Abstract
Phyllosphere microorganisms are sensitive to fluctuations in wind, temperature, solar radiation, and rain. However, recent explorations of patterns in phyllosphere communities across time often focus on seasonal shifts and leaf senescence without measuring the contribution of environmental drivers and leaf traits. Here, we focus on the effects of rain on the phyllosphere bacterial community of the wetland macrophyte broadleaf cattail (Typha latifolia) across an entire year, specifically targeting days before and 1, 3, and 5 days after rain events. To isolate the contribution of precipitation from other factors, we covered a subset of plants to shield them from rainfall. We used targeted Illumina sequencing of the V4 region of the bacterial 16S rRNA gene to characterize phyllosphere community composition. Rain events did not have a detectable effect on phyllosphere community richness or evenness regardless of whether the leaves were covered from rain or not, suggesting that foliar microbial communities are robust to such disturbances. While climatic and leaf-based variables effectively modeled seasonal trends in phyllosphere diversity and composition, they provided more limited explanatory value at shorter time scales. These findings underscore the dominance of long-term seasonal patterns related to climatic variation as the main factor influencing the phyllosphere community.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The diversity and composition of the plant microbiome are a critical determinant in plant survival and success in the face of challenging conditions [1]. Similar to other host-associated microbial communities, bacterial populations on the plant surface (the phyllosphere) are subject to different environmental stressors depending on their location. These conditions can disrupt or encourage both beneficial and pathogenic taxa. Historically, research of phyllosphere microorganisms has centered on the spread and establishment of plant pathogens. However, a diverse community of leaf-associated microorganisms may protect against plant disease, and thus, the ecology of this microhabitat is important to promoting plant health [2,3,4] (as reviewed by Stone et al. [1]).
Conditions at the leaf surface are more variable than in the soil environment and impose several limitations on microbial populations, such as an abundance of ultraviolet (UV) radiation and frequent lack of available moisture. Microorganisms isolated from the phyllosphere have demonstrated many ways to mitigate these stressors including the production of pigments [5, 6], formation of biofilms [7,8,9], and localization to protected microenvironments [10, 11]. The unprotected nature of the leaf means that microorganisms on the aboveground portion of the plant are also subject to movement from wind and rain [12, 13]. These fluxes create local patterns in bacterial atmospheric composition (potentially influencing immigration to the leaf surface) [10, 14] and large-scale movement [15, 16]. Wind-induced migration from the leaf surface to the atmosphere is readily apparent from the dominance of phyllosphere-associated lineages in even high-elevation atmosphere samples [17]. In contrast, rain is thought to be principally a mechanism of downward immigration wherein organisms travel with precipitation [18]. Although splash dispersal can remove and transport leaf epiphytes throughout a plant canopy at small scales [13, 19], deposition of bacterial cells on the leaf surface and, especially, population growth following rain seem to be of far more importance [18].
Much of our understanding of the influence of climatic variables on the phyllosphere microbiome come from studies at the organismal or population level [10]. Further, most such studies have been developed around a small group of cultural microorganisms, usually plant pathogens (e.g., [19,20,21,22,23,24]). Little is known about the effects of climate on the collective microbial community. Temporal variation in phyllosphere community composition likely represents the combined influence of leaf age and ecological succession, coupled with environmental changes [10] and changes in the microbial composition of the atmosphere [25]. However, few, if any, investigations have disentangled the effects of climatic variables and changes from leaf characteristics that follow seasonal trends. Furthermore, because many phyllosphere studies focus on agricultural systems, these studies are concluded at the time of harvest or at the end of the growing season—before seasonal changes in both leaf characteristics and phyllosphere composition may become pronounced [1]. This is compounded by the lack of recent studies on the species-area and species-time relationships in the phyllosphere. Earlier work on fungal phyllosphere communities has suggested that no such relationship exists between leaf size and species count [10] but a recent study of bacterial communities using modern sequencing methods has found a significant relationship [26] and such relationships have been observed for bacteria in soils, sediments, and aquatic settings both culture-based identification and molecular methods [27].
To address these topics, we characterized the bacterial community of broadleaf cattail (Typha latifolia) across an entire year in a natural setting, with specific sampling on days before and after rain events. T. latifolia was chosen as the target species because its aboveground tissues remain standing after senescence, allowing us to observe bacterial community development following the shift to senescence. Further, T. latifolia leaves emerge from a well-mixed aqueous environment, rather than from soil, which may produce variation in initial community composition due to soil heterogeneity. To understand the effect of rain more completely, we established a treatment group of plants covered by protective canopies on rainy days to compare against uncovered plants. We collected climatic data pertaining to precipitation, UV radiation, photosynthetically active radiation (PAR), temperature, relative humidity, and wind speed from NOAA weather station databases and relate them to patterns in bacterial alpha and beta diversity. Thus, we explore the relationship between leaf size, senescence, seasonal patterns, and short-term weather impacts. We expected that reductions in bacterial diversity would occur following leaf senescence, aligning with previous findings of fungal dominance during this shift [28]. As most bacterial diversity exists at low abundance and is therefore sensitive to demographic stochasticity, we predicted that bacterial diversity would be lower initially after rainfall and subsequently increase, but only in plants exposed to rain (i.e., those not under canopies). We expected that larger leaf size would lead to higher bacterial alpha diversity. We decomposed bacterial beta diversity into differences driven by either species turnover (loss of one species followed by the gain of another) or by richness differences (asymmetric loss or gain). Lastly, we predicted that bacterial communities will differentiate primarily by leaf senescence and that this difference will be driven by richness differences in bacterial communities rather than by bacterial turnover per se. As environmental variables are often strongly correlated, we utilized machine learning, specifically random forest methodology, to model these variables against bacterial diversity.
Materials and Methods
Sample Collection
Aerial (i.e., non-submerged) leaves from standing T. latifolia plants were collected from the University of Mississippi Field Station (UMFS; 34.4237 N, 89.3859 W) pond 80 using sterile sampling procedures. Leaves selected did not have visible signs of herbivory or disease, were cut from point that they became separated from other leaves on the plant or the point that the leaf became submerged underwater, and placed into sterile sampling bags. Gloves and shears were sterilized between samples with 70% ethyl alcohol. The average amount of leaf material collected was 141 ± 69.43 cm2 (mean ± 1 SD). Sampling began on April 29th, 2015 and ended on April 25th, 2016. Collection of T. latifolia leaves occurred at regular time intervals (semi-monthly) as well as before and after rain events forecasted to produce > 2.5 cm of rain. For forecasted rain events, samples were collected 1 day before, 1 day after, 3 days after, and 5 days after. On regularly scheduled sampling days, the 1628-m2 pond was mapped into nine subsections (~ 180 m2 each) and leaf material was removed from a single plant in four random subsections. On days when rain was forecasted, 1-m2 greenhouse polyethylene film rain canopies (152 μm thick) were erected using a PVC frame over T. latifolia plants in four randomly chosen subsections (selection was independent between covered and non-covered samples, allowing sampling to occur twice in one subsection) which were sampled along with non-covered subsections (i.e., eight total samples collected). The canopies remained in place until 5 days following the rain event. Because some rain events coincided with semi-monthly sampling, a total of 84 samples were produced from 21 semi-monthly sampling dates. No sampling was conducted when rain failed to occur as predicted. Five rain events were sampled, totaling 19 days and 152 samples (two rain events occurred back-to-back, allowing the fifth day of one rain event to serve as the day-before-rain of the succeeding rain event). Thus, a total of 236 samples were collected on 40 sampling dates during the yearlong sampling period. Samples collected from different sections on each day were not pooled and instead remained separate.
Following collection, leaf samples were immediately taken to the laboratory and the phyllosphere community removed by scrubbing in 6 mL of sterile 1 mM sodium bicarbonate buffer with sterile, single-use toothbrushes. The buffer suspension was centrifuged (7000 rcf × 2 min) and DNA extracted from the resulting pellet using MoBio PowerSoil DNA extraction kits (MoBio Laboratories, Carlsbad, CA) following standard protocols. The V4 region (253 base pairs) of the bacterial 16S rRNA gene was amplified with barcoded targeted primers (515F forward and 806R reverse) [29]. Amplification of the V4 region was conducted with an initial denaturation step of 95 °C (2 min) followed by 30 cycles each consisting of 95 °C (20 s), 55 °C (15 s), and 72 °C (2 min) and with a final elongation step of 72 °C (10 min). Amplified fragments were purified and concentrations standardized using SequalPrep Normalization Plates (Life Technologies, Grand Island, NY), pooled, and paired-end sequenced on the Illumina MiSeq platform at the University of Mississippi Medical Center Molecular and Genomics Core Facility. After microbial community removal, leaves were frozen (− 20 °C) until imaged as jpeg files using the digital camera of a Samsung Galaxy S4 at 2048 × 1152 resolution from a fixed height. Total leaf area (cm2) was quantified using ImageJ 1.51 [30] while green/brown pixel ratio, and thus green and brown surface area, was obtained by calculating the proportion of green to brown pixels in each image in R using the jpeg package [31]. Freezing had no observable effect on the ratio of green to brown tissue on each leaf.
Sequence Data Processing
Raw 16S rRNA gene sequence data (FASTQ) were deposited into the NCBI Sequence Read Archive (SRA) under accession number PRJNA487794. The initial 9,217,422 sequences were processed, clustered into operational taxonomic units (OTUs), and taxonomically assigned using the mothur bioinformatics pipeline (v.1.39.5) following standard protocols [32,33,34] to remove erroneous sequences from downstream analyses. Remaining sequences were aligned against the SILVA database (version 128) of bacterial sequences [35], screened to remove Eukarya and Archaea, and grouped to OTUs defined by 97% sequence similarity and then classified against SILVA. The resulting 2,885,169 sequences were classified to 6871 OTUs. Of these, 3436 OTUs were represented only once across the dataset and were removed leaving 3435 bacterial OTUs for subsequent analyses. The average number of reads per sample was 13,642 ± 6720 (mean ± 1 SD). To reduce the impact of samples with poor sequencing depth on diversity analyses, 27 samples with low sequence counts (< 2500 sequences) were removed leaving 209 samples. The following analyses were conducted on the remaining samples which were not rarefied [36] although the key results were identical to those from rarefied samples.
Climatic Data Acquisition and Processing
Precipitation data were accessed from the National Oceanic and Atmospheric Administration (NOAA) land-based station located at the UMFS (Abbeville, MS; Station ID: GHCND:US1MSLY0004) from April 1st 2015 through April 30th 2016. Radiation and atmospheric data, including direct and diffuse solar radiation, UV-B, infrared (IR), photosynthetically active radiation (PAR), temperature, relative humidity, and wind speed, were obtained from the NOAA surface radiation network station in Goodwin Creek, MS (34.2547° N, 89.8729° W; 48 km from the UMFS) for 2015 and 2016. Radiation data (recorded every minute) were subject to quality filtering (any measurement flagged as poor quality was coded as not available, or NA) and combined from separate daily files into one dataset. Missing climatic data from some variables spanned multiple days (up to 29) and were imputed from non-missing data using loess trendlines with the loess function in R version 3.5.1 [37].
To understand the extent that prior climatic conditions play in driving bacterial community patterns, additional variables were constructed by calculating means for all climate variables at 1, 3, 5, 7, and 14-day intervals leading into each sampling day. Net infrared and total net radiation were removed due to poor data quality, and to retain some information about hourly fluctuations, radiation features were aggregated with daily maximum and minimum values as well as daily means. Along with total precipitation, rain incidence (rain or no rain) and heavy rain incidence (> 2.5 cm) variables were coded as either 0 or 1 indicating either the absence or presence of rain on a given day. From these, the number of days since the last rain event (or heavy rain event) was calculated for each date. Both sums and means were then generated for precipitation features across the window intervals as above. Samples that were experimentally covered from the rain had all precipitation values set to 0 before these summaries were calculated, and whether or not a sample was covered was included as a factor in statistical analyses.
Statistical Analyses
Non-bioinformatic statistical analyses were conducted in R version 3.5.1, with the exception of the indicator function in mothur which was used to identify OTUs significantly associated with either living or senesced leaves. Alpha diversity was summarized by the Simpson evenness measure (Dsimp) and Chao estimated richness (Schao). To test the effect of rain on alpha diversity in our experimental setting, we fit quadratic regression models of bacterial richness and evenness (both log-transformed) against the number of days since the last rain event (from − 1 to 5) as well as the interaction between rain and covering treatment (covered or uncovered). Here, the lm function in R was used, where the term describing the number of days since the last rain event was squared. To test the effect of rain on the abundance of individual OTUs, the same quadratic model was repeatedly run using the abundance of each OTU, in turn, as the response variable. To control for the type I error rate across multiple statistical comparisons, the p values from all individual OTU models were extracted, ranked from lowest to highest, and cumulatively added. Only those OTUs whose p values fell within the cumulative 0.05 threshold were considered to significantly respond to rain. The effect of covering on species accumulation following rain events was tested by setting species richness to zero on the day preceding rain, calculating the number of novel species gained from that day until day 5, as if samples were pooled, and generating a regression model with an interaction term between time and covering. Separately from the above models, the effect of leaf senescence on (log-transformed) bacterial richness and evenness was tested with one-way ANOVAs.
Random forests were used to relate alpha diversity (Schao and Dsimp) patterns to climatic variables (precipitation, radiation, temperature, and wind), leaf data (total leaf area and green/brown ratio), and covering by rain shelters. Although issues of multicollinearity do not affect random forest performance, to aid in the interpretability of which aspects of climate are most predictive of bacterial richness and evenness, random forest models were computed against a reduced dimension dataset of seven variables (composite precipitation, composite radiation, composite temperature, composite wind, total leaf area, green/brown ratio, and rain shelter covering; Supplementary Table S1). To perform dimensionality reduction, precipitation, radiation, temperature, and wind variables were each subjected to principal component analysis (PCA). The first principal component was enough to explain a high proportion of variance (≥ 0.9) for variables related to precipitation, radiation, temperature, and wind resulting in a single, composite variable summarizing each group. The random forest methodology, briefly, builds a single predictive model from multiple individual regression trees which recursively partition samples based on logical assignments of predictor variables and whose performance is measured by the mean squared error between observed values of the response with the means of groups created by those partitions [38]. To limit the influence of correlations between variables and to attain better predictive capacity, many regression tree models are created, each containing a small subset of the predictor variables available, and individual functions are averaged to generate a single ensemble model [39]. Model fitting was implemented using the randomForest package for R and train/test validation through caret [40, 41]. To minimize correlations between features, individual trees (500 per test) were limited to a subset of three parameters at a time. Train/test validation was performed on an initial, but unfixed (i.e., increasing each test) window of 16 training samples and progressing chronologically by four samples yielding 114 tests total. There was no difference between random forests performance when using all features individually or on condensed features.
Beta diversity patterns were visualized using non-metric multidimensional scaling (NMDS) on Hellinger-transformed Bray-Curtis (βbc), Jaccard (βj) dissimilarity measures using the metaMDS function in the vegan package [42]. To understand the effect of environmental variation and leaf senescence on species composition, independent of differences in alpha diversity, Bray Curtis, and Jaccard measures were decomposed into replacement and richness difference fractions using the Podani method [43] provided by the beta.div.comp function in the adespatial package [44]. Permutational multivariate analysis of variance (PERMANOVA) was used to assess the statistical difference between living and senesced leaf composition using the adonis function in the vegan package. To assess the degree that living and senesced leaves had different degrees of community dispersion, independent of differences in richness, the betadisper and permutest functions were used from the vegan package. Local contributions to beta diversity (LCBD) values were calculated for each sample on Bray-Curtis and Jaccard replacement and richness components, as well as on total beta diversity measures. Random forest models were then run, as above, using LCBD components as the response variables. All code and associated data for statistical modeling has been made available at http://github.com/bramstone.
Results
Experimental Mitigation of Rain Through Covering
Contrary to our predictions, we did not observe a significant loss and regain of bacterial diversity in the phyllosphere following rain events. This was the case for both richness and evenness where no patterns were detected applying either quadratic or linear trends to each measure. The effect of experimentally covering plants was included as an interaction term between covering and rain although no significant interaction was observed (pchao = 0.50, psimp = 0.69; Fig. 1a, b). Similarly, we observed no difference in novel species accumulation following rain events between covered and uncovered plants (p = 0.82; Fig. 1c). These community-level patterns were consistent at the level of individual bacterial taxa where we observed no effect of leaf covering on either quadratic or linear responses of OTU frequencies to rain (data not shown here).
Bacterial community diversity in the phyllosphere of Typha latifolia leaves in response to rain. Points represent means of daily subsamples (four per day for each category) across five separate rain events superimposed against predicted values (lines) based on linear and quadratic models. Light green elements represent data from leaves covered from rain, dark green elements represent data from uncovered samples. a Estimated species richness (Schao) of covered and uncovered leaves. b Community evenness (inverse of Simpson’s metric) of covered and uncovered leaves. c Bacterial species accumulation of covered and uncovered leaves
Leaf Senescence and the Bacterial Community
Aligning with our initial prediction, we found that both phyllosphere bacterial richness and evenness decreased following senescence of T. latifolia leaves (Fchao = 12.0, pchao < 0.001; Fsimp = 62.6, psimp < 0.001; Fig. 2) as determined by the loss of green leaf tissue from sampled plants (dates after December 15, 2015; Supplemental Figure S1). PERMANOVA analyses demonstrated significant clustering of bacterial communities on living and senescent leaves using both Bray-Curtis (F1,207 = 56.6, p < 0.001) and Jaccard indices (F1,207 = 13.6, p < 0.001) (Fig. 3a, b). However, this pattern was reduced or non-existent when community composition was visualized using replacement-component dissimilarities (βbc-repl: F1,207 = 34.6, p < 0.001; βj-repl: F1,207 = 0, p = 1) which account for differences in alpha-diversity between samples (Fig. 3c, d), supporting, at least partially, our original prediction that compositional changes in beta-diversity were driven by alpha-diversity differences. Similarly, permutational tests of dispersion on replacement-component dissimilarities showed that community composition was more dynamic in the spring and summer, and more consistent following senescence (βbc: F1,207 = 91.1, p < 0.001; βj: F1,207 = 13.5, p = 0.002) (Fig. 3a, b); however, this pattern was not apparent with either replacement component measure (βbc-repl: F1,207 = 60.1, p < 0.001; βj-repl: F1,207 = 0.29, p = 0.62) (Fig. 3c, d), indicating that higher beta diversity during spring and summer months was driven by greater variation in alpha diversity.
Changes in bacterial alpha diversity in the phyllosphere of Typha latifolia leaves from April 29, 2015 to April 25, 2016 and aggregated by leaf senescence. Time series plots consist of points representing diversity measures from individual samples and lines representing monthly average diversity measures while down-facing arrows represent measured rain events where sampling frequency was increased to 2-day intervals. Boxplots represent average diversity on living leaves (green) and senesced leaves (brown). a Estimated species richness (Schao) over time. b Community evenness (inverse of Simpson’s dominance metric) over time
Non-metric multidimensional scaling (NMDS) ordinations of bacterial composition in the phyllosphere of Typha latifolia leaves from April 29, 2015 to April 25, 2016. Green points represent leaves with at least some green tissue while brown points represent fully senescent leaves (dates after December 15, 2015). a Bray-Curtis abundance-based dissimilarities (stress = 0.15). b Jaccard presence-absence dissimilarities (stress = 0.11). c Bray-Curtis replacement component dissimilarities (stress = 0.34). d Jaccard replacement component dissimilarities (stress = 0.39)
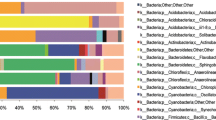
Bacterial OTUs significantly associated with both early-seasonal (i.e., living) and late-seasonal (i.e., senesced) leaves grouped predominantly into the Proteobacteria (which accounted for 44% and 48% or all sequences, respectively, Fig. 4). On senescent leaves, a higher proportion of OTUs identified as Actinobacteria and Bacteroidetes (5% and 28% of all sequences, respectively) than on living leaves (3% and 10%), which had more representation from the Acidobacteria and Verrucomicrobia (8% and 3% from living leaves, respectively, compared to 3% and 1% in senescent leaves). Specific OTUs that were associated with living leaves significantly more so than with senescent leaves included sequence types identified as Acinetobacter, Clostridium, Gemmata, and Pseudomonas (p < 0.001 in all cases). In contrast, senesced leaves harbored OTUs that included the genera Sphingomonas, Roseomonas, Hymenobacter, and Spirosoma.
Comparison of changes in the relative abundance of major bacterial phyla occurring in the phyllosphere of Typha latifolia leaves across the sampling period against seasonal changes in precipitation and temperature. Vertical black bars represent the time of leaf senescence as determined by the loss of green leaf tissue from sampled plants. a Bar heights represent weekly averages of relative abundance, across all leaves sampled during a given week, of the six most abundant bacterial phyla accounting for > 95% of sequences on average. Remaining phyla were combined into “Other” category. b Daily precipitation (mm) and mean daily temperature (degrees C) occurring at the field sampling site. Bacterial community composition shows a clear shift following leaf senescence, with greater proportional representation from the Bacteroidetes. Down-facing arrows represent rain events where sampling frequency was increased to two-day intervals
Climate and the Bacterial Community
Climatic variables were consistently the more informative predictors of phyllosphere bacterial richness, while T. latifolia leaf traits were more informative predictors of bacterial evenness. Random forest models were able to predict 40.7% and 35.6% of bacterial richness and evenness, respectively (determined from model R2 values; Supplemental Figure S2) when modeled against composite precipitation, composite radiation, composite temperature, composite wind, total leaf area, green/brown ratio, and rain shelter covering (Supplemental Table S1). Precipitation was more important to bacterial richness than evenness and whether or not a sample was covered from rain provided minimal explanatory power (Fig. 5).
Proportional predictor variable importance across all regression tree iterations in random forest models of a estimated bacterial species richness (Schao) and b Simpson evenness in the phyllosphere of Typha latifolia leaves. “Green Leaf %” indicates the proportion of total leaf area covered by living green tissue. Leaf area represents total leaf area (cm2). All other variables represent composite variables extracted from a single principal component of several related variables
Random forest models were less effective predictors of LCBD values, predicting 31.8% of local Bray-Curtis contributions and 30.3% of Jaccard contributions (with similar fits for individual beta diversity components). Aligning with alpha diversity models, green leaf area, and precipitation were the driving variables of LCBD patterns, although some differences emerged between different components of each beta diversity measure (Fig. 6). Bray-Curtis and its replacement component were driven strongly by how much green area was present on T. latifolia leaves, indicating a strong influence of this variable on the relative abundances of bacterial OTUs. The Bray-Curtis richness component emphasized precipitation and temperature – variables with strong seasonal trends—as much as leaf condition (Fig. 6). Total Jaccard dissimilarities—which emphasize changes in presence or absence—were strongly influenced by precipitation and temperature, while individual replacement and richness components were more strongly influenced by the percentage of T. latifolia leaves that remained green (Fig. 6). This suggests that leaf senescence drove Jaccard dissimilarities through changes in alpha diversity (richness component) as well as turnover in the composition of bacterial OTUs (replacement component). Similar to alpha diversity patterns, whether or not a sample was isolated from rain by experimental covers offered little predictive value to differences in LCBD values (Fig. 6).
Proportional predictor variable importance across all regression tree iterations in random forest models of local contributions to beta diversity (LCBD) using a Bray-Curtis and b Jaccard dissimilarity measures of bacterial communities on the phyllosphere of Typha latifolia leaves. Dissimilarity measures are further broken down into richness difference (light blue), replacement (red), and total (sum; purple) dissimilarity components. “Green Leaf %” indicates the proportion of total leaf area covered by living green tissue. Leaf area represents total leaf area (cm2). All other variables represent composite variables extracted from a single principal component of several related variables
Discussion
Taken together, our results support previous conclusions of strong seasonal signals in the phyllosphere microbiome [23, 45,46,47,48]. Bacterial community richness corresponded more to environmental parameters than did community evenness. Evenness, in contrast, remained low following senescence, suggesting that bacterial communities were dominated by fewer species than when leaves were alive. Individual rain events exerted little influence on bacterial diversity, patterns of individual OTUs, or composition. Consistent with these findings, multivariate ordinations of bacterial community composition demonstrated that after accounting for differences in alpha-diversity [43], clear grouping based on senescence was highly diminished, confirming that differences in alpha-diversity were important in driving distinction between bacterial communities on living and senesced leaves. Notably, Jaccard-based composition on living and senescent leaves was virtually identical after accounting for differences in alpha diversity. This suggests that differences remain in relative abundances between living and senescent leaves, after accounting for alpha-diversity differences, as captured by Bray-Curtis dissimilarities. By contrast, Jaccard dissimilarities, which lack information on change in relative abundance, could differentiate living and senesced leaves only by their difference in alpha diversity. Overall, random forest predictive performance was not particularly high, indicating that other, non-climatic variables—such as those associated with the quality of the leaf habitat—may be important in predicting bacterial diversity in the phyllosphere.
In contrast to longer seasonal patterns, short term changes did not seem to have a strong effect on bacterial community diversity or composition. Specifically, with regard to precipitation, experimentally covering plants did not change the accumulation of new bacterial species to the T. latifolia phyllosphere or the level of local diversity therein. Although rain was an important predictor variable in random forest models of bacterial diversity, the lack of response to individual rain events supports our conclusion that seasonal patterns collectively exert a stronger effect on bacterial communities. Previous work has demonstrated a surge in abundance of some microbial taxa following rain [20], but this phenomenon does not appear to lead to an appreciable change in diversity at the level of the whole community. Another seasonal phyllosphere study drew similar conclusions with respect to rain [48], with strong seasonal trends and a high initial variability in community composition followed by convergence. While the possibility of rain aiding the succession process was highlighted, there were no other clear consequences of rain on either bacterial community diversity or composition [48]. A subsequent study of the effect of rain on the phyllosphere communities throughout the tree canopy found similar results [26]. Thus, it appears that the bacterial community response to rain in the phyllosphere may not be strongly affected by the response of individual constituents.
Important taxa associated with living and senescent leaves were those generally found in the phyllosphere and soil. Prominent OTUs indicative of senescent leaves were typically aerobic chemoheterotrophs, likely functioning as decomposers. Bacterial taxa in the phyllosphere may respond purely to changes in the living vs. senescent leaf surface over time, but seasonal patterns also appear in the phyllosphere of plants that do not show seasonal senescence [45, 46], suggesting that there is at least some climatic influence. Similarly, progressive fungal colonization occurs in the phyllosphere also following seasonal trends and leaf senescence [10, 49]. Generally, fungal composition transitions to filamentous lineages to capitalize on plant cellulose on senesced leaves [28]. These fungal groups are the principal drivers of microbial decomposition of aerial leaf material which can proceed quickly. In other Typha spp., up to 50 or 60% of leaf carbon may be converted to fungal biomass and CO2 by the summer following leaf senescence [50, 51]. It is likely that bacteria play some role into this process, although their exact relationship with fungi or the degradation of leaf litter is unknown.
Several bacteria in this dataset were significantly associated with senescent leaves, occurring in high abundance. Here, Sphingomonas was associated with senescent leaves and was the single most abundant OTU. Given that Sphingomonas is a common phyllosphere constituent [52] and can utilize a large spectrum of carbon sources [53], this prominence is not surprising. Hymenobacter has been isolated from soil and airborne settings and like Spirosoma, which is phototrophic [54], has been isolated in habitats with high UV radiation, a useful trait in the phyllosphere [55, 56]. Moreover, Hymenobacter has been observed to degrade fungal chitin, and likely utilizes fungal carbon as an energy source [57]. Roseomonas is a ubiquitous environmental organism, isolated from freshwater sediment, air, water, contains bacteriochlorophyll-a, and includes psychrophilic lineages [58,59,60,61,62]. Similarly, Spirosoma has been isolated from Arctic permafrost soils [63]. As senescent leaf sampling coincided with colder winter months, these bacteria likely reach high abundances due to their adaptations to cold and radiation intense environments.
Our conclusions demonstrate that seasonal contributions to phyllosphere bacterial diversity and composition are detectable at multiple ecological levels. More importantly, the separation of isolated weather patterns from broad-scale seasonal trends is an important distinction that is often overlooked in surveys of phyllosphere communities. R2 values of random forest predictor models were low compared to those built on very large datasets (e.g. such as for commercial purposes), suggesting either the need for more samples or that other factors not measured in the present study may be important drivers of phyllosphere diversity. We conclude that bacterial diversity in the phyllosphere is largely unaffected by short-term weather patterns but is particularly sensitive to climatic and leaf-associated changes that occur during seasonal progression.
References
Stone BWG, Weingarten EW, Jackson CR (2018) The role of the phyllosphere microbiome in plant health and function. Annu Plant Rev Online 1:1–24. https://doi.org/10.1002/9781119312994.apr0614
Balint-Kurti P, Simmons SJ, Blum JE, Ballaré CL, Stapleton AE (2010) Maize leaf epiphytic bacteria diversity patterns are genetically correlated with resistance to fungal pathogen infection. Mol Plant-Microbe Interact 23:473–484. https://doi.org/10.1094/MPMI-23-4-0473
Innerebner G, Knief C, Vorholt JA (2011) Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol 77:3202–3210. https://doi.org/10.1128/AEM.00133-11
Fernando WGD, Nakkeeran S, Zhang Y, Savchuk S (2007) Biological control of Sclerotinia sclerotiorum (Lib.) de Bary by Pseudomonas and Bacillus species on canola petals. Crop Prot 26:100–107. https://doi.org/10.1016/j.cropro.2006.04.007
Stout JD (1960) Bacteria of soil and pasture leaves at Claudelands Showgrounds. New Zeal J Agric Res 3:413–430. https://doi.org/10.1080/00288233.1960.10426626
Sundin GW (2002) Ultraviolet radiation on leaves: its influence on microbial communities and their adaptations. In: Lindow SE, Hecht-Poinar EJ, Elliot V (eds) Phyllosphere microbiology. APS Press, St Paul, pp 27–38
Ophir T, Gutnick DL (1994) A role for polysaccharides in the protection of microorganisms from desiccation. Appl Environ Microbiol 60:740–745
Wilson M, Lindow SE (1994) Inoculum density-dependent mortality and colonization of the phyllosphere by Pseudomonas syringae. Appl Environ Microbiol 60:2232–2237
Morris CE, Barnes MB, McLean RJC (2002) Biofilms on leaf surfaces: implications for the biology, ecology and management of populations of epiphytic bacteria. In: Lindow SE, Hecht-Poinar EJ, Elliot V (eds) Phyllosphere microbiology. APS Press, St Paul, pp 317–339
Kinkel LL (1997) Microbial population dynamics on leaves. Annu Rev Phytopathol 35:327–347. https://doi.org/10.1146/annurev.phyto.35.1.327
Baldotto LEB, Olivares FL (2008) Phylloepiphytic interaction between bacteria and different plant species in a tropical agricultural system. Can J Microbiol 54:918–931. https://doi.org/10.1139/W08-087
Lindemann J, Constantinidou HA, Barchet WR, Upper CD (1982) Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl Environ Microbiol 44:1059–1063
Lindemann J, Upper CD (1985) Aerial dispersal of epiphytic bacteria over bean plants. Appl Environ Microbiol 50:1229–1232
Maignien L, DeForce EA, Chafee ME et al (2014) Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. MBio 5:1–10. https://doi.org/10.1128/mBio.00682-13
Morris CE, Sands DC, Vinatzer BA, Glaux C, Guilbaud C, Buffière A, Yan S, Dominguez H, Thompson BM (2008) The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J 2:321–334. https://doi.org/10.1038/ismej.2007.113
Bowers RM, McLetchie S, Knight R, Fierer N (2011) Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J 5:601–612. https://doi.org/10.1038/ismej.2010.167
Šantl-Temkiv T, Finster K, Dittmar T, Hansen BM, Thyrhaug R, Nielsen NW, Karlson UG (2013) Hailstones: a window into the microbial and chemical inventory of a storm cloud. PLoS One 8:e53550. https://doi.org/10.1371/journal.pone.0053550
Constantinidou HA, Hirano SS, Baker LS, Upper CD (1990) Atmospheric dispersal of ice nucleation-active bacteria: the role of rain. Phytopathology 80:934–937
Cevallos-Cevallos JM, Danyluk MD, Gu G et al (2012) Dispersal of Salmonella Typhimurium by rain splash onto tomato plants. J Food Prot 75:472–479. https://doi.org/10.4315/0362-028X.JFP-11-399
Hirano SS, Baker LS, Upper CD (1996) Raindrop momentum triggers growth of leaf-associated populations of Pseudomonas syringae on field-grown snap bean plants. Appl Environ Microbiol 62:2560–2566
Duveiller E (1994) A study of Xanthomonas campestris pv. undulosa populations associated with symptomless wheat leaves. Parasitica 50:109–117
Jacobs JL, Carroll TL, Sundin GW (2005) The role of pigmentation, ultraviolet radiation tolerance and leaf colonization strategies in the epiphytic survival of phyllosphere bacteria. Microb Ecol 49:104–113. https://doi.org/10.1007/s00248-006-9175-0
Thompson IP, Bailey MJ, Fenlon JS, Fermor TR, Lilley AK, Lynch JM, McCormack PJ, McQuilken MP, Purdy KJ, Rainey PB, Whipps JM (1993) Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar-beet (Beta vulgaris). Plant Soil 150:177–191
Woody ST, Ives AR, Nordheim EV, Andrews JH (2007) Dispersal, density dependence, and population dynamics of a fungal microbe on leaf surfaces. Ecology 88:1513–1524. https://doi.org/10.1890/05-2026
Pedgley DE (1991) Aerobiology: the atmosphere as a source and sink for microbes. In: Andrews JH, Hirano SS (eds) Microbial ecology of leaves. Springer, New York, pp 43–59
Stone BWG, Jackson CR (2019) Canopy position is a stronger determinant of bacterial community composition and diversity than environmental disturbance in the phyllosphere. FEMS Microbiol Ecol 95:1–11. https://doi.org/10.1093/femsec/fiz032
Green J, Bohannan BJM (2006) Spatial scaling of microbial biodiversity. Trends Ecol Evol 21:501–507. https://doi.org/10.1016/j.tree.2006.06.012
Voriskova J, Baldrian P (2013) Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J 7:477–486. https://doi.org/10.1038/ismej.2012.116
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Rasband W (1997) ImageJ
Urbanek S (2019) Jpeg: read and write JPEG images
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16s rRNA-based studies. PLoS One 6. https://doi.org/10.1371/journal.pone.0027310
Jackson C, Stone B, Tyler H (2015) Emerging perspectives on the natural microbiome of fresh produce vegetables. Agriculture 5:170–187. https://doi.org/10.3390/agriculture5020170
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. https://doi.org/10.1093/nar/gkm864
McMurdie PJ, Holmes S (2014) Waste not want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. https://doi.org/10.1371/journal.pcbi.1003531
Team RC (2018) R: a language and environment for statistical computing
Breiman L (2017) Classification and regression trees. Routledge, Abingdon
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2:18–22
Kuhn M, Wing J, Westion S, et al (2017) Caret: classification and regression training
Oksanen J, Blanchet F, Kindt R, et al (2018) Vegan: community ecology package
Podani J, Schmera D (2011) A new conceptual and methodological framework for exploring and explaining pattern in presence - absence data. Oikos 120:1625–1638. https://doi.org/10.1111/j.1600-0706.2011.19451.x
Dray S, Blanchet G, Borcard D, et al (2017) adespatial: multivariate multiscale spatial analysis
Redford AJ, Fierer N (2009) Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol 58:189–198. https://doi.org/10.1007/s00248-009-9495-y
Jackson CR, Denney W (2011) Annual and seasonal variation in the phyllosphere bacterial community associated with leaves of the southern Magnolia (Magnolia grandiflora). Microb Ecol 61:113–122
Williams TR, Moyne AL, Harris LJ, Marco ML (2013) Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce Phyllosphere. PLoS One 8:1–14. https://doi.org/10.1371/journal.pone.0068642
Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS (2015) Seasonal community succession of the phyllosphere microbiome. Mol Plant-Microbe Interact 28:274–285. https://doi.org/10.1094/MPMI-10-14-0331-FI
Last FT (1955) Seasonal incidence of Sporobolomyces on cereal leaves. Trans Br Mycol Soc 38:221–239. https://doi.org/10.1016/S0007-1536(55)80069-1
Kuehn KA, Ohsowski BM, Francoeur SN, Neelyb RK (2011) Contributions of fungi to carbon flow and nutrient cycling from standing dead Typha angustifolia leaf litter in a temperate freshwater marsh. Limnol Oceanogr 56:529–539. https://doi.org/10.4319/lo.2011.56.2.0529
Su R, Kuehn KA, Phipps SW (2015) Fungal contributions to carbon flow and nutrient cycling during decomposition of standing Typha domingensis leaves in a subtropical freshwater marsh. Freshw Biol 60:2100–2112. https://doi.org/10.1111/fwb.12635
Vacher C, Hampe A, Porté AJ, Sauer U, Compant S, Morris CE (2016) The phyllosphere: microbial jungle at the plant–climate interface. Annu Rev Ecol Evol Syst 47:1–24. https://doi.org/10.1146/annurev-ecolsys-121415-032238
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci 106:16428–16433. https://doi.org/10.1073/pnas.0905240106
Atamna-Ismaeel N, Finkel O, Glaser F, von Mering C, Vorholt JA, Koblížek M, Belkin S, Béjà O (2012) Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ Microbiol Rep 4:209–216. https://doi.org/10.1111/j.1758-2229.2011.00323.x
Buczolits S, Busse H-J (2011) Hymenobacter. In: Krieg NR, Ludwig W, Whitman W et al (eds) Bergey’s manual of systematic bacteriology, vol 4. 2nd edn. Springer, New York, pp 397–404
Buczolits S, Denner EBM, Vybiral D, Wieser M, Kämpfer P, Busse HJ (2002) Classification of three airborne bacteria and proposal of Hymenobacter aerophilus sp. nov. Int J Syst Evol Microbiol 52:445–446. https://doi.org/10.1099/00207713-52-2-445
Buczolits S, Denner EBM, Kämpfer P, Busse HJ (2006) Proposal of Hymenobacter norwichensis sp. nov., classification of “Taxeobacter ocellatus”, “Taxeobacter gelupurpurascens” and “Taxeobacter chitinovorans” as Hymenobacter ocellatus sp. nov., Hymenobacter gelipurpurascens sp. nov. and Hymenobacter chitinivo. Int J Syst Evol Microbiol 56:2071–2078. https://doi.org/10.1099/ijs.0.64371-0
Antibus DE, Leff LG, Hall BL, Baeseman JL, Blackwood CB (2012) Cultivable bacteria from ancient algal mats from the McMurdo dry valleys, Antarctica. Extremophiles 16:105–114. https://doi.org/10.1007/s00792-011-0410-3
Venkata Ramana V, Sasikala C, Takaichi S, Ramana CV (2010) Roseomonas aestuarii sp. nov., a bacteriochlorophyll-a containing alphaproteobacterium isolated from an estuarine habitat of India. Syst Appl Microbiol 33:198–203. https://doi.org/10.1016/j.syapm.2009.09.004
Zhang YQ, Yu LY, Wang D, Liu HY, Sun CH, Jiang W, Zhang YQ, Li WJ (2008) Roseomonas vinacea sp. nov., a gram-negative coccobacillus isolated from a soil sample. Int J Syst Evol Microbiol 58:2070–2074. https://doi.org/10.1099/ijs.0.65789-0
Gallego V, Sánchez-Porro C, García MT, Ventosa A (2006) Roseomonas aquatica sp. nov., isolated from drinking water. Int J Syst Evol Microbiol 56:2291–2295. https://doi.org/10.1099/ijs.0.64379-0
Jiang CY, Dai X, Wang BJ et al (2006) Roseomonas lacus sp. nov., isolated from freshwater lake sediment. Int J Syst Evol Microbiol 56:25–28. https://doi.org/10.1099/ijs.0.63938-0
Finster KW, Herbert RA, Lomstein BA (2009) Spirosoma spitsbergense sp. nov. and Spirosoma luteum sp. nov., isolated from a high Arctic permafrost soil, and emended description of the genus Spirosoma. Int J Syst Evol Microbiol 59:839–844. https://doi.org/10.1099/ijs.0.002725-0
Acknowledgments
We would like to acknowledge Kristin Warner and Jessie Smith for their help in the collection and laboratory work associated with this project.
Sequence Data
The sequence data reported are available in the SRA database under the project accession number PRJNA487794.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Significance
This research considers the importance of both short-term environmental variation and seasonal patterns in phyllosphere bacterial assembly more directly than in previous work. Recent next-generation sequencing studies of the phyllosphere microbial community across seasonal boundaries have shown clear shifts in composition and diversity following leaf senescence. However, these studies have not effectively investigated how factors such as rain, wind, solar radiation, and temperature interact with leaf senescence in driving community composition changes. This work advances the field by shedding light into these causative factors as they affect the foliar plant microbiome. Understanding the development of bacterial epiphyte communities in response to these drivers forms a significant and original contribution to our understanding of microbial dynamics in the plant phyllosphere. In addition, we apply a machine learning regression model to determine the importance of multiple correlated climatic variables.
Electronic supplementary material
ESM 1
(PDF 455 kb)
Rights and permissions
About this article
Cite this article
Stone, B.W.G., Jackson, C.R. Seasonal Patterns Contribute More Towards Phyllosphere Bacterial Community Structure than Short-Term Perturbations. Microb Ecol 81, 146–156 (2021). https://doi.org/10.1007/s00248-020-01564-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01564-z