Abstract
Evidence has shown that the altered osteogenic differentiation of human bone marrow stromal cells (BMSCs) under pathological conditions, such as osteoporosis, lead to the imbalance of bone tissue generation and destruction. Recent studies have indicated that long noncoding RNAs may play a role in regulating BMSCs osteogenic differentiation. This contributed to our impetus to move forward with the investigation of the function of lncRNA SERPINB9P1 in osteogenic differentiation of BMSCs and the potential mechanisms involved. Osteogenic differentiation of BMSCs was induced by osteogenic medium. Relative expression of lncRNA SERPINB9P1 and miR-545-3p were tested by qRT-PCR. Osteogenic mineralization was examined by Alizarin S Red staining, ALP staining, and ALP activity assay. Expression of osteoblastic markers were detected by Western blot. RNA-binding protein immunoprecipitation and dual-luciferase reporter assays were performed to test the interaction between lncRNA SERPINB9P1 and miR-545-3p. BMSCs osteogenic differentiation resulted in LncRNA SERPINB9P1 overexpression while miR-545-3p inhibition. Functional assays suggest that knockdown of lncRNA SERPINB9P1 or overexpression of miR-545-3p both inhibit BMSC osteogenic differentiation. lncRNA SERPINB9P1 was proven to regulate the osteogenic differentiation of BMSCs by altering SIRT6 expression through its suppressive effects on miR-545-3p. lncRNA SERPINB9P1 promotes osteogenic differentiation of BMSCs through the miR-545-3p/SIRT6 pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bone marrow stromal cells (BMSCs), initially isolated by Friedenstein et al., are a group of stem cells with multipotential differentiation in bone marrow [1, 2]. BMSCs support hematopoiesis and bone regulation giving them a wide range of applications in cellular and genetic engineering [3, 4]. BMSCs have a decreased ability to differentiate during aging and preferentially differentiate into adipocytes rather than osteoblasts in older individuals. Pathological conditions, such as osteoporosis, are associated with altered a BMSCs osteogenic differentiation leading to an imbalance of bone tissue generation and destruction [5, 6]. The mechanism of osteogenic differentiation in BMSCs has yet to be delineated.
Osteoporosis is a common metabolic bone disease, which frequently leads to osteoporotic fractures seriously increasing the morbidity and mortality in the elderly population [7, 8]. The disruption of bone balance leads to the pathogenesis of osteoporosis. During advanced aging and in other pathological conditions, increased osteoclast activity and decreased osteoblast function lead to a net loss of bone mass. Osteogenic deficiency is associated with the altered differentiation of BMSCs in patients with osteoporosis. BMSCs preferentially differentiate into adipose tissue and less frequently into osteogenic differentiation. Their differentiation direction is regulated by BMP/Smad, Wnt, Notch, Hedgehog, and other signaling pathways [9,10,11], which are involved in transcriptional regulation, posttranscriptional regulation, and epigenetic regulation mechanisms. Future research will focus on finding the key factors that determine the differentiation pathways of BMSCs, providing the development of novel ideas for the treatment of osteoporosis.
Normal metabolism of bone maintains a balanced state of “generation and apoptosis,” which is regulated by a series of signaling pathways [12,13,14]. Current studies show that lncRNA and miRNA regulate a variety of pathways and change the function of related proteins [15,16,17]. Recently, reports of the synergistic regulation of BMSCs osteogenic differentiation by lncRNA and miRNA have increased significantly. LncRNA and miRNA regulate BMSCs differentiation as competing endogenous RNAs (ceRNAs) [18]. A study on the mechanism of bone-specific lncRNA on related cytokines and signaling pathways is conducive to further reveal the role of lncRNA in the mechanism of bone metabolism [19, 20]. LncRNA SERPINB9P1 is associated with cell cycle, apoptosis, invasion and metastasis, cell differentiation and other important physiological, and pathological processes [21, 22]. Recent studies have revealed that lncRNA SERPINB9P1 is down regulated in patients with osteoporosis [22]. However, the functional involvement of lncRNA SERPINB9P1 in osteoporosis and its mechanism remains unclear.
The level of miR-545-3p was found to be reduced during MC3T3-E1 cell osteogenic differentiation, and this process was also repressed by miR-545-3p overexpression [23]. Through the prediction of the Starbase database, we found miR-545-3p binding sites in lncRNA SERPINB9P1, which led to the speculation that lncRNA SERPINB9P1 may affect the osteogenic differentiation of BMSCs by regulating miR-545-3p. The specific mechanism involved in lncRNA SERPINB9P1 and miR-545-3p effects on regulating osteogenic differentiation has not been clarified.
Previous studies have proven that SIRT6 could promote BMSCs osteogenic differentiation and inhibit adipogenesis [24,25,26], suggesting that SIRT6 is associated with BMSC differentiation [27, 28]. However, whether miR-545-3p affects the osteogenic differentiation of BMSCs through SIRT6 has yet to be elucidated.
Our goal is to study the functions of lncRNA SERPINB9P1 in osteoporosis pathogenesis and explore whether lncRNA SERPINB9P1 can promote the osteogenic differentiation of BMSCs by the miR-545-3p/SIRT6 pathway. It is our hope that our research will ultimately reveal new regulatory pathways in osteoporosis and provide the identification of potential biomarkers and novel therapeutic targets in the treatment of osteoporosis.
Materials and Methods
BMSCs Isolation
The bone marrow samples were isolated from the proximal femur of three individuals (1 male and 2 females, ages 3, 2, and 7 years old) who were previously diagnosed with developmental dysplasia of the hip or a femoral neck fracture and subsequently underwent surgery in the Affiliated Children’s Hospital of Nanchang University. All people have consented to the study, and the procedures were approved by the Ethics Committee of the Affiliated Children's Hospital of Nanchang University (JXSETYY-YXKY-20220183). BMSCs in cell suspension were isolated by adherence method. Around 5 ml fresh bone marrow was collected from the end of osteotomy or fracture during surgery. The fresh bone marrow was centrifuged and then resuspended in α-MEM (Gibco, MA0216), supplemented with fetal bovine serum (10%, Gibco, 10099), gentamicin (50 μg/mL, Gibco, 15710064), fungizone (0.3 μg/mL, Gibco, 15290-018), ascorbic acid (5 μg/L, GLPBIO, 50-81-7) for 72 h. Following the removal of the culture medium and non-adherent cells, the cells were then maintained in α-MEM complete medium for cell passage. The BMSCs were characterized by flow cytometry using positive markers CD44, CD90, CD105) and negative markers (CD45/CD34). Anti-CD44 (397517), anti-CD90 (328107), anti-CD105 (323203), anti-CD45 (304007), and anti-CD34 (343505) were purchased from BioLegend.
Osteogenic Differentiation of BMSCs
BMSCs differentiation was induced by osteogenic medium (OM) (Gibco, A1007201), which contains Dexamethasone (Sigma, D1756-25MG), β-glycerophosphate (Sigma, G5422-25G), and ascorbic acid (GLPBIO, 50-81-7).
Quantitative Reverse Transcription (RT)-PCR Assay
For the detection of lncRNA SERPINB9P1 and miR-545-3p, total RNA of BMSCs was prepared via TRIzol (Invitrogen, 15596-026), and the extracted RNA (2–3 μg) was reverse transcribed into cDNA using a synthesis kit (Sigma, 5081955001). RT-PCR was performed with the help of a SYBR Green Mix kit (Thermo Fisher Scientific, 4309155) [29]. Primers were synthesized by Invitrogen (USA) and sequences were showed below. SIRT6 (CCC ACG GAG TCT GGA CCAT and CTC TGC CAG TTT GTC CCT G); lncRNA SERPINB9P1 (ACG CAC AAG CCC CAT AAA GA and GGA AGA CCT TCC GCA GAA CA). miR-545-3p (GCC GAG TCA GCA AAC ATT TAT TGT GTG C and GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACG CAC AC); U6 (CTC GCT TCG GCA GCA CA and AAC GCT TCA CGA ATT TGC GT).
Western Blot Assay
Treated BMSCs were lysed in the lysis buffer (Cell Signaling Technologies) with the presence of protease inhibitors (Roche). The protein concentration was calculated based on a BCA kit Pierce (Rockford). The protein sample (50 μg) was isolated by 10% SDS-PAGE, and then transferred onto the PVDF membranes (Millipore, Bedfordshire). The PVDF membranes were incubated with 5–8% milk for 2 h. followed by the incubation overnight with appropriate antibodies against GAPDH (1:10,000, ab181602, abcam), OCN (1:5000, ab133612, abcam), SIRT6 (1:2000, ab191385, abcam), Runx2 (1:1000, ab76956, abcam), ALP (1:500, ab82846, abcam), RANKL (1:1000, ab9957, Abcam), and OPG (1:500, ab73400, Abcam). Then, the membranes were washed again with PBST followed by the incubation with secondary antibodies (IgG-HRP, abcam, 1:2000) for 2 h. Signals were detected after washing the membranes three times with PBST using the ECL imaging system (Bio-Rad, CA, USA).
Cell Transfection and Infection
miR-545-3p mimics, and miR-545-3p inhibitor, including corresponding controls, were supplied by Vigenebio (Shanghai, China). Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was adopted to complete the cell transfection processes.
For lentivirus infection, lentiviruses carrying lncRNA SERPINB9P1 overexpression (oe-SERPINB9P1), lncRNA SERPINB9P1 knockdown (sh-SERPINB9P1), SIRT6 overexpression (oe-Sirt6), and corresponding controls were obtained from Genepharma (Shanghai, China). BMSCs were seeded in a six-well plate at a density of 105 per well and transduced with the lentiviruses at MOI 30 in serum-free α-MEM with HitransG A (GeneChem, Shanghai, China) for 4 h and supplemented with medium containing 20% FBS. After 24 h of culture, the cells were replaced with α-MEM containing 10% FBS.
Alizarin Red S Staining
Alizarin S Red (Beyotime, China) was widely used in the determination of calcium deposition in cell culture. The steps are outlined: culture the BMSCs for 21 days with OM, remove the medium and wash cells 1–2 times with PBS. Afterward, fix the cells with 4% formaldehyde for 20 min, and wash the cells again with PBS three times. Remove the PBS completely and then add Alizarin Red S to each well. After 10–20 min of incubation with gentle shaking, cells were washed with PBS and evaluated for signal detection.
ALP Staining
Remove the cell culture medium and wash the cells once or twice with PBS. Cells were fixed in 4% formaldehyde for 20 min, after which they were washed three times with PBS and stained using the Alkaline Phosphatase Kit (Sigma-Aldrich). Stained cells were examined under a microscope (BX51; Olympus, Tokyo, Japan).
ALP Activity Assay
After 7 days of culture in OM medium, BMSCs were harvested and washed three times with cold PBS. The cells were then resuspended using the ALP kit assay (abcam, USA) buffer, and the supernatant was centrifuged at high speed for about l0 minutes. The supernatant was processed according to the kit instructions. The results were determined by a microplate reader at 405 nm.
Dual-Luciferase Reporter Assay
The wild-type and mutant segments of lncRNA SERPINB9P1/SIRT6 3′UTR were inserted into pmirGLO vector (Promega, USA) to establish lncRNA SERPINB9P1-WT/lncRNA SERPINB9P1-MUT and SIRT6-WT/SIRT6-MUT recombinant vectors. The miRNAs (miR-545-3p mimics or miR-NC) and the recombinant reporters (WT or Mut) were cotransfected into 293 T cells and cultured for 48 h, followed by the detection of luciferase activity.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
The EZ-Magna RIP kit (EMD Millipore, USA) was used in the RIP assay. In brief, BMSCs (1 × 107) were lysed in RIP lysis buffer, the cell extracts were then incubated with magnetic beads coupled with antibodies against Ago2 (1:50, ab32381, abcam, USA) or IgG (1:50, ab109761, abcam). Immunoprecipitated RNAs were extracted using TRIzol reagent, then qRT-PCR was performed to detect the expression levels of lncRNA SERPINB9P1 and miR-545-3p.
Statistical Analysis
Data was presented as mean ± standard deviation. GraphPad Prism 8.0 was employed to perform statistical analysis through one-way analysis of variance or student’s t test. Difference was considered significant if p < 0.05.
Results
lncRNA SERPINB9P1 was Increased and miR-545-3p was Decreased During BMSCs Osteogenic Differentiation
First, we examined the expression levels of lncRNA SERPINB9P1 and miR-545-3p in isolated BMSCs. The classification of the isolated BMSCs is determined according to the characteristics of the cell surface antigens. The relative specific molecules of surface antigens detected by flow cytometry for BMSCs phenotypic identification, including CD44, CD90, CD105, CD45, and CD34 (Supplemental Fig. 1A). The protein levels of ALP, Runx2, and OCN in BMSCs on days 1, 5, 10, and 15 were also tested. The results show that all three markers increased gradually with the prolongation of the culture time. The results also showed that the induction of osteogenic differentiation by OM was successful (Supplemental Fig. 1B). We then tested the relative levels of lncRNA SERPINB9P1 and miR-545-3p during osteoblast differentiation. During osteoblast differentiation, lncRNA SERPINB9P1 was overexpressed and miR-545-3p was inhibited with the prolongation of culture time, suggesting that lncRNA SERPINB9P1 and miR-545-3p may be linked with the differentiation of BMSCs (Supplemental Fig. 1C).
Knockdown of SERPINB9P1 Inhibited the Differentiation of BMSCs
To clarify whether lncRNA SERPINB9P1 participates in BMSC osteogenic differentiation, the lncRNA SERPINB9P1 expression was knocked down by sh-SERPINB9P1 and overexpressed by SERPINB9P1 lentiviruses in BMSCs. After infection of sh-SERPINB9P1 or oe-SERPINB9P1 into BMSCs cells for about 48 h, cell samples were collected and the knockdown and overexpression efficiency was tested. Compared with control cells, the level of lncRNA SERPINB9P1 in BMSCs infected with sh-SERPINB9P1 decreased significantly and the expression of lncRNA SERPINB9P1 dramatically increased with oe-SERPINB9P1 lentiviruses (Fig. 1A). The ability of osteogenic differentiation was evaluated with Alizarin Red S staining and the activity of ALP. Compared with control cells, the expression of ALP decreased when lncRNA SERPINB9P1 was knocked down. Accordingly, the expression of ALP increased when lncRNA SERPINB9P1 was overexpressed (Fig. 1B). These results showed that knockdown of lncRNA SERPINB9P1 repressed BMSC osteogenic differentiation, and overexpression of lncRNA SERPINB9P1 facilitated it. After 21 days of incubation, Alizarin Red S staining (Fig. 1B) was observed in all five groups (Control group, sh-vector group, sh-SERPINB9P1 group, oe-vector group, oe-SERPINB9P1 group). After lncRNA SERPINB9P1 knockdown, the calcium salt nodules were comparatively weaker than those from the control group. The ALP activity assay showed that compared with control cells, lncRNA SERPINB9P1 knockdown significantly decreased ALP activity. Accordingly, oe-SERPINB9P1 group increased ALP activity (Fig. 1C). The ALP activity assay also proved that lncRNA SERPINB9P1 facilitated BMSC osteogenic differentiation and knockdown of lncRNA SERPINB9P1 reduced the expression levels of ALP, OCN, and Runx2, while overexpression of lncRNA SERPINB9P1 increased the expression levels of these markers (Fig. 1D). Moreover, lncRNA SERPINB9P1 overexpression was found to decrease the expression of RANKL and increased the expression of OPG (Supplemental Fig. 2A).
lncRNA SERPINB9P1 regulated osteogenic differentiation of BMSCs. The cells were infected with sh-SERPIB9P1 or oe-SERPINB9P1. A SERPINB9P1 level was detected after 48 h of infection. B After the cells were cultured in OM for 21 days, the ability of osteogenic differentiation was evaluated with the activity of ALP and Alizarin Red S staining after knocking down lncRNA SERPINB9P1. The cells were cultured in OM for 7 days. C The ALP activity was evaluated. D Relative protein levels of ALP, OCN, and Runx2 in each group. GAPDH as the loading control. Data indicate mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001
lncRNA SERPINB9P1 Binds to miR-545-3p Directly
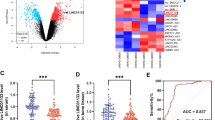
The relative miR-545-3p level was increased or decreased after lncRNA SERPINB9P1 knockdown or overexpression, respectively (Fig. 2A). Ago2 RIP assay was used to test whether lncRNA SERPINB9P1 and miR-545-3p occupied the same RNA-induced silencing complex (RISC) in BMSCs. Our results indicated that lncRNA SERPINB9P1 and miR-545-3p were enriched in the Ago2 group compared to that in the IgG group (Fig. 2B). The interaction between lncRNA SERPINB9P1 and miR-545-3p was further tested by dual-luciferase reporter assay. The results showed that the miR-545-3p mimic reduced the firefly luciferase activity in 293 T cells transfected with lncRNA SERPINB9P1-WT. In contrast, the firefly luciferase activity in 293 T cells transfected with lncRNA SERPINB9P1-MUT not reduced by the miR-545-3p mimic (Fig. 2C, D). These results indicated that lncRNA SERPINB9P1 sponged miR-545-3p.
lncRNA SERPINB9P1 binds to miR-545-3p. A The cells were infected with sh-SERPIB9P1 or oe-SERPINB9P1, relative miR-545-3p expression in each group. B RIP assay verified the interaction of lncRNA SERPINB9P1 and miR-545-3p. C The binding site between miR-545-3p and lncRNA SERPINB9P1. D Verification of the interplay between miR-545-3p and lncRNA SERPINB9P1 using dual-luciferase reporter assay. Data indicate mean ± SD, n = 3. *p < 0.05, **p < 0.01, and ***p < 0.001
lncRNA SERPINB9P1 Regulated BMSC Osteogenic Differentiation Via miR-545-3p
On the basis of knocking down lncRNA SERPINB9P1, we added the inhibitor of miR-545-3p to test the functional relationship between lncRNA SERPINB9P1 and miR-545-3p in BMSC osteogenic differentiation. As predicted, the sh-SERPINB9P1 infection led to an elevation of miR-545-3p in BMSCs, which was abrogated by introducing the miR-545-3p inhibitor (Fig. 3A). Subsequently, the ability of osteogenic differentiation was evaluated. Compared with control cells, the staining area of ALP and Alizarin Red S decreased when knocking down lncRNA SERPINB9P1. When the inhibitor of miR-545-3p was added into the BMSCs cells infected with sh-SERPINB9P1, the decreased staining area of ALP and Alizarin Red S was reversed (Fig. 3B). The ALP activity assay showed that compared with control cells, lncRNA SERPINB9P1 knocking down significantly decreased ALP activity. Accordingly, the inhibitor of miR-545-3p rescued the decrease of ALP activity (Fig. 3C). Western blot analysis also showed that when knocking down lncRNA SERPINB9P1, the expression levels of ALP, OCN, and Runx2 were decreased, and this phenomenon was blocked after miR-545-3p inhibitor treatment (Fig. 3D). Inhibition of miR-545-3p was also found to decrease the expression of RANKL and increase the expression of OPG (Supplemental Fig. 2B). The evidence above proved that lncRNA SERPINB9P1 regulated BMSC osteogenic differentiation via miR-545-3p.
lncRNA SERPINB9P1 regulated BMSC osteogenic differentiation by miR-545-3p. The cells were treated with sh-SERPIB9P1 and miR-545-3p inhibitor. A Relative miR-545-3p level in each group. B After the cells were cultured with OM for 21 days, the expression of osteogenic differentiation was evaluated with the ALP and Alizarin Red S staining assay. The cells were cultured in OM for 7 days. C The ALP activity was evaluated. D Western blot assays were carried out to test the expression level of β-catenin, ALP, OCN, and Runx2. GAPDH as loading control. Data indicate mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001
SIRT6 Binds to miR-545-3p Directly
To determine the interaction between miR-545-3p and SIRT6, the relative expression of SIRT6 was evaluated after transfection with miR-545-3p mimic or inhibitor. We found that miR-545-3p overexpression or knockdown led to a downregulation or upregulation of SIRT6, respectively (Fig. 4A, B). We also found that miR-545-3p mimic sharply attenuated the luciferase activity driven by SIRT6 WT (Fig. 4C). Additionally, lncRNA SERPINB9P1 knockdown led to a reduction of SIRT6 in BMSCs, and miR-545-3p inhibitor abrogated the reduction of SIRT6 caused by lncRNA SERPINB9P1 knockdown (Fig. 4D, E).
SIRT6 binds directly to miR-545-3p. A, B The cells were treated with miR-545-3p mimics or miR-545-3p inhibitor, relative mRNA and protein expression of SIRT6 (A). Impact of lncRNA SERPINB9P1 knockdown or/and miR-545-3p overexpression on the level of SIRT6 (B). C Verification of the interplay between miR-545-3p and SIRT6 using dual-luciferase reporter assay. D, E The cells were treated with sh-SERPIB9P1 and miR-545-3p inhibitor, relative mRNA and protein levels of SIRT6 in treated BMSCs. Data presented as mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001
Overexpression of SIRT6 Rescues the Influences of miR-545-3p Overexpression on BMSCs
To determine if the impact of miR-545-3p on BMSC osteogenic differentiation was mediated by SIRT6, BMSCs were treated with miR-545-3p mimic and oe-SIRT6. Overexpression miR-545-3p caused a reduction of SIRT6, and the decreased expression of SIRT6 were blocked by overexpression of SIRT6 (Fig. 5A). Subsequently, the ability of osteogenic differentiation was evaluated. Compared with control cells, miR-545-3p overexpression led to reductions of ALP and Alizarin Red S staining area, and this phenomenon was blocked after oe-SIRT6 infection (Fig. 5B). ALP activity assay showed that miR-545-3p mimic decreased ALP activity, and SIRT6 overexpression could rescue the decrease of ALP activity (Fig. 5C). Western blot results revealed that SIRT6, ALP, OCN, and Runx2 were decreased after miR-545-3p overexpression, and SIRT6 overexpression abrogated the reduction of ALP, OCN, and Runx2 of BMSCs (Fig. 5D).
Overexpression of SIRT6 can rescue the influences of miR-545-3p overexpression on BMSC. The cells were treated with miR-545-3p mimics and oe-SIRT6. A SIRT6 level detected in treated BMSCs. B After the cells were cultured with OM for 21 days, the ability of osteogenic differentiation was evaluated with ALP and Alizarin Red S staining. The cells were cultured in OM for 7 days. C The ALP activity was evaluated. D The expressions of SIRT6, β-catenin, ALP, OCN, and Runx2 were detected. GAPDH as the loading control. Data presented as mean ± SD, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
BMSCs are the precursor stem cells of osteoblasts and adipocytes, and their differentiation plays a remarkable role in bone balance [30]. Reduced capacity of BMSC osteogenic differentiation has been well demonstrated to be one of the most important factors contributing the development of osteoporosis (OP) [31,32,33]. It is generally believed that inhibiting the adipogenic differentiation of BMSCs and correcting the imbalance of bone metabolism are one of the most promising directions for the treatment of OP [34,35,36]. Considering that an increasing number of lncRNAs have been confirmed to be linked with the osteogenic differentiation process of BMSCs. Targeting lncRNAs may further define the pathogenesis of OP, and lead to the development of more effective therapeutic targets for OP.
An increasing number of studies have shown that noncoding RNAs participate in the regulation of the differentiation and function of osteoblasts, chondrocytes, and osteoclasts, suggesting that noncoding RNAs are a key group of regulators in the process of bone formation, absorption, remodeling, and repair [12]. Upon completion of human genome sequencing, an increasing amount of evidence has confirmed the critical role of lncRNAs in promoting osteogenic differentiation of BMSCs [37]. For example, lncRNA PART1 was reported to modulate BMSC osteogenic differentiation by regulating the miR-185-5p/RUNX3 axis [38]. Moreover, lncRNA SNHG1 was revealed to be a repressor of BMSC osteogenic differentiation via the regulation of Dkk1 by miR-101 [39]. LncRNA SERPINB9P1 was previously reported to be downregulated in postmenopausal osteoporosis patients [22], however, its role in osteogenic differentiation has not yet been determined. Herein, we reported that the expression of lncRNA SERPINB9P1 was increased during the differentiation of BMSCs. Functional experiments indicated that knockdown of lncRNA SERPINB9P1 could inhibit the osteogenic differentiation of BMSCs. MiRNAs have also been reported to participate in the BMSC osteogenic differentiation through multiple mechanisms. For example, miR-486-3p has been demonstrated to facilitate BMSC osteogenic differentiation by regulating CTNN and activating the Wnt/Catenin pathway [40]. Moreover, miR-129-5p could modulate BMSC osteogenic differentiation and bone regeneration by repressing the expression of Dkk3 [41]. Recently, SP1-stimulated miR-545-3p was found to repress osteogenesis by targeting the LRP5-activated Wnt/Catenin axis [23]. Herein, we reported a decreased miR-545-3p level during BMSC osteogenic differentiation, and found that miR-545-3p overexpression could inhibit the osteogenic differentiation of BMSCs.
LncRNA and miRNA inhibit each other and form a precise regulatory network to regulate the target genes of miRNA [42,43,44]. SIRT6 was found to modulate BMSC osteogenic differentiation through multiple mechanisms, including the interaction with miRNAs. For example, miR-186 was reported to regulate rat BMSC osteogenic differentiation by targeting SIRT6 [45]. miR-128 was found to repress BMSC osteogenic differentiation by reducing the expression of SIRT6 [46]. In our study. lncRNA SERPINB9P1 was proven to act as a sponger of miR-545-3p to regulate SIRT6. We determined that either knockdown of lncRNA SERPINB9P1 or overexpression of miR-545-3p could decrease SIRT6 expression. We also found that the miR-545-3p inhibitor reversed the inhibitory effect of decreased lncRNA SERPINB9P1 expression on BMSC osteogenic differentiation. We present the novel evidence that lncRNA SERPINB9P1 can sponge miR-545-3p to participate in the regulation of osteogenic differentiation of BMSCs through SIRT6.
In conclusion, the evidences provided by our study supported that lncRNA SERPINB9P1 could facilitate BMSC osteogenic differentiation through the miR-545-3p/SIRT6 pathway. The novel lncRNA SERPINB9P1/miR-545-3p/SIRT6 axis may offer a novel perspective for OP treatment. Nevertheless, we have not conducted animal experiments to verify the SERPINB9P1/miR-545-3p/SIRT6 pathway because the genome sequence of human lncRNA SERPINB9P1 had very low homology with mouse and rat genome sequences by UCSC homology prediction (Supplementary Fig. 3). In the future, we will further collect more clinical samples to validate this pathway.
Abbreviations
- lncRNA:
-
Long non-coding RNA
- MiR:
-
MicroRNA
- MSC:
-
Marrow stem cell
- BMSC:
-
Bone marrow stem cell
- OP:
-
Osteoporosis
- OB:
-
Osteoblasts
- OM:
-
Osteogenic medium
- ALP:
-
Alkaline phosphatase
References
Li H, Ghazanfari R, Zacharaki D, Lim HC, Scheding S (2016) Isolation and characterization of primary bone marrow mesenchymal stromal cells. Ann N Y Acad Sci 1370(1):109–118
Derubeis AR, Cancedda R (2004) Bone marrow stromal cells (BMSCs) in bone engineering: limitations and recent advances. Ann Biomed Eng 32(1):160–165
Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD et al (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284(5411):143–147
Woodbury D, Schwarz EJ, Prockop DJ, Black IB (2000) Adult rat and human bone marrow stromal cells differentiate into neurons. J Neurosci Res 61(4):364–370
Nuttall ME, Gimble JM (2004) Controlling the balance between osteoblastogenesis and adipogenesis and the consequent therapeutic implications. Curr Opin Pharmacol 4(3):290–294
Kha HT, Basseri B, Shouhed D, Richardson J, Tetradis S, Hahn TJ et al (2004) Oxysterols regulate differentiation of mesenchymal stem cells: pro-bone and anti-fat. J Bone Miner Res 19(5):830–840
Cole ZA, Dennison EM, Cooper C (2008) Osteoporosis epidemiology update. Curr Rheumatol Rep 10(2):92–96
Xia WB, He SL, Xu L, Liu AM, Jiang Y, Li M et al (2012) Rapidly increasing rates of hip fracture in Beijing, China. J Bone Miner Res 27(1):125–129
Tang QQ, Otto TC, Lane MD (2004) Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci USA 101(26):9607–9611
Jacobsen CM, Schwartz MA, Roberts HJ, Lim KE, Spevak L, Boskey AL et al (2016) Enhanced Wnt signaling improves bone mass and strength, but not brittleness, in the Col1a1(+/mov13) mouse model of type I Osteogenesis Imperfecta. Bone 90:127–132
Ramasamy SK, Kusumbe AP, Wang L, Adams RH (2014) Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature 507(7492):376–380
Johnson ML, Kamel MA (2007) The Wnt signaling pathway and bone metabolism. Curr Opin Rheumatol 19(4):376–382
Maeda K, Kobayashi Y, Koide M, Uehara S, Okamoto M, Ishihara A et al (2019) The regulation of bone metabolism and disorders by Wnt signaling. Int J Mol Sci 20(22):5525
Lindsey RC, Rundle CH, Mohan S (2018) Role of IGF1 and EFN-EPH signaling in skeletal metabolism. J Mol Endocrinol 61(1):T87–T102
Zhao S, Chen H, Ding B, Li J, Lv F, Han K et al (2019) Construction of a transcription factorlong noncoding RNAmicroRNA network for the identification of key regulators in lung adenocarcinoma and lung squamous cell carcinoma. Mol Med Rep 19(2):1101–1109
Yan H, Wang Q, Shen Q, Li Z, Tian J, Jiang Q et al (2018) Identification of potential transcription factors, long noncoding RNAs, and microRNAs associated with hepatocellular carcinoma. J Cancer Res Ther 14(Supplement):S622–S627
Jiang L, Yu X, Ma X, Liu H, Zhou S, Zhou X et al (2019) Identification of transcription factor-miRNA-lncRNA feed-forward loops in breast cancer subtypes. Comput Biol Chem 78:1–7
Song WQ, Gu WQ, Qian YB, Ma X, Mao YJ, Liu WJ (2015) Identification of long non-coding RNA involved in osteogenic differentiation from mesenchymal stem cells using RNA-Seq data. Genet Mol Res 14(4):18268–18279
Luo M, Huang HX, Huang H, Li ZT, Lai YY (2014) Hedgehog signaling pathway and osteoporosis. Zhongguo Gu Shang 27(2):169–172
Li B (2018) MicroRNA regulation in osteogenic and adipogenic differentiation of bone mesenchymal stem cells and its application in bone regeneration. Curr Stem Cell Res Ther 13(1):26–30
Liu L, Wang H, Zhang X, Chen R (2020) Identification of potential biomarkers in neonatal sepsis by establishing a competitive endogenous RNA network. Comb Chem High Throughput Screen 23(5):369–380
Wang S (2020) Investigation of long non-coding RNA expression profiles in patients with post-menopausal osteoporosis by RNA sequencing. Exp Ther Med 20(2):1487–1497
Li L, Qiu X, Sun Y, Zhang N, Wang L (2019) SP1-stimulated miR-545-3p inhibits osteogenesis via targeting LRP5-activated Wnt/beta-catenin signaling. Biochem Biophys Res Commun 517(1):103–110
Kim SJ, Piao Y, Lee MG, Han AR, Kim K, Hwang CJ et al (2020) Loss of Sirtuin 6 in osteoblast lineage cells activates osteoclasts, resulting in osteopenia. Bone 138:115497
Shen X, Chen X, Huang J, Xu R, Cheng J, Jiang H (2020) Age-dependent role of SIRT6 in jawbone via regulating senescence and autophagy of bone marrow stromal cells. J Mol Histol 51(1):67–76
Xiao F, Zhou Y, Liu Y, Xie M, Guo G (2019) Inhibitory effect of Sirtuin6 (SIRT6) on osteogenic differentiation of bone marrow mesenchymal stem cells. Med Sci Monit 25:8412–8421
Wei W, Guo X, Gu L, Jia J, Yang M, Yuan W et al (2021) Bone marrow mesenchymal stem cell exosomes suppress phosphate-induced aortic calcification via SIRT6-HMGB1 deacetylation. Stem Cell Res Ther 12(1):235
Pan XH, Chen YH, Yang YK, Zhang XJ, Lin QK, Li ZA et al (2019) Relationship between senescence in macaques and bone marrow mesenchymal stem cells and the molecular mechanism. Aging (Albany NY) 11(2):590–614
Ban K, Noyan-Ashraf MH, Hoefer J, Bolz SS, Drucker DJ, Husain M (2008) Cardioprotective and vasodilatory actions of glucagon-like peptide 1 receptor are mediated through both glucagon-like peptide 1 receptor-dependent and -independent pathways. Circulation 117(18):2340–2350
Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q et al (2016) Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ 23(7):1128–1139
Kim J, Ko J (2014) A novel PPARgamma2 modulator sLZIP controls the balance between adipogenesis and osteogenesis during mesenchymal stem cell differentiation. Cell Death Differ 21(10):1642–1655
Huang J, Zhao L, Xing L, Chen D (2010) MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells 28(2):357–364
Liao L, Yang X, Su X, Hu C, Zhu X, Yang N et al (2013) Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis 4:e600
Kim M, Lee YJ, Jee SC, Choi I, Sung JS (2016) Anti-adipogenic effects of sesamol on human mesenchymal stem cells. Biochem Biophys Res Commun 469(1):49–54
An Q, Wu D, Ma Y, Zhou B, Liu Q (2015) Suppression of Evi1 promotes the osteogenic differentiation and inhibits the adipogenic differentiation of bone marrow-derived mesenchymal stem cells in vitro. Int J Mol Med 36(6):1615–1622
Wang C, Meng H, Wang X, Zhao C, Peng J, Wang Y (2016) Differentiation of bone marrow mesenchymal stem cells in osteoblasts and adipocytes and its role in treatment of osteoporosis. Med Sci Monit 22:226–233
Jiang S, Xia M, Yang J, Shao J, Liao X, Zhu J et al (2015) Novel insights into a treatment for aplastic anemia based on the advanced proliferation of bone marrowderived mesenchymal stem cells induced by fibroblast growth factor 1. Mol Med Rep 12(6):7877–7882
Zhang J, Xu N, Yu C, Miao K, Wang Q (2021) LncRNA PART1/miR-185-5p/RUNX3 feedback loop modulates osteogenic differentiation of bone marrow mesenchymal stem cells. Autoimmunity 54(7):422–429
Xiang J, Fu HQ, Xu Z, Fan WJ, Liu F, Chen B (2020) lncRNA SNHG1 attenuates osteogenic differentiation via the miR101/DKK1 axis in bone marrow mesenchymal stem cells. Mol Med Rep 22(5):3715–3722
Zhang Z, Jiang W, Hu M, Gao R, Zhou X (2021) MiR-486-3p promotes osteogenic differentiation of BMSC by targeting CTNNBIP1 and activating the Wnt/beta-catenin pathway. Biochem Biophys Res Commun 566:59–66
Zhao C, Gu Y, Wang Y, Qin Q, Wang T, Huang M et al (2021) miR-129-5p promotes osteogenic differentiation of BMSCs and bone regeneration via repressing Dkk3. Stem Cells Int 2021:7435605
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146(3):353–358
Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y et al (2015) The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget 6(23):19759–19779
Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K et al (2013) Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 4:2939
Xiao J, Qin S, Li W, Yao L, Huang P, Liao J et al (2020) Osteogenic differentiation of rat bone mesenchymal stem cells modulated by MiR-186 via SIRT6. Life Sci 253:117660
Zhao J, Liu S, Zhang W, Ni L, Hu Z, Sheng Z et al (2019) MiR-128 inhibits the osteogenic differentiation in osteoporosis by down-regulating SIRT6 expression. Biosci Rep. https://doi.org/10.1042/BSR20191405
Acknowledgements
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Min Wu, Min Dai, Xuqiang Liu, Qunqun Zeng, and Yingjie Lu declare that they have no conflict of interest.
Ethics approval and consent to participate
Human bone marrow was collected in orthopaedic surgery. All people have consented to the study, and the procedures were approved by the Ethics Committee of the Affiliated Children's Hospital of Nanchang University (JXSETYY-YXKY-20220183).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
223_2022_1034_MOESM1_ESM.tif
Supplemental Figure 1. lncRNA SERPINB9P1 was elevated and miR-545-3p was decreased during BMSC osteogenic differentiation. (A) The phenotype of BMSCs were tested by flow cytometry. (B) Relative expression of ALP, Runx2, and OCN were detected on days 0, 1, 5, 10, and 15. GAPDH as the loading control. (C) Relative expression of lncRNA SERPINB9P1 and miR-545-3p during BMSC osteogenic differentiation on days 0, 1, 5, 10, and 15. Data indicate mean ± SD, n = 3. *p < 0.05, **p < 0.01, and ***p < 0.001. Supplementary file1 (TIF 9702 KB)
223_2022_1034_MOESM2_ESM.tif
Supplemental Figure 2 lncRNA SERPINB9P1 overexpression and miR-545-3p inhibition decreased the expression of RANKL and increased the expression of OPG. (A and B) Western blot was performed to test the protein expression of RANKL and OPG in BMSCs treated with lncRNA SERPINB9P1 overexpression plasmid (oe-SERPINB9P1) or miR-545-3p inhibitor. Supplementary file2 (TIF 832 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, M., Dai, M., Liu, X. et al. lncRNA SERPINB9P1 Regulates SIRT6 Mediated Osteogenic Differentiation of BMSCs via miR-545-3p. Calcif Tissue Int 112, 92–102 (2023). https://doi.org/10.1007/s00223-022-01034-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00223-022-01034-3