Abstract
Byers Peninsula (Livingston Island, Antarctica), the largest seasonally ice-free region of the Maritime Antarctica, holds a large number of lakes, ponds, and streams. The prokaryotic structure and bacterial diversity in sediment samples collected during the 2008–2009 austral summer from five inland lakes, two coastal lakes, and an estuarine site were analyzed by Catalyzed Reporter Deposition Fluorescence In Situ Hybridization (CARD-FISH) and 16S rRNA 454 tag pyrosequencing techniques, respectively. Differently from inland lakes, which range around the oligotrophic status, coastal lakes are eutrophic environments, enriched by nutrient inputs from marine animals. Although the prokaryotic abundances (estimated as DAPI stained cells) in sediment samples were quite similar among inland and coastal lakes, Bacteria always far dominated over Archaea. Despite the phylogenetic analysis indicated that most of sequences were affiliated to a few taxonomic groups, mainly referred to Proteobacteria, Bacteroidetes, and Actinobacteria, their relative abundances greatly differed from each site. Differences in bacterial composition showed that lacustrine sediments were more phyla rich than the estuarine sediment. Proteobacterial classes in lacustrine samples were dominated by Betaproteobacteria (followed by Alphaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria), while in the estuarine sample, they were mainly related to Gammaproteobacteria (followed by Deltaproteobacteria, Epsilonproteobacteria, Alphaproteobacteria, and Betaproteobacteria). Higher number of sequences of Alphaproteobacteria, Cyanobacteria, Verrucomicrobia, and Planctomycetes were observed in sediments of inland lakes compared to those of coastal lakes, whereas Chloroflexi were relatively more abundant in the sediments of coastal eutrophic lakes. As demonstrated by the great number of dominant bacterial genera, bacterial diversity was higher in the sediments of inland lakes than that in coastal lakes. Ilumatobacter (Actinobacteria), Gp16 (Acidobacteria), and Gemmatimonas (Gemmatimonadetes) were recovered as dominant genera in both inland and coastal lakes, but not in the estuarine sample, indicating that they may be useful markers of Antarctic lakes. The proximity to the sea, the different lake depths and the external or internal origin of the nutrient sources shape the bacterial communities composition in lacustrine sediments of Byers Peninsula.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The Maritime Antarctic region, which comprises the western side of the Antarctic Peninsula and the nearby Subantarctic islands, is characterized by a less extreme climate with higher mean temperatures and more precipitation than southern Antarctic areas. Byers Peninsula, the largest seasonally ice-free region of the Maritime Antarctica, includes a large number of lakes, ponds, and streams [1]. Most lakes are shallow and unstratified, or the deepest ones (up to 9 m in Midge Lake), mainly inland lakes located in the central plateau, are cold monomictic, with a summer mixing ice-free period and winter stratification below the ice cap [2].
The harsh environmental conditions in Antarctic lakes explain the dominance of microorganisms [3]. Culture-independent molecular approaches, mainly based on PCR amplification of small subunit ribosomal RNA sequences, demonstrated that the microbial communities inhabiting the water column in Maritime Antarctic lakes consist of relatively low number of taxa, including a variety of viruses, Bacteria, Archaea, heterotrophic protists, and algae, compared to temperate lakes [4–6]. Bacterioplankton diversity of some Antarctic lakes, investigated by using genetic fingerprinting techniques, showed a close relationship between geographical (proximity to the sea), physical (depth), and chemical features (inorganic dissolved nutrients and chlorophyll-a), and the influence of lake’s catchment processes on bacterial diversity [7, 8].
Even under the summer homogeneous physical conditions, the presence of benthic mosses (Drepanocladus longifolius) covering the bottom of the deepest lakes can be an important source of bacterial diversity in these aquatic systems [1, 9]. Benthic mosses may favor a “biological stratification,” causing strong differences in the relative abundance of the dominant bacterial taxa within the water column [7, 10]. The development of distinct microbial populations in the deep part of the lakes of Byers Peninsula compared to surface waters has been previously reported [8, 11].
To gain more information on the microbiota of lacustrine ecosystems of Byers Peninsula, in our study, sediments were collected from five inland lakes located in the central plateau (Limnopolar, Somero, Domo, Chica, and Turbio), and from two coastal lakes (Maderos and Refugio). The studied lakes represent the lacustrine environmental heterogeneity in the Peninsula [7], since the trophic status of inland lakes ranges from ultraoligotrophic to mesotrophic, whereas coastal lakes display eutrophic conditions because marine animals enrich the waters with organic materials [1, 7]. Similar trophic conditions were described in other coastal lakes of the Maritime Antarctica (e.g., the Pingüi Pond, located in the Hope Bay) [4]. Lake sediments are one of the most complex microbial habitats, where prokaryotes give the main contribution to the transformation of organic carbon, sulfur, nitrogen compounds, and metals, and therefore, they play an important role in nutrient cycling and food webs. In studies of the structure and function of aquatic ecosystems, reliable estimates of microbial numbers, diversity, and activity are critical [12].
The aim of the present work is to describe, for the first time, the prokaryotic community structure and bacterial composition in the sediments from both inland and coastal Maritime Antarctic lakes, ranging from oligotrophic to eutrophic conditions, and to relate the Bacteria associated with sediments with the main ecological features of the studied environments, including the sediment biogeochemical features.
Materials and Methods
Study Area and Sampling
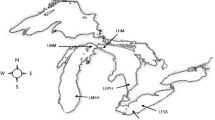
Byers Peninsula (Livingston Island, South Shetland Islands) is a 60.6-km2 area that remains ice-free during the summer period. It is located between latitudes 62° 34′ 35″–62° 40′ 35″ S and longitudes 60° 54′ 14″–61° 13′ 07″ W and has been designed as the Antarctic Specially Protected Area (ASPA) no. 126 due to important natural and historical values [13]. In the framework of the International Polar Year and coexisting with a high number of parallel studies in the area [14], surface sediment samples were collected during the austral summer 2008–2009 from five oligotrophic, inland lakes located in the central plateau of the Peninsula, coded as previously designed [1] (L1, Lake Limnopolar; L2, Lake Somero; L8, Lake Domo; L11, Lake Chica; L15, Lake Turbio) and from two coastal lakes (L5, Lake Maderos, and L6, Lake Refugio) located at the President Beaches and South Beaches, respectively. An estuarine sediment sample (S1) was collected as well at the mouth of the Petreles stream into the ocean, whose sediments are regularly affected by tides (Fig. 1).
Sampling was performed manually by using acid-washed, alcohol-sterilized polycarbonate corers (15 mm in diameter, 5 cm length) and 2-cm depth sediment cores were obtained. For each sample, several sediment cores were immediately placed in acid-washed plastic bags (Whirlpak) or in PE acid-washed bottles, then pretreated in situ within 5 h after sampling, as indicated below, and later shipped to our labs in Europe under the selected storage conditions. Subsamples for microbial abundance estimation were fixed with formaldehyde 2 % (v/v; final concentration), whereas subsamples for chemical analyses and DNA extraction were directly stored at −20 °C, and a replicate for DNA analysis was added by RNA-later stabilization reagent (Qiagen) until further processing.
Chemical Characterization of Water and Sediment Samples
Some variables were determined in situ, including maximum depth, temperature, pH, and conductivity, that were measured using the appropriate sensors, as previously described [2, 10]. The concentrations of dissolved nitrogen, nitrite plus nitrate, measured by the cadmium reductive method, and ammonium, phosphorus (orthophosphate), and soluble silica compounds in water were determined in the lab on in situ-filtered samples following standard methods [15]. Total nitrogen (TN) and total phosphorus (TP) in water were determined on nonfiltered samples after digestion, using the same method described below for the sediments, but in this case on 20 ml water samples. All these analyses were performed on samples frozen immediately after sampling, then melt in the lab in Europe prior to the analyses. For the dissolved organic carbon (DOC) determinations, water was filtered in situ through 0.2-μm cellulose nitrate filters and then fixed with 0.2 ml of 1 N HCl; these samples were stored in acid-washed glass bottles at 4 °C and analyzed using a Shimadzu TOC-V CSN analyzer. Seston (that includes phytoplankton) samples were collected through filtration of a certain water volume on GF/F glass fiber filters (47 mm in diameter, Whatman) for the determination of chlorophyll-a concentration. Filters were kept frozen until the moment of the high-performance liquid chromatography (HPLC) analyses, which were performed on acetone extracts as previously described [16].
Moisture, organic matter, and carbonate content of the sediments were determined using gravimetric analysis [15]. Dry weight was determined after drying the sediments at 105 °C for 3 h. Subsequently, the weight of organic matter was obtained after ignition of the sample at 460 °C for 6 h by subtracting the weight loss from the previously determined dry weight. Then, the same procedure was followed to determine the carbonate content after ignition at 960 °C. For elemental (TN and TP) analyses, melted samples were first dried at 50 °C until obtaining a stable weight, and subsequently, they were ground to powder in a mortar. Total nitrogen was determined by adding NaOH–persulfate digestion reagent (6 g l−1 NaOH and 6 g l−1 K2S2O8 final concentration), then digested for 2 h at 150 °C. The resulting nitrate was determined by the UV spectroscopy technique [17]. Total phosphorus was determined after a persulfatic-acid hydrolysis (0.072 N sulfuric acid and 12 g l−1 potassium peroxodisulfate final concentrations) of samples at 150 °C for 2 h. The obtained orthophosphate was determined using an ascorbic acid reduction of the phosphomolybdate complex following the Murphy and Riley method as for orthophosphate in water samples [15]. Chemical oxygen demand (COD) in the sediments was determined after oxidation of the organic matter in a strong sulfuric acid medium by K2Cr2O7 at 150 °C, with back titration. The content of photosynthetic pigments in the sediments (i.e., chlorophylls and taxa-specific carotenoids) was assessed by HPLC [16]. Pigments were extracted from a suitable amount of sediment from each sample in pure acetone by vortexing and sonication. This procedure was repeated several times until the absorbance of the last extract did not exceed the 1 % of the first one. Peaks were identified by comparing them with those of pure standards purchased from DHI (Denmark). The potential hydrolytic capability in the sediments was explored by measuring fluorescently the degradative activity of endo-1,4-β-glucanase (cellulase). Accordingly, the fluorogenic molecule 4-methyl-umbelliferyl-beta-cellobiose served as substrate [18] for enzymatic activities determined at 4 and 14 °C, each in duplicates.
Prokaryotic Cells Abundance
Cell Detachment from Sediments by Density Gradient Centrifugation
To detach cells from the slurry sediment fixed with formaldehyde, chemical and physical treatments were performed as previously described [19] with some minor modifications. Briefly, 1 g of wet slurry sediment was mixed with sodium pyrophosphate (0.1 %, v/v; Sigma-Aldrich) and Tween 20 (0.5 %, v/v; Research Organics Inc.), and then sonicated for 20 cycles of 30 s in ice by using a Brandelin SonoPlus HD 200 (Probe MS 72/D). Density gradient centrifugation was performed using Hystodenz™ as density nonionic gradient medium (Sigma-Aldrich). One volume in Histodenz concentration (1.310 g ml−1 of sterile distilled water) was carefully added to one volume of each slurry sediment by using a syringe needle with adequate length to reach the bottom of a tube. Samples were centrifuged (14,000g for 45 min at 4 °C) to allow the formation of four distinct layers (i.e., supernatant, cell layer, Histodenz cushion, and sediment pellet). The cell layer was carefully collected by an Eppendorf tip and used for subsequent analyses.
Abundance of Prokaryotic Cells Determined by DAPI Staining
To determine the abundance of total prokaryotic cells (total counts, TC), harvested cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) fluorochrome (1 μg ml−1, final concentration) and filtered through black polycarbonate membrane filters (0.2-μm pore size, 25-mm diameter, Nuclepore Corporation, Pleasanton, USA). TC were evaluated by using epifluorescence microscopy (Olympus BX-60M, at ×1000 magnification) [20].
Enumeration of Bacteria and Archaea by CARD-FISH
Catalysed Reporter Deposition-Fluorescence In Situ Hybridization (CARD-FISH) was used on harvested cells to estimate the abundance of microorganisms ascribed to Bacteria and Archaea domains, according to a protocol previously reported [21] with slight modifications. Probes EUB338 I, EUB338 II, and EUB338 III were combined in a mixture (EUB338 I–III) to enumerate bacterial cells, and ARCH915 probe [22] was used to evaluate archaeal cells. Oligonucleotides labeled with the cyanine dye Cy3 were purchased from ThermoHybaid (Interactiva Division, Ulm, Germany). Briefly, filter sections were prepared for hybridization by first embedding the cells with 0.2 % (w/v) low-gelling point agarose and drying at 37 °C, then inactivating the endogenous peroxidases by submerging filters 10 min in 0.01 M HCl. Permeabilization of cells was conducted in a lysozyme solution (10 mg ml−1 lysozyme; 0.05 M EDTA; 0.1 M Tris–HCl, pH 8) at 37 °C for 60 min, followed by washing in deionized sterile water and absolute ethanol. The hybridization was carried out at 46 °C for 2.5 h in a 300:1 mix of hybridization buffer (0.36 M NaCl; 8 mM Tris-HCl, pH 8; 40 mg ml−1 dextran sulfate; 35 % formamide; 0.4 % Roche Blocking Reagent; 0.08 % SDS) and horseradish peroxidase-conjugate probes (working solutions 50 ng μl−1; Biomers, Ulm, Germany). For signal amplification with catalyzed reporter deposition, filter sections were incubated in the dark for 15 min at 37 °C in a mix of amplification buffer (1× PBS, 0.1 % Roche Blocking Reagent, 2 M NaCl and 0.1 g/ml dextran sulfate), H2O2 (0.015 %) and the fluorescently labeled tyramide. Filter sections were then counterstained with DAPI at a final concentration of 1 μg ml−1 and mounted onto microscope slides with a drop of a 4:1 mixture of Citifluor-VectaShield. For each sample, between 50 and 200 cells were then counted under epifluorescence, using Olympus BX60 microscope, equipped with an appropriate filter set for Cy3.
Bacterial Community Composition Analysis
DNA Extraction
DNA was extracted from 5 g of sediment by employing the MoBio PowerMax Soil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA) according to the manufacturer’s instructions. DNA concentrations and purity were quantified by using a NanoDrop ND-1000 UV-vis Spectrophotometer (NanoDrop Technologies, USA).
Amplification of 16S rRNA Genes and Pyrosequencing
The V3-V4 region of the 16S rRNA genes was amplified by PCR. In order to reduce bias in massive sequencing, the “two-step PCR” protocol was applied [23], consisting in a first step of 20 PCR cycles with conventional PCR primers and then using 1 μl of first reaction amplicon for 5 cycles PCR with barcoded primers. For each sample, triplicate PCR reactions were set up at 0 °C under a PCR cabin by using 2.5 U of Taq Fast Start High Fidelity PCR System (Roche) in a reaction buffer containing 25 mM MgCl2, 10 mM dNTPs, 10 mg ml−1 BSA (New England Biolabs), and 0.4 μM of the two universal bacterial primers Bact341F (5′-CCTACGGGAGGCAGCAG-3′) and Bact805R (5′-GACTACCAGGGTATCTAAT-3′). The three reactions were pooled and used for the second PCR with the same conditions. Amplicons were then purified using a 0.8 % agarose gel (w/v) and a QIAquick Gel Extraction Kit (Qiagen) following manufacturer’s instructions. The following thermal cycling scheme was used: 94 °C for 3′, 20 cycles (5 cycles for the second PCR) of 94 °C 15″, 55 °C 45″, 72 °C 60″, final extension at 72 °C for 8′. The sequences of the partial 16S rRNA genes were determined by using a Roche GS-FLX 454 pyrosequencer (Roche, Mannheim, Germany), following the instructions of the manufacturer for amplicon sequencing.
Postrun Analysis
All the raw reads were treated with the Pyrosequencing Pipeline Initial Process [24] of the Ribosomal Database Project (RDP) in order to sort those exactly matching the specific barcodes into different samples, to trim off the adapters, barcodes and primers using the default parameters, and to remove sequences containing ambiguous “N” or shorter than 150 bp [25]. Selected reads were denoised using the “pre.cluster” command in Mothur platform [26] to remove sequences that are likely due to pyrosequencing errors [27]. The average length of all bacterial sequences without the primers was 515 bp. Chimeric sequences were excluded by using USEARCH (http://www.drive5.com/usearch/) [28].
Analyses of Bacterial Communities
To generate taxonomic profiles, sequences were assigned to taxonomic groups by using the Naïve Bayesian classifier v.2.1 [29] from RDP, with a bootstrap cutoff of 80 %. Reads were clustered in operational taxonomic units (OTUs) at 97 % pairwise identities using UCLAST [28]. Diversity analyses were conducted using the software program Mothur (http://www.mothur.org/wiki/Download_mothur) [26]. The observed richness (OTUs at 97 % similarity level), the nonparametric estimators of richness (abundance-based coverage estimator, ACE) and Chao1, and the Shannon diversity index (H′) were computed for all recovered high-quality sequences from each sample.
To compare the bacterial community compositions across groups of samples, distinguished as inland and coastal lakes, Bray–Curtis similarity analyses were performed and similarity matrices were used to obtain dendrograms and nonmetric multidimensional scaling (NMDS) plots by using PRIMER 6.1.12 (Primer-E, Ltd).
Principal component analyses (PCAs) were also performed on data from selected physical and chemical properties of sediments and lake waters, and the relative abundance of significant bacterial groups. Environmental variables used in these analyses were as follows: lake depth (Depth), water electrical conductivity (Cond), and, for sediments, concentrations of chlorophyll-a (Chl-a), phaeophytin (Phaeo), organic matter (OM), total nitrogen (TN), and total phosphorus (TP), as well as the evaluation of 1,4-β-glucanase activity at 4 °C (Cellu4). Nonparametric Mann–Whitney tests were performed to evaluate differences in the relative abundance of bacterial groups. Differences were considered significant when p value was less than 0.05. Prior to this analysis, the data were log-transformed to linearize the relationships and avoid the influence of magnitude.
Results
Chemical Characterization of the Main Limnological Variables
The water of all the systems has low mineralization, with coastal lakes (L5 and L6) showing higher saline content though they are still freshwater lakes (Table 1).
Trophic status of inland lakes generally ranges around ultraoligotrophic (L8) to oligotrophic conditions, whereas coastal lakes are eutrophic (L5 and L6), as shown by the chlorophyll-a, COD, and nutrient (N and P) concentrations (Table 1).
Sediment features (Table 2) appeared in consonance with the trophic status registered on lakes’ waters. The higher abundances of TN, TP, and organic matter (COD and % OM) were recorded in sediments from the coastal lakes (L5 and L6) and from the shallowest inland Lake Somero, as well as in the Petreles river mouth where a slight accumulation of materials occurred. Sediments of Lake Somero, which are partly covered by microbial mats, also showed the highest pigment (chlorophyll-a and phaeophytin) content, while in the coastal lakes, where microbial mats were not so extensive, relatively high pigment concentrations were observed in comparison with those of the rest of the inland lakes. Chlorophyll-a to phaeophytin ratios were higher for the coastal lakes (L5 and L6) and the lowest for the estuarine S1 site. N/P molar ratios in the sediments ranged from 4.6 to 13.7, and they were always below the Redfield ratio (16:1). 1,4-β-glucanase (cellulase) activity, a measure of the degradative capacity of the sediment’s microbiota, always yielded higher rates for both the coastal lakes and the estuarine sample compared to the inland lakes, regardless the temperature at which the enzymatic activity was determined, although this activity consistently increased at 14 °C compared to 4 °C.
Prokaryotic Abundance
The abundance of total prokaryotic cells (TC) in the analyzed sediments, obtained after DAPI staining, ranged from 64.8 (L8, the ultraoligotrophic lake) to 184.0 × 105 cells g−1 (L6, coastal eutrophic lake), and was lower in all cases in comparison to those values retrieved from the estuarine S1 site (Table 3).
The abundances of cells hybridized with probes for Bacteria (EUB338 I-III) and Archaea (ARCH915) are reported in Table 3. The recovery of Bacteria by CARD-FISH related to TC (ranging from 65.27 to 74.47 % of TC) was higher in the eutrophic coastal lakes and in S1 site than that retrieved from the oligotrophic inland lakes (46.3–61.2 % of TC) (Table 3). Archaeal contribution to TC in the lakes ranged from 0.4 (L11) to 1.2 % (L8) and was 1.1 % in S1 site. Archaea to Bacteria ratio was higher (3:100) in the ultraoligotrophic inland Lake Domo than in the other lakes, and was also higher than that observed in the estuarine sediment (1:100).
Phylogenetic Diversity of Bacteria
Pyrosequencing Reads
The pyrosequencing-based analysis of the V3-V4 region of the 16S rRNA genes for Bacteria produced 335,477 total sequences. After quality check within the RDP pyrosequencing pipeline and removing chimeras, 287,297 high-quality sequences were obtained. The highest number of high-quality reads was in sediment from lake L11 (39,661 reads), whereas the lowest value was found in the estuarine sediment (29,915 reads) (Table 4). H′ reached the highest value (4.0) in the inland L15 site and the lowest (1.9) in the coastal L6 site. Chao1 and ACE estimators predicted that the highest richness was in L1 and the lowest in L6 site. Total coverage (TCov) ranged from 47.6 (L15) to 52.7 % (L8) as estimated by Chao1, and from 31.3 (L1) to 69.4 % (L2) by ACE. The diversity index and the observed richness (OTUs) did not show a general symmetric pattern, since L15, with the highest H′ showed the lowest number of OTUs, even though L6, with the lowest H′, displayed the lowest number of OTUs.
Bacterial Taxa
A total number of 23 different bacterial phyla were retrieved, of which 16 were common to all samples. The estuarine sediment was less phylum rich than those from lacustrine sites, since four taxa (i.e., Clamydiae, OP10, BRC1, and Lentisphaerae) were absent in S1 (Table S1 in the supplemental material). Overall, sequences of the dominant taxonomic groups (abundance ≥1 %) across all sediment samples were mostly affiliated with Proteobacteria (range 15.6–40.0 % of high-quality sequences), Actinobacteria (range 8.6–27.8 %), Bacteroidetes (range 7.5–38.0 %), and Verrucomicrobia (1.2–6.1 %). However, the relative abundance at phylum level varied considerably across the different samples, determining different bacterial assemblages (Fig. 2; Table S1). Proteobacteria was the predominant phylum in L1, L2, L11, L5, and S1, whereas Actinobacteria was the predominant group in L8, L15, and L6. Differences in relative abundances were also observed for sequences affiliated to proteobacterial classes: in the lacustrine samples, they were mainly referred to Betaproteobacteria (followed by Alphaproteobacteria, Deltaproteobacteria, Gammaproteobacteria, and Epsilonproteobacteria), while in the estuarine samples, they were mainly related to Gammaproteobacteria (followed by Deltaproteobacteria, Epsilonproteobacteria, Alphaproteobacteria, and Betaproteobacteria).
Comparison of bacterial community composition in sediments collected from inland lakes (L1, L2, L8, L11, and L15), coastal lakes (L5 and L6), and in the estuarine sediment (S1). Others included the following taxa: Nitrospira, WS3, Spirochaetes, SR1, Fusobacteria, Chlamydiae, OP10, BRC1, Lentisphaerae, Fibrobacteres, and Deinococcus-Thermus
A very different community composition was observed in the sediment from S1 site, since sequences affiliated to Proteobacteria, Bacteroidetes, and Actinobacteria covered almost all classified bacterial sequences (Fig. 2). Firmicutes were more abundant (3.4 %) in the estuarine S1 site than in lacustrine ones (<1.8 %). Acidobacteria (6.5–20.9 %), Gemmatimonadetes (2.2–3.8 %), and Verrucomicrobia (3.0–6.1 %) were abundant in the lacustrine sediments, while they constituted a minor component in the estuarine sample. Exception was Lake Chica, where the lowest abundance of Actinobacteria was retrieved (Fig. 2).
The bacterial community composition of sediments of both coastal lakes differed from those determined for inland lakes, and also greatly differed each other (Fig. 2). In fact, Proteobacteria were there less abundant than in the inland lakes, but they resulted slightly more abundant than the Actinobacteria and Bacteroidetes in L5. Conversely, Actinobacteria resulted more abundant than the Proteobacteria and Bacteroidetes in L6.
Cyanobacterial abundance varied considerably between estuarine and lacustrine sediments, where they were more abundant in inland (range 2.5–10.2 %) than in coastal lakes (range 0.3–0.7 %). Cyanobacterial sequences were particularly abundant in sediments from L2, L11, and L15, whose bed was partly covered by microbial mats as lake L2 or presented partial mat coverage in the shores and higher picocyanobacterial abundance in the water column (L11 and L15). In lacustrine samples, relatively high abundant phyla also included TM7, OD1, Planctomycetes, and Chloroflexi. All the latter were particularly scarce in the estuarine site.
Among the low abundant phyla (abundance <1 %), Nitrospira, WS3, Spirochaetes, and SR1 occurred across all sediment samples, even if at different relative abundances. Sequences related to Chlamydiae and OP10 were recovered in lacustrine but not in the estuarine sediments. Sequences affiliated with Fibrobacteres and Deinococcus-Thermus occurred in all samples but were not retrieved in L6 and L2, respectively. Sequences related to Fusobacteria were present only in samples from L5 and S1. BRC1 and Lentisphaerae were absent in samples L1, L8, and S1, while Lentisphaerae were only retrieved in L11 (Table S1). NMDS diagram, representing similarities in the bacterial community composition (phyla and proteobacterial classes) of the Antarctic lacustrine sediment samples, grouped all inland lakes except Lake Chica, whereas the coastal lakes (L5 and L6) did not cluster together because of the big differences among their respective bacterial communities (Fig. 3).
PCA based on selected physico-chemical properties of lake sediments and waters, as well as on the relative abundance of the main bacterial groups (Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, Cyanobacteria, Verrucomicrobia, and Chloroflexi), was performed to identify groups of samples with similar community compositions and to find their relationships with the environmental variables (Fig. 4). The PCA showed that the bacterial communities from inland lakes differed from those of coastal lakes, confirming results obtained by NMDS analysis that examined only the bacterial composition (Fig. 3). The two main components explained 76.1 % of the total variance. Axis 1 (explaining 55.6 % of the variance) was strongly associated with the relative abundance of Acidobacteria and with a combination of physical and chemical variables (TP, Cellu4, OM, depth, Chl-a, and Cond) that are strictly related to the trophic status of the lakes. Instead, Axis 2 (20.5 % of the variance) was mainly related to the abundances of Bacteroidetes, Deltaproteobacteria, and Chlorofexi and to TN concentrations.
Principal component analysis based on selected physico-chemical variables and the most significant bacterial populations associated with sediment samples from inland lakes (L1, L2, L8, L11, and L15) and coastal lakes (L5 and L6). Variable examined were as follows: lake depth (Depth), water electrical conductivity (Cond) and, for the sediments, chlorophyll-a (Chla) and phaeophytin (Phaeo) concentrations, relative organic matter content (OM), total nitrogen (TN), total phosphorus (TP), and 1,4-β-glucanase (cellulase) activity at 4 °C (Cellu4). Bacterial phyla/classes: Alphaproteobacteria (Alpha), Betaproteobacteria (Beta), Deltaproteobacteria (Delta), Acidobacteria (Acidob), Actinobacteria (Actinob), Bacteroidetes (Bactero), Cyanobacteria (Cyanob), Chloroflexi (Chlorof), and Verrucomicrobia (Verruc)
The two coastal lakes (L5 and L6), with high factor scores for the Axis 1 and Axis 2, appeared clearly separated from the other lakes by Axis 1. Although distantly, the shallowest inland L2 appeared closer to the coastal lakes in Axis 1, mainly due to the influence of the high values of its nutrient concentrations and chlorophyll-a content (Table 2), which are markers of higher trophic status. The negative side of Axis 1 was associated to Verrucomicrobia and Cyanobacteria, and that of Axis 2 to Actinobacteria and Betaproteobacteria. Deeper inland lakes L1, L15, and L8 grouped together, as occurred in the NMDS plot (Fig. 3), and were distinct from L11, which showed the highest factor score for Axis 2 (Fig. 2).
Bacterial Genera
Of the total high-quality bacterial sequences, about 64 % were not classified at genus level. A total of 1107 genera were resolved from the rest, 40 of which were ubiquitous to all samples. The dominant bacterial genera (41), which occurred in ≥1 % of the total bacterial sequences at least in one of the eight samples, are shown in Table 5. Almost all the dominant genera retrieved from the estuarine sediment (14/15) were not retrieved in lacustrine sediments, with the only exception of OD1 that was also collected in L8 and L11 sites. Differently from lacustrine sediments, genera identified in the estuarine sediment were characteristically related to Gammaproteobacteria, Epsilonproteobacteria, and Bacteroidetes. Genera within Alphaproteobacteria were only retrieved from inland lacustrine sediments, among which Sphingomonas was particularly abundant in L8 and L15 sites. Ilumatobacter (Actinobacteria), Gp16 (Acidobacteria), and Gemmatimonas (Gemmatimonadetes) were recovered in all inland and coastal lakes, but not in the estuarine sample. Ferruginibacter (Bacteroidetes) was retrieved in all inland lakes and only in one of the two coastal lakes (L6). Nine genera were retrieved only in one of the seven lakes and may be considered distinctive of each lake. Prolixibacter (Bacteroidetes) and GpI (Cyanobacteria) were present only in L2 site. Within Actinobacteria, genera referred to Marmoricola and Cryobacterium were unique in L8, as well as Conexibacter in L15, and Gp3 (Acidobacteria) in L5. Hymenobacter (Bacteroidetes) was only retrieved in L15, as Verrucomicrobium in L6, and Longilinea (Chloroflexi) in L5.
Discussion
The water bodies studied in the present work are representative of the main lake types from Maritime Antarctica, differing in their trophic status, morphological features, and their distance to the sea, which in turn is linked to the relative influence of sea animals causing lake eutrophication [7]. Limnological properties (evaluated as chlorophyll-a and nutrient concentrations), recorded in water and sediment samples, distinguished those inland lakes as oligotrophic, with two opposite exceptions (L2 and L8), and the coastal ones as eutrophic.
The estuarine S1 site represented a totally different environment, since sediments are directly exposed to the effects of the sea, such as the diel fluctuations in the salinity. Among the inland lakes, the shallowest Lake Somero, partly covered by biofilms and microbial mats and surrounded by mosses, showed limnological features more similar to those of eutrophic coastal lakes (L5 and L6) than the other inland lakes. In contrast, the inland Lake Domo was considered ultraoligotrophic, since very low chlorophyll concentrations were always recorded, and inorganic nutrients were almost undetectable. Chlorophyll-a to phaeophytin ratios, which are indicative of the physiological status and photosynthetic capacity of primary producers, were also higher for the coastal lakes, in concordance with their much higher external nutrient inputs. Differently, this ratio was the lowest for the estuarine sample, indicating higher degradation in the populations of photosynthetic organisms or even a major abundance of detritus. The external sources of organic matter (mainly from sea animals) greatly influenced the trophic status of the coastal lakes sediments. When considering N/P ratios in the studied lakes, a relative low abundance of N was observed, especially in L11 and L15 sites, indicating that the sediments could represent a relatively better source of phosphorus than of nitrogen. According to the organic matter availability in sediments, the 1,4-β-glucanase (cellulase) activity (i.e., a measure of the degradative capacity of the sediment’s microbiota) was higher in the coastal lakes and the estuarine sample than in the inland lakes, with the only exception of the shallow Lake Somero, which showed intermediate characteristics among both types of lakes. The cellulase activity at 14 °C was higher than that at 4 °C, and it was positively correlated (p < 0.01) with bacterial abundances, suggesting that mesophilic microorganisms could be, at least partially, responsible for this activity.
Microorganisms may greatly influence the functioning of environments, including those characterized by very harsh conditions as the Antarctic lacustrine sediments. Beside similar prokaryotic abundance in inland and coastal lakes, Archaea represented 1–3 % of the total community. In comparison with the other lakes, the sediment from the ultraoligotrophic Lake Domo displayed the lowest prokaryotic abundances and the highest Archaea to Bacteria ratio, which may suggest that bacterial and archaeal populations were differently affected by the trophic conditions. It is consistent with data reported a negative correlation between total archaeal rRNA gene levels and chlorophyll a concentration from other environments in Antarctica [30].
Despite the high-throughput sequencing efforts, the values for the ACE and Chao1 estimators indicated that the diversity was not totally well covered for the bacterial community associated with the studied sediment samples (Table 4). Sequence reads unclassified at phylum level greatly varied among samples (10 % of total reads in the estuarine site, about 20 % in inland lakes and >30 % in coastal lakes), and they may be considered as representatives of novel, not yet described bacterial taxa.
As estimated by the Shannon H′ index, bacterial diversity showed a large variation among the examined sites (Table 4). As generally accepted [31], moderately disturbed conditions often result in high diversity of communities, like those in the estuarine S1 site, where the large fluctuations of the key physico-chemical parameters may allow for the coexistence of more diverse bacterial assemblages. The proximity to the sea may also differently affect the bacterial diversity and richness, as observed in the coastal Maderos and Refugio lakes. This was sustained by the multivariate analysis, since variables other than total phosphorous and nitrogen content in sediments, such as lake depth and salinity, were also important determinants of bacterial diversity. Differently from L5, the extreme eutrophic conditions registered at L6 site can selectively favor numerically fewer bacterial taxa, best dealing with these features. Among inland lakes, bacterial community with the highest degree of diversity and the lowest richness was from the deepest lake Turbio (L15). This was substantiated by taxonomic analyses of the different bacterial phylogenetic lineages that yielded the greatest number of genera. Compared to the other inland lakes, Lake Turbio is highly turbid because of a high wind fetch, which can be a selective factor, but also shows a relatively higher level of disturbance by the stronger wind effects.
Even if the phylogenetic analysis indicated that most of sequences were affiliated with few taxonomic groups dominated by Proteobacteria, Bacteroidetes, and Actinobacteria, their relative abundances greatly differed from each site. The same groups have been previously reported as the major phyla in the planktonic bacterial communities in both Arctic and Antarctic lakes [32–34]. However, sequences related to these three phyla covered ~83 % of total bacterial sequences in the estuarine sediment, while only ~50 % in lacustrine sediments, indicating that more phylotypes are involved in the lacustrine than in the estuarine bacterial composition. In addition, sequences referred to Acidobacteria, Cyanobacteria, and Verrucomicrobia constituted a relevant fraction of total retrieved sequences from lacustrine samples. Representative sequences of Verrucomicrobia and Bacteroidetes phyla were relatively more abundant in sediments of the lakes, and both groups were also observed in the surface waters of Lake Limnopolar [8]. Acidobacteria are commonly retrieved from some Antarctic terrestrial ecosystems [35]. Acidobacteria were not found in the clone libraries obtained from the deep water sample collected from the same lake in a previous study [8].
Although Proteobacteria was the largest phylum, its major classes and their proportions varied greatly among the different sites. Proteobacterial classes in lacustrine samples were dominated by Betaproteobacteria (followed by Alpha-, Delta- and Gammaproteobacteria), while in the estuarine samples, they were mainly related to Gammaproteobacteria (followed by Deltaproteobacteria, Epsilonproteobacteria, Alphaproteobacteria, and Betaproteobacteria). The presence of Gammaproteobacteria, traditionally associated with polar marine sediments [36, 37] in the lacustrine sediments, might suggest that these bacteria are transported into inland Antarctic lakes in aerosols moved by winds, and/or by fecal pellets of seabirds [8, 10], whereas the more direct marine influence by tides most probably accounts for the dominance of Gammaproteobacteria in the estuarine site.
Concerning the affiliation of the sequences from lacustrine sediments to lower taxonomic levels, dominant retrieved phyla contained genera that have been described for polar freshwater environments. Sequences related to the genera Ilumatobacter (Actinobacteria), Gemmatimonas (Gemmatimonadetes), and Gp16 (Acidobacteria) were recovered as dominant in both inland and coastal lakes, but not in the estuarine sample, indicating that they may represent markers of sediments from Antarctic lakes. Several sequences were affiliated with the Flavobacterium genus (within the phylum Bacteroidetes) that is considered a common heterotrophic member of the Antarctic bacterial communities in both aquatic and terrestrial environments [7, 38]. Novel Flavobacterium species related to the genus have been often reported from Antarctica [39–42], with interesting biotechnological properties [35]. On the other hand, the detection of sequences affiliated with the Geobacter genus (Deltaproteobacteria), previously retrieved in a wide variety of pristine environments [35], in Antarctic inland lakes might reflect their still pristine conditions.
Sequences related to the nonphotosynthetic Longilinea genus (within the phylum Chloroflexi), including new cultured mesophilic, strictly anaerobic, heterotrophic filamentous bacteria in the class of Anaerolineae [43], were only retrieved in the coastal L5 site.
Only in the sediments of Lake Somero which are partly covered by microbial mats, dominant sequences were assigned to GpI (Cyanobacteria), which includes species capable of fixing nitrogen, suggesting an inner load of nutrients to fed phototrophic and heterotrophic organisms. The microbial mats represent a common characteristic of vast areas in Byers Peninsula where they play important trophic functions also for lakes, as its relatively high rate of primary productivity may convert them in sources of nutrients to the microorganisms in the water column, regardless they are located in the own lake bed or in the lake’s catchment area [44, 45].
Some sequences that distinguished bacterial populations of lakes L1, L8, and L15 from L11 were affiliated to potentially photosynthetic members belonging to the order Rhodobacterales (Alphaproteobacteria). Other sequences were affiliated with the genus Sphingomonas, which is a widely distributed group in aquatic (both freshwater and marine) and terrestrial environments in Antarctica and in temperate zones. This genus was also previously reported to be a dominant genus in water samples of Limnopolar and Somero lakes [1]. Among Betaproteobacteria, the most abundant group in polar freshwater ecosystems [46], several sequences were assigned to the genus Methylibium, which includes highly specialized bacteria able to use C1 compounds as a carbon source. This genus, retrieved in most of the examined lakes, was also detected in sediments from Arctic lakes by using pyrosequencing techniques [47].
Other highly abundant phyla (≥1 % of total reads) retrieved in Antarctic lacustrine sediments included also Planctomycetes members, previously found in Antarctic soils [48], suggesting that they may come from the surrounding terrestrial habitat in the highly dynamic lake catchments. At the tail of bacterial diversity, comprising <70 reads in the samples, members of the Nitrospira, Fibrobacteres, Fusobacteria, and of several other candidate divisions (WS3, OP10, BRC1) originally detected in other extreme, low-diversity environments were also found in the present study. Considerable numbers of sequence reads related to candidate divisions included TM7 and OD1, that are found in a wide range of environments [49, 50], but of which no cultivated representatives exist to date. Thus, these Antarctic sites represent interesting environments to perform culture efforts to recover more species and diversity.
Our results demonstrated that in sediments of Byers peninsula, a close relationship exists between bacterial community compositions and the limnological characteristics. Geographical location and particularly the proximity to the sea, where marine animals provide the main external nutrient source, as well as the lake depths, which are related to the higher nutrient remobilization from sediments in the shallower lakes (internal load), may greatly affect their bacterial community composition.
Particular attention is required to inland Maritime Antarctic lakes, that as remote environments in a relatively mild polar climate may be considered as sentinels of climate change, and that as a main stressor, can modify the bacterial community structure and composition.
References
Toro M, Camacho A, Rochera C, Rico E, Bañón M, Fernández-Valiente E, Marco E, Justel A, Avendaño MC, Ariosa Y, Vincent WF, Quesada A (2007) Limnological characteristics of the freshwater ecosystems of Byers Peninsula, Livingston Island, in maritime Antarctica. Polar Biol 30:635–649
Rochera C, Toro M, Rico E, Fernández-Valiente E, Villaescusa JA, Picazo A, Quesada A, Camacho A (2013) Structure of planktonic microbial communities along a trophic gradient in lakes of Byers Peninsula, South Shetland Islands. Antarct Sci 25:277–287
Ellis-Evans JC, Laybourn-Parry J, Bayliss PR, Perriss SJ (1998) Physical, chemical and microbial community characteristics of lakes of the Larsenmann Hills, Continental Antarctica. Arch Hydrobiol 141:209–230
Izaguirre I, Allende L, Marinone MC (2003) Comparative study of the planktonic communities of three lakes of contrasting trophic status at Hope Bay (Antarctic Peninsula). J Plankton Res 25:1079–1097
Laybourn-Parry J, Quayle WC, Henshaw T, Ruddell A, Marchant HJ (2001) Life on the edge: the plankton and chemistry of Beaver lake, an ultra-oligotrophic epishelf lake, Antarctica. Freshw Biol 46:1205–1217
López-Bueno A, Tamares J, Velázquez D, Moya A, Quesada A, Alcamí A (2009) High diversity of the viral community from an Antarctic lake. Science 326:858–861
Villaescusa JA, Casamayor EO, Rochera C, Velázquez D, Chicote A, Quesada A, Camacho A (2010) A close link between bacterial community composition and environmental heterogeneity in maritime Antarctic lakes. Int Microbiol 13:67–77
Villaescusa JA, Casamayor EO, Rochera C, Quesada A, Michaud L, Camacho A (2013) Unexpected heterogeneous vertical structure in the bacterioplankton community of a non-stratified Antarctic lake. Antarct Sci 25:229–238
Pearce DA, Butler HB (2002) Short term stability of the microbial community structure in a maritime Antarctic lake. Polar Biol 25:479–487
Villaescusa JA, Rochera C, Velázquez D, Rico E, Quesada A, Camacho A (2013) Bacterioplankton summer dynamics in a maritime Antarctic lake. Limnetica 32:253–268
Camacho A (2006) Planktonic microbial assemblages and the potential effects of metazooplankton predation on the food web of lakes from the maritime Antarctica and sub-Antarctic islands. Rev Environ Sci Biotechnol 5:167–185
Sjoling S, Cowan DA (2003) High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles 7:275–282
López-Martínez J, Thomson MRA, Thomson JW (eds) (1996) Geomorphological map of Byers Peninsula, Livingston Island. BAS GEOMAP Series, Sheet 5-A. British Antarctic Survey, Cambridge
Quesada A, Camacho A, Lyons WB (2013) Multidisciplinary research on Byers Peninsula, Livingston Island. Antarct Sci 25:121–122
APHA–AWWA–WEF (1992) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, Washington, D.C
Picazo A, Rochera C, Vicente E, Miracle MR, Camacho A (2013) Spectrophotometric methods for the determination of photosynthetic pigments in stratified lakes: a critical analysis based on comparisons with HPLC determinations in a model lake. Limnetica 32:139–158
Ferree MA, Shannon RD (2001) Evaluation of a second derivative UV/Visible spectroscopy technique for nitrate and total nitrogen analysis of wastewater samples. Water Res 35:327–332
Boscher HTS, Cappenberg TE (1994) A sensitive method using 4-methylumbelliferyl-3-cellobiose as a substrate to measure (1,4)-3-glucanase activity in sediments. Appl Environ Microbiol 60:3592–3596
Amalfitano S, Fazi S (2008) Recovery and quantification of bacterial cells associated with streambed sediments. J Microbiol Methods 75:237–243
Maugeri TL, Acosta Pomar MLC, Bruni V (1990) Picoplancton. In: Innamorati M, Ferrari I, Marino D, Ribera D’Alcalà M (eds) Metodi per lo studio del plancton marino, vol 11. Nova Thalassia, Trieste: Lint, pp 199–205
Pernthaler A, Pernthaler J, Amann R (2002) Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68:3094–3101
Ishii K, Mußmann M, MacGregor BJ, Amann R (2004) An improved fluorescence in situ hybridization protocol for the identification of Bacteria and Archaea in marine sediment. FEMS Microbiol Ecol 50:203–212
Berry DH, Mahfoudh KB, Wagner M, Loy A (2011) Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol 77:7846–7849
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37, D141eD145
Claesson M, O'Sullivan O, Wang Q, Nikkila J, Marchesi J, Smidt H, De Vos W, Ross R, O'Toole P (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4, e6669. doi:10.1371/journal.pone.0006669
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB et al (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12:1889–1898
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–1246
Wang QG, Garrity M, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K, DeLong EF (1998) Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol 64:2585–2595
Connell JH (1979) Tropical rain forests and coral reefs as open non-equilibrium systems. In: Anderson RM, Turner BD, Taylor LR (eds) Population dynamics. Symposium of the British Ecological Society. Blackwell Scientific Publications, Oxford, UK, pp 141–163
Møller AK, Søborg DA, Al-Soud WA, Sørensen SJ, Kroer N (2013) Bacterial community structure in High-Arctic snow and freshwater as revealed by pyrosequencing of 16S rRNA genes and cultivation. Polar Res 32:17390. doi:10.3402/polar.v32i0.17390
Mosier AC, Murray AE, Frisen CH (2007) Microbiota within the perennial ice cover of Lake Vida, Antarctica. FEMS Microbiol Ecol 59:274–288
Pearce DA, van der Gast CJ, Lawley B, Ellis-Evans JC (2003) Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol Ecol 45:59–70
Pearce DA, Hodgson DA, Thorne MAS, Burns G, Cockell CS (2013) Preliminary analysis of life within a former subglacial lake sediment in Antarctica. Diversity 5:680–702
Bowman JP, McCammon SA, Dann AL (2005) Biogeographic and quantitative analyses of abundant uncultivated gamma-proteobacterial clades from marine sediment. Microb Ecol 49:451–460
Ravenschlag K, Sahm K, Amann R (2001) Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl Environ Microbiol 67:387–395
Michaud L, Caruso C, Mangano S, Interdonato F, Bruni V, Lo Giudice A (2012) Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol Ecol 82:391–404
McCammon SA, Bowman JP (2000) Taxonomy of Antarctic Flavobacterium species: description of Flavobacterium gillisiae sp. nov., Flavobacterium tegetincola sp. nov., and Flavobacterium xanthum sp. nov., nom. rev. and reclassification of Flavobacterium salegens as Salegentibacter salegens gen. nov., comb. nov. Int J Syst Evol Microbiol 50:1055–1063
Peeters K, Hodgson DA, Convey P, Willems A (2011) Culturable diversity of heterotrophic bacteria in Forlidas Pond (Pensacola Mountains) and Lundström Lake (Shackleton Range), Antarctica. Microb Ecol 62:399–413
Van Trappen S, Vandecandelaere I, Mergaert J, Swings J (2005) Flavobacterium fryxellicola sp. nov. and Flavobacterium psychrolimnae sp. nov., novel psychrophilic bacteria isolated from microbial mats in Antarctic lakes. Int J Syst Evol Microbiol 55:769–772
Yi H, Chun J (2006) Flavobacterium weaverense sp. nov. and Flavobacterium segetis sp. nov., novel psychrophiles isolated from the Antarctic. Int J Syst Evol Microbiol 56:1239–1244
Yamada T, Imachi H, Ohashi A, Harada H, Hanada S, Kamagata Y, Sekiguchi Y (2007) Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int J Syst Evol Microbiol 57:2299–2306
Camacho A, Rochera C, Villaescusa JA, Velázquez D, Toro M, Rico E, Fernández-Valente E, Justel A, Banon M, Quesada A (2012) Maritime Antarctic lakes as sentinels of climate change. Int J Des Nat Ecodyn 7:239–250
Fernández-Valiente E, Camacho A, Rochera C, Rico E, Vincent WF (2007) Community structure and physiological characterization of microbial mats in Byers Peninsula, Livingston Island (South Shetland Islands, Antarctica). FEMS Microbiol Ecol 59:377–385
Pearce DA, Galand PE (2008) Microbial biodiversity and biogeography. In: Vincent W, Laybourn-Parry J (eds) Polar lakes and rivers, limnology of Arctic and Antarctic aquatic ecosystems. Oxford University Press, New York, pp 213–230
He R, Wooller MJ, Pohlman JW, Quensen J, Tiedje JM, Leigh MB (2012) Shifts in identity and activity of methanotrophs in Arctic lake sediments in response to temperature changes. Appl Environ Microbiol 78:4715–4723
Niederberger TD, McDonald IR, Hacker AL, Soo RM, Barrett JE, Wall DH, Cary SG (2008) Microbial community composition in soils of northern Victoria Land, Antarctica. Environ Microbiol 10:1710–1724
Harris JK, Kelley ST, Pace NR (2004) New perspective on uncultured bacterial phylogenetic division OP11. Appl Environ Microbiol 70:845–849
Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL (2001) Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol 67:411–419
Acknowledgments
This work was supported by grant from the National Antarctic Research Program (PNRA), Italian Ministry of Education and Research (project PNRA 2010/C1.04): “Studio della diversità microbica (Batteri ed Archea) nel continente antartico” to Dr. Valeria Lentini, and by grant CGL2005-06549-C02-02/ANT from the Spanish Ministry of Education and Science to AC (co-financed by European FEDER funds). We thank Ciro Rappazzo for his help in sediment analyses in our lab at the University of Valencia. Logistics for sample collection was also partly supported by the IPY Byers Peninsula/Limnopolar Project (POL2006-06635) funded by the Spanish Ministry of Science and Technology. We are very grateful to the rest of members of the Limnopolar Research Team for their invaluable help in the field site, as well as to the UTM (Maritime Technology Unit, CSIC) and Las Palmas crew (Spanish Navy) who provided us with strong logistical support to make possible this expedition.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is dedicated to the memory of our wonderful colleague and friend, Dr. Luigi Michaud, who dramatically passed away in Antarctica.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Table S1
(DOC 29 kb)
Rights and permissions
About this article
Cite this article
Gugliandolo, C., Michaud, L., Lo Giudice, A. et al. Prokaryotic Community in Lacustrine Sediments of Byers Peninsula (Livingston Island, Maritime Antarctica). Microb Ecol 71, 387–400 (2016). https://doi.org/10.1007/s00248-015-0666-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0666-8