs and 67 sequences and Nitrosopumilus cluster contained only 1 OTU and 6 sequences. The environmental sequences that showed the highest similarities with the representative sequences of the 23 OTUs were also included in the phylogenetic analysis. The most similar environmental sequences were retrieved from hot springs, estuary sediments, intertidal sediments, lake or river sediments, reservoir sediments, wetland sediments, deep-sea sediments, and calcareous fluvo-aquic soils. In order to determine which AOA species the 23 OTUs belonged to, the similarities were compared between the representative sequences of the 23 OTUs and some of the known AOA strains from different clusters (Table S3). The similarities between OTU30, OTU41, OTU47, OTU64, OTU65, OTU70, and Nitrososphaera gargensis were 90.8, 85.8, 86.0, 86.2, 88.2, and 90.2 %, separately. All the similarities were more than 85 %. As previously recommended that amoA sequences with less than 85 % nucleic acid sequence identity are likely to represent two different AOA species (Pester et al. 2012), the 6 OTUs were belonged to the species of Nitrososphaera gargensis. The rest of the 17 OTUs might be new AOA species since their similarities with the known AOA strains were all below 85 %.
Environmental factors that influence the diversities and communities of AOA and AOB
To assess the potential relationship between the community structure of ammonia-oxidizing microorganisms and environmental factors in soil samples from water-level-fluctuating zones in Three Gorges Reservoir, a CCA test was performed for AOA and a RDA test was performed for AOB using the Canoco software based on the AOA and AOB community structures and environmental factors (Fig. 5). Of all the 11 investigated environmental factors, ORP had the most significant effect on the community distribution of AOA (p = 0.009). None of the environmental factors was found to significantly influence the community distribution of AOB. In the CCA analysis of AOA, the two axes totally explained 69.8 % of all the data. In the RDA analysis of AOB, totally 91.7 % of all the data was explained by the two axes.
Linear relationships between different environmental factors and amoA gene abundance, OTU numbers, and AOA and AOB diversity indices were characterized using the Pearson correlation coefficient, as shown in Table S4. Soil pH was found to be significantly positively correlated with AOA OTU numbers (p < 0.01) and Shannon index of AOA (p < 0.05), negatively correlated with Simpson index of AOA (p < 0.05). ORP was significantly negatively correlated with AOA OTU numbers (p < 0.05), ratio of OTU numbers (AOA:AOB) (p < 0.01), and ratio of amoA gene abundances (AOA:AOB) (p < 0.05). ORP was also significantly positively correlated with AOB abundance (p < 0.05).
Discussion
The water-level-fluctuating zones in Three Gorges Reservoir were quite special environments where the soils in the zones were half-year inundated and half-year exposed. Compared to the non-flooded soils, the soils in the water-level-fluctuating zones went through oxygenated/hypoxic alternant conditions and had lower oxidation-reduction potential (Table 1). As previously reported, the ORP was positively related with the logarithm of the oxygen concentration (Prosser and Nicol 2012). What more, the non-flooded soils were exposed to air and the oxygen concentration in the soils were nearly saturated. However, the periodically flooded soils were in the depth of 30 m when reservoir started to impound water and it was difficult for oxygen to permeate to the flooded soils. As previously reported, the dissolved oxygen was decreasing steadily from the surface to below the detection level at 20–25 m (Parsons et al. 2014). So, it was reasonable that the oxygen concentrations in the periodically flooded soils were relatively lower than in the non-flooded soils when the water-level-fluctuating zones were underwater.
The oxygenated/hypoxic alternant conditions in the water-level-fluctuating zones did affect the abundances and communities of ammonia-oxidizing microbes. Both the results of qPCR and diversity analysis in this study demonstrated that AOA might be more adaptable in the hypoxic environments compared to AOB. AOA would survive better than AOB in the water-level-fluctuating zones. This was in accordance with the previous study that wet tropical soil AOA are tolerant of extended periods of anoxia (Pett-Ridge et al. 2013).
Among all the environmental factors, the oxidation-reduction potential (ORP) was found to be significantly positively correlated with the AOB abundance (p < 0.05) (Table S4). The AOB abundance in the periodically flooded soils was at least an order of magnitude lower than that in the non-flooded soils (Fig. 2). This could be explained by the relatively low affinity of AOB for oxygen which has been certified by the previous study (Park and Noguera 2007). In the periodically flooded soils where the oxygen concentration were low, some species of AOB could not live under the hypoxic conditions and disappeared after the soils were flooded for more than 6 months. However, the growth of AOB in the periodically flooded soils could not be excluded anyway. Nitrosomonas europaea was found to be capable of nitrite denitrification in anoxic conditions, and this had been recognized as a process of high importance for anaerobic growth (Bock 1995), but most of the AOB sequences obtained in this study were assigned to Nitrosospira cluster. The oxidation-reduction potential (ORP) was also found to be significantly negatively correlated with the ratio of the amoA gene abundance (AOA:AOB) (p < 0.05) (Table S4). Although AOA abundance was decreased along with decreased ORP, the decrement of AOA abundance was lower than that of AOB, leading to the higher ratio of amoA gene abundance (AOA:AOB) in the periodically flooded soils where the oxidation-reduction potential was lower. The fact that the ratio of amoA gene abundance (AOA:AOB) increased in the periodically flooded soils indicated that AOA was more tolerant or adaptable to the hypoxic environments compared to AOB. This could be explained by the higher affinity of AOA for oxygen compared to AOB. The K m value of Nitrosopumilus maritimus SCM1 for oxygen was 3.90 μM, which was 47 times lower than the K m value of Nitrosomonas europaea C-31 (Martens-Habbena et al. 2009; Park et al. 2010). Other than the kinetic tests for oxygen using some isolated or enriched AOA or AOB strains (Park and Noguera 2007; Martens-Habbena et al. 2009; Kim et al. 2012), the fact that AOA was more tolerant or adaptable to the hypoxic environments than AOB has also been discovered in some previous ecological studies. In the depth of 0–30 cm of agricultural soils, the ratio of AOA:AOB was below 1000, while in the 30–40 cm where the condition became hypoxic, the ratio went up to more than 3000 (Leininger et al. 2006). In two oxygen-deficient ecosystems of the eastern south pacific: the oxygen minimum zone off Peru and northern Chile, AOA are an abundant community (up to 105 amoA copies ml−1), while bacterial amoA genes were undetected (Molina et al. 2010). In the two oxygen minimum zones (OMZ) in Arabian Sea and the ETSP, the abundance of archaeal amoA gene in the core of the OMZ was higher than that in the surface waters (Peng et al. 2013).
As for the diversity, ORP was found to be significantly negatively correlated with AOA OTU numbers (p < 0.05) and ratio of OTU numbers (AOA:AOB) (p < 0.01). AOA OTU numbers increased in the periodically flooded soils compared to that in the non-flooded soils, while AOB OTU numbers decreased after the soils were flooded for almost 6 months (Fig. 3, Table S1). Similar to the results of qPCR, the change of diversity in AOA and AOB also suggested that AOA was more adaptable than AOB in the hypoxic conditions. The diversity of AOA increased in the hypoxic conditions has been proved in serious studies. In the ESP-permanent OMZ, the AOA OTU numbers were lower than 4 in the well-oxygenated zones where the oxygen concentrations were more than 164 μM; on the contrary, the AOA OTU numbers could reach to 10 in the oxygen-deficient zones where the oxygen concentrations were below 10 μM (Molina et al. 2010). In the Gulf of California, the AOA OTU numbers increased from 2 in the oxygen-deficient zones (oxygen concentrations about 10 μM) to 12 in the well-oxygen zones (oxygen concentrations about 150 μM) (Beman et al. 2008). The similar results were also found in the Monterey Bay and the North Pacific Subtropical Gyre, where the oxygen-deficient zones (oxygen concentrations about 32 μM) presented higher diversity than the well-oxygen zones (Mincer et al. 2007).
In the periodically flooded soils, 23 newly emerged AOA species were detected. They showed high similarities (≥98.0 %) to the sequences achieved from anoxic conditions, such as hot springs (GQ226117.1) (Jiang et al. 2010), wetland sediments (KJ645341.1) (Yang et al. 2014) and deep-sea sediments (GU270254.1) (Dang et al. 2010). Based on the threshold before (Pester et al. 2012), 6 of the 23 OTUs might be the Nitrososphaera gargensis species which was enriched in the thermal spring with low oxygen concentration (Hatzenpichler et al. 2008).
The increased AOA OTU numbers and the higher ratios of abundance (AOA:AOB) in the periodically flooded soils where the oxygen concentration were lower suggested AOA have better adaptability in oxygenated/hypoxic alternant conditions in the water-level-fluctuating zones in the Three Gorges Reservoir and probably responsible for the ammonia oxidation there. Both the AOA and AOB amoA gene copy numbers decreased in the periodically flooded soils compared to the non-flooded soils which might weaken the ammonia oxidation process in the water-level-fluctuating zones.
References
Beman JM, Popp BN, Francis CA (2008) Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J 2:429–441
Blainey PC, Mosier AC, Potanina A, Francis CA and Quake SR (2011) Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6: e16626
Bock ESI, Stuven R, Zart D (1995) Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Arch Microbiol 163:16–20
Dang H, Luan XW, Chen R, Zhang X, Guo L, Klotz MG (2010) Diversity, abundance and distribution of amoA-encoding archaea in deep-sea methane seep sediments of the Okhotsk Sea. FEMS Microbiol Ecol 72:370–385
de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818
Fang F, Yang Y, Guo J, Zhou H, Fu C, Li Z (2011) Three-dimensional fluorescence spectral characterization of soil dissolved organic matters in the fluctuating water-level zone of Kai County, Three Gorges Reservoir. Front Environ Sci Eng China 5:426–434
Gorman-Lewis D, Martens-Habbena W, Stahl DA (2014) Thermodynamic characterization of proton-ionizable functional groups on the cell surfaces of ammonia-oxidizing bacteria and archaea. Geobiology 12:157–171
Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW (2011) Niche specialization of terrestrial archaeal ammonia oxidizers. Proc Natl Acad Sci U S A 108:21206–21211
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A 105:2134–2139
Hu BL, Shen LD, Liu S, Cai C, Chen TT, Kartal B, Harhangi HR, Op den Camp HJ, Lou LP, Xu XY, Zheng P, Jetten MS (2013) Enrichment of an anammox bacterial community from a flooded paddy soil. Environ Microbiol Rep 5:483–489
Hu BL, Liu S, Wang W, Shen LD, Lou LP, Liu WP, Tian GM, Xu XY, Zheng P (2014a) pH-dominated niche segregation of ammonia-oxidising microorganisms in Chinese agricultural soils. FEMS Microbiol Ecol 90:290–299
Hu BL, Shen LD, Lian X, Zhu Q, Liu S, Huang Q, He ZF, Geng S, Cheng DQ, Lou LP, Xu XY, Zheng P, He YF (2014b) Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci U S A 111:4495–4500
Jiang H, Huang Q, Dong H, Wang P, Wang F, Li W, Zhang C (2010) RNA-based investigation of ammonia-oxidizing archaea in hot springs of Yunnan Province, China. Appl Environ Microbiol 76:4538–4541
Jung MY, Park SJ, Min D, Kim JS, Rijpstra WI, Sinninghe Damste JS, Kim GJ, Madsen EL, Rhee SK (2011) Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol 77:8635–8647
Kim JG, Jung MY, Park SJ, Rijpstra WI, Sinninghe Damste JS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I.1b from an agricultural soil. Environ Microbiol 14:1528–1543
Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA (2005) Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546
Lam P, Jensen MM, Lavik G, McGinnis DF, Muller B, Schubert CJ, Amann R, Thamdrup B, Kuypers MM (2007) Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci U S A 104:7104–7109
Laura E. Lehtovirta-Morley KS, Vilcinskas A, Prosser JI, Nicol GW (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci U S A 108: 15892–15897
Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C (2006) Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809
Liu S, Shen L, Lou L, Tian G, Zheng P, Hu B (2013) Spatial distribution and factors shaping the niche segregation of ammonia-oxidizing microorganisms in the Qiantang River, China. Appl Environ Microbiol 79:4065–4071
Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA (2009) Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979
Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF (2007) Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol 9:1162–1175
Molina V, Belmar L, Ulloa O (2010) High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ Microbiol 12:2450–2465
Park HD, Noguera DR (2007) Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J Appl Microbiol 102:1401–1417
Park BJ, Park SJ, Yoon DN, Schouten S, Sinninghe Damste JS, Rhee SK (2010) Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol 76:7575–7587
Parsons RJ, Nelson CE, Carlson CA, Denman CC, Andersson AJ, Kledzik AL, Vergin KL, McNally SP, Treusch AH and Giovannoni SJ (2014) Marine bacterioplankton community turnover within seasonally hypoxic waters of a subtropical sound: Devil’s Hole, Bermuda. Environ Microbiol. doi:10.1111/1462-2920.12445
Peng X, Jayakumar A, Ward BB (2013) Community composition of ammonia-oxidizing archaea from surface and anoxic depths of oceanic oxygen minimum zones. Front Microbiol 4:177
Pester M, Rattei T, Flechl S, Grongroft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M (2012) amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539
Pett-Ridge J, Petersen DG, Nuccio E, Firestone MK (2013) Influence of oxic/anoxic fluctuations on ammonia oxidizers and nitrification potential in a wet tropical soil. FEMS Microbiol Ecol 85:179–194
Pitcher A, Villanueva L, Hopmans EC, Schouten S, Reichart GJ, Sinninghe Damste JS (2011) Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone. ISME J 5:1896–1904
Prosser JI, Nicol GW (2008) Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol 10:2931–2941
Prosser JI, Nicol GW (2012) Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol 20:523–531
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66:5368–5382
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structure gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shen JP, Zhang LM, Zhu YG, Zhang JB, He JZ (2008) Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10:1601–1611
Shen LD, Liu S, Lou LP, Liu WP, Xu XY, Zheng P, Hu BL (2013) Broad distribution of diverse anaerobic ammonium-oxidizing bacteria in Chinese agricultural soils. Appl Environ Microbiol 79:6167–6172
Shen LD, Huang Q, He ZF, Lian X, Liu S, He YF, Lou LP, Xu XY, Zheng P, Hu BL (2014) Vertical distribution of nitrite-dependent anaerobic methane-oxidising bacteria in natural freshwater wetland soils. Appl Microbiol Biotechnol 99:349–357
Stewart FJ, Ulloa O, DeLong EF (2012) Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14:23–40
Verhamme DT, Prosser JI, Nicol GW (2011) Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J 5:1067–1071
Yang Y, Shan J, Zhang J, Zhang X, Xie S, Liu Y (2014) Ammonia- and methane-oxidizing microorganisms in high-altitude wetland sediments and adjacent agricultural soils. Appl Microbiol Biotechnol 98:10197–10209
Zhang B, Fang F, Guo J, Chen Y, Li Z, Guo S (2012) Phosphorus fractions and phosphate sorption-release characteristics relevant to the soil composition of water-level-fluctuating zone of Three Gorges Reservoir. Ecol Eng 40:153–159
Acknowledgments
The authors wish to thank the National Science Foundation for Young Scientists of China (No. 51108408), the Fundamental Research Funds for the Central Universities (No. 2015QNA6012), and the Foundation from State Environmental Protection Key Laboratory of Microorganism Application and Risk Control of Tsinghua University (No. SMARC2013D004).
Conflict of interest
The authors declare that they have no competing interests.
Compliance with ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
ESM 1.
(PDF 216 kb)
Rights and permissions
About this article
Cite this article
Liu, S., Hu, B., He, Z. et al. Ammonia-oxidizing archaea have better adaptability in oxygenated/hypoxic alternant conditions compared to ammonia-oxidizing bacteria. Appl Microbiol Biotechnol 99, 8587–8596 (2015). https://doi.org/10.1007/s00253-015-6750-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6750-7